Abstract
We review current advances in experimental as well as computational modeling and simulation approaches to structural systems biology, whose overall aim is to build quantitative models of signaling networks while retaining the crucial elements of molecular specificity. We briefly discuss the current and emerging experimental and computational methods, particularly focusing on hybrid and multiscale methods, and highlight several applications in cell signaling with quantitative and predictive capabilities. The scope of such models range from delineating protein–protein interactions to describing clinical implications.
Keywords: Systems biology, Structural biology, Multiscale modeling, Signaling networks, Molecular resolution
INTRODUCTION
Human genome-sequencing has enabled the creation of an exhaustive parts-list in mammalian cellular systems. The primary challenge has now shifted towards functional relationships between the parts and mechanistic description of how the parts function as modules and as a whole. Systems biology lies at the heart of addressing this grand challenge60 and focuses mainly on synthesizing these relationships when they are considered together. Pathways have long represented a convenient way of summarizing the results of many hundreds of experiments in order to chart the flow of signals or metabolites in a cell. The past decade has prompted the creation of several databases of metabolic and signaling pathways, including the Kyoto Encyclopedia of Genes and Genomes, BioCarta, and Signal Transduction Knowledge Environment.3 In general, these resources represent the relationships between molecules in a cell either as reactions or as activation or inhibition events. The specificity of cellular responses is decoded by spatial and temporal signals propagating through intracellular signaling pathways. Computational models and network analysis tools continue to provide insights into the complex relationships between the stimuli, cellular responses, and cell fate.7,25,57
A full understanding of how molecules interact, however, can be derived only from three-dimensional (3D) structures, which provide atomic details about the specificity of binding and molecular recognition. However, structural biology remains limited in terms of informing systems biology because of challenges in determining macromolecular (protein–protein and protein–nucleic acid) interfaces and in dealing with large protein complexes. Studying molecular complexes and how they propagate at the cellular signaling scale can be complementarily achieved through multiscale computational modeling. The task of multiscale modeling is to bridge the scales of structural and systems biology to elucidate the effects of perturbations to macromolecular structure on downstream signaling events initiated by multiprotein complexes. As the robustness of biological systems hinges upon the efficient transfer of information across multiple spatial and temporal scales, the application of multiscale modeling methods is an effective way to bridge the scales and provide more insights than would be gleaned at any single scale.58
METHODS AND MODELS
Experimental Methodologies
New techniques in crystal structure determination have recently emerged topredict and model the structures of interacting proteins.3 These include improvement in overexpression and purification procedures to obtain sufficient material for structural analysis, the ability to express the subunits of a complex in model organisms, and improvements in crystallization techniques and synchrotron radiation facilities to utilize smaller sample amounts to solve structures of complexes. Techniques such as cryoelectron microscopy routinely reconstruct structures of large complexes at lower-resolution using much smaller amounts of material. Many efforts have been undertaken to provide comprehensive lists of protein–protein interactions. The yeast two-hybrid system and affinity purification methodologies remain the most widely used systems, although other experimental methods, including chemical crosslinking, chemical foot-printing, protein arrays, fluorescence resonance energy transfer, and fluorescence cross-correlation spectroscopy, are becoming increasingly popular. Despite the improvements in these experimental systems, there is still a large gap between the number of inferred complexes and those for which 3D structures are available.3
Computational Methodologies
Protein–protein interactions are also predicted computationally, complementing the efforts of high-throughput experiments.57 Statistical approaches are based on the comparison of complete genome sequences and the more established criteria of complementarity, as demonstrated by the extensive literature in yeast genetics. Other statistical methods are based on co-evolution patterns across several species.3 Structures of interacting proteins are also modeled computationally through homology modeling (if structures have been previously determined for suitable homologous proteins).18 In the absence of known structural information, domain or motif-based modeling methods are available. These methods have predicted several domain–domain and domain–motif interactions.3 Computational chemistry approaches based on the potential energy (or force-field) of molecular interactions to predict atomic details for a pair of interacting proteins are also available.24 Docking techniques attempt to find the best-docked complex on the basis of shape or electrostatic complementarity between protein surfaces.3,10,11,14 Suites of techniques that model interacting structures by homology (i.e., utilize protein–protein complexes for which coordinate data are available to model interactions between their homologues) are also becoming popular.3
Hybrid Methodologies
A general limitation of the modeling approaches is the availability and requirement of protein crystal structures, although techniques such as homology modeling can overcome these issues by generating unresolved structures based on closely-related protein structures. However, the construction of accurate macromolecular complexes remains a challenge, as the orchestrated assembly of such complexes is difficult to capture under most experimental conditions. New hybrid techniques integrating multiple scales for prediction of macromolecular complex structure are becoming increasingly available. Hybrid multiscale methods,59 in which multiple lower-resolution techniques are combined to generate atomic models of protein complexes, represent one approach to molecular complex prediction. These hybrid techniques, which may integrate methods such as X-ray crystallography, crosslinking studies, and cryoelectron microscopy, provide a way to combine the accuracy of atomic level models with the computational speed allowed by coarse-grained (CG) models. For example, the 3D structure of the small nuclear ribonucleoprotein particles that bind to pre-messenger RNA to form the spliceosome has been proposed by integrating different data types50: high-resolution structures were determined by X-ray crystallography, and the relative positions by crosslinking studies, the RNA pieces were modeled according to the known binding interactions, the total volume was determined by cryoelectron EM, and all of the available 3D models were fitted into the cryoelectron microscopy map. Such approaches have been successfully applied to several biological systems, including myosin/actin dynamics,53 the assembly of virus capsids,37,52 and the nucleosome-pore complex.2,17
Saunders and Voth41 divide hybrid methods into two categories: mapping methods, which use information from one scale of representation to inform or create (e.g., through model parameterization) a lower-resolution model, and bridging methods, which involve bidirectional information flow between scales. Mapping methods have been applied to study the self-assembly of viral capsids,12,23 including the HIV capsid.20 These models represent capsid subunit geometry using CG techniques, in which several protein subunits comprise a single CG site, to investigate the effect of subunit shape on capsid assembly. However, in order to efficiently explore the parameter space of the CG capsid models (e.g., by varying subunit interaction strength), simulation results derived from high-resolution models of subunit interactions can help to quantitatively narrow the parameter space, as has also been done for models of actin and tubulin dynamics.19
Bridging methods are more difficult to implement than mapping methods, as it is a challenge to attain high accuracy for each scale of representation. However, several groups have applied bridging techniques to specific biological systems of interest.4,13 In one study,5 CG simulations of the immature HIV virion were performed to identify the key molecular interactions responsible for maintaining the capsid structure, and these highlighted interactions then guided high-resolution molecular dynamics (MD) simulations of the capsid, in order to provide detailed hypotheses on the effect of perturbations (such as mutation) on the capsid structure.
A more advanced multiscale modeling approach is to semi-automate the iterations of information flow between models, i.e., rather than map the results of a high-resolution MD simulation to a CG simulation, construct the system such that that the models inform each other during simulation in real-time.41 The CG component will progress with large time steps, and the high-resolution component will proceed with faster steps. There is growing interest in extending these semi-automated methods to specific systems of interest, in order to reduce the need for manual transfer of information between scales.
Multiscale Modeling Approaches
In recent works, researchers describe a new multiscale modeling approach for studying functional interactions in signaling networks through multiscale hybrid physical/systems approaches. Specifically, they adopt a multiscale strategy for constructing models of intracellular signaling networks with the ability to encode the resolution of molecular decisions at key nodes (see applications in sections “Predictive Multiscale Modeling of the ErbB Mutational Landscape”, “Predicting the Effects of Molecular Perturbations on Receptor Kinase Activation”, “Transcribing the Effects of Molecular Alterations into Downstream Signal Activation”, “Clinical Implications of Multiscale Modeling of ErbB Receptor Signaling”, and “Multiscale Modeling of the ErbB3/HER3 Signaling Network”). Network models of cellular signaling pathways represent the highest level of modeling in this approach and are used to monitor the global behavior of both wild type and oncogenically activated systems. Network analysis techniques are used to represent information flow in regulatory information cascades.
The approach to study the comparative signaling dynamics of complex networks in pairs of related systems is based on the hypothesis that that they differ in a defined set of molecular species. Based on this hypothesis, a pair of networks (for wild type versus oncogenically activated cells) is constructed, and the differential behavior of the systems is analyzed by comparing the properties within the pair of networks. Alterations in network behavior are inferred based on the application of a multiscale strategy to resolve how key structural differences translate into altered topologies in the network. That is, in the multiscale approach, molecular modeling or structure-based experimentation are adopted to quantify altered topologies of interactions as well as to provide the missing topologies/parameters for network models. Multiscale computational methodologies (including those discussed in “Hybrid Methodologies” section) offer a powerful, quantitative, and complimentary avenue for the study of intracellular signaling, which if utilized correctly can predict effects of mutations on receptor tyrosine kinase (RTK) activation mechanisms. As discussed in “Applications” section, such analyses can also be extended to gain insight into the clinical aspects (see “Clinical Implications of Multiscale Modeling of ErbB Receptor Signaling” and “Multiscale Modeling of the ErbB3/HER3 Signaling Network” sections) of oncogenic signaling.
APPLICATIONS
Signaling Specificity in the Fibroblast Growth Factor Receptor (FGFR) Family
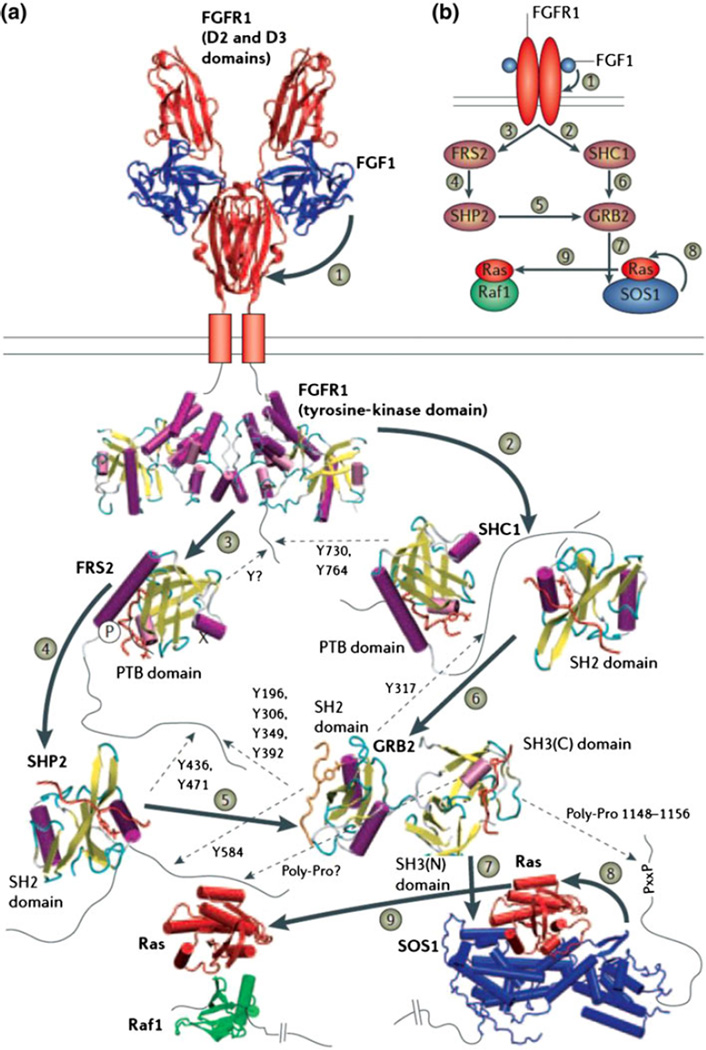
The binding of ligands to FGFRs leads to receptor dimerization and the subsequent activation of the kinase domain. Autophosphorylation of the FGFR kinase results in the recruitment of several downstream proteins such as FRS2 (FGFR substrate-2) and SHC1 (SH2 domain-containing transforming protein-1) through their phosphotyrosine binding (PTB) or SH2 domains, and in the subsequent phosphorylation of sites on these proteins by the RTK. These sites are then recognized by the SH2 domains of other proteins, including GRB2 (growth-factor-receptor-bound protein-2) and SHP2 (SH2-domain-containing tyrosine phosphatase-2). Combining such pathways with structural details renders them more useful for systems biology (see Fig. 1). Crystal structures are available for the dimeric form of the intracellular tyrosine-kinase domain of FGFR; crystal structures or models are also available for many of these PTB and SH2 domains,30–32 but modeling on the basis of other PTB-or SH2-domain structures that have been solved in complex with their peptide substrates is required to infer how they interact with their substrates. GRB2 and SHP2 then bind to other proteins further downstream in the cascade. For example, the C-terminal SH3 domain of GRB2 binds to proline-rich segments in the C terminus of son-of-sevenless-1 (SOS1), an interaction that can be modeled on the basis of other SH3–peptide complexes. SOS1 binds to Ras, and a crystal structure is available for this interaction. The Ras–Raf interaction can be modeled on the basis of a related crystal structure.3 The model reveals that the binding of Raf and SOS1 to Ras are mutually exclusive, because they bind to the same segment of the Ras molecule. Similar details can be gleaned for the rest of this pathway, from the receptor all the way to the nucleus. The quest for structural understanding can also highlight important missing details such as location of phosphorylation sites and other sites of posttranslational modifications.
FIGURE 1.
(Reproduced with permission from Aloy and Russell3). Structural basis for specificity and interactions in the FGFR signaling pathway.
Modeling the Structural Basis of Signaling in the Epidermal Growth Factor Receptor (EGFR) Family
ErbB family receptors (named because of their homology to the erythroblastoma viral gene product, v-erbB, and comprising the epidermal growth factor receptor or EGFR/ErbB1/HER1, ErbB2/HER2, ErbB3/HER3, and ErbB4/HER4) signal by activating crucial pathways in response to stimulation by ligands such as epidermal growth factor (EGF) and other related peptide growth factors. Through ligand-stimulated formation of various homodimeric and heterodimeric complexes, ErbB receptors are activated, leading to the phosphorylation of multiple tyrosine residues on the C-terminal tail of the receptors as well as on other substrate proteins. Through specific interactions between the phosphotyrosine sites with protein binding domains, the receptors bind to cytosolic partners that are responsible for the recruitment and activation of multiple downstream cascades.15,42,61 Activation through the mitogen-activated protein kinase (MAPK) cascades of the extracellular signal-regulated kinases (ERKs) is functionally linked to cellular proliferation. The phosphoinositide 3-kinase (PI3K) pathway leads to the activation of the serine/threonine protein kinase Akt (cellular homologue of the viral oncogene v-Akt), which is linked to cell survival. Other significant pathways mediated by ErbB signaling include activation and nuclear translocation of signal transducers and activators of transcription proteins (STATs)27 and clathrin mediated endocytosis.26,33,35,40,49 Even though detailed network models have been developed,8 the molecular context in which ErbB receptors activate and regulate signaling has not been fully recognized.
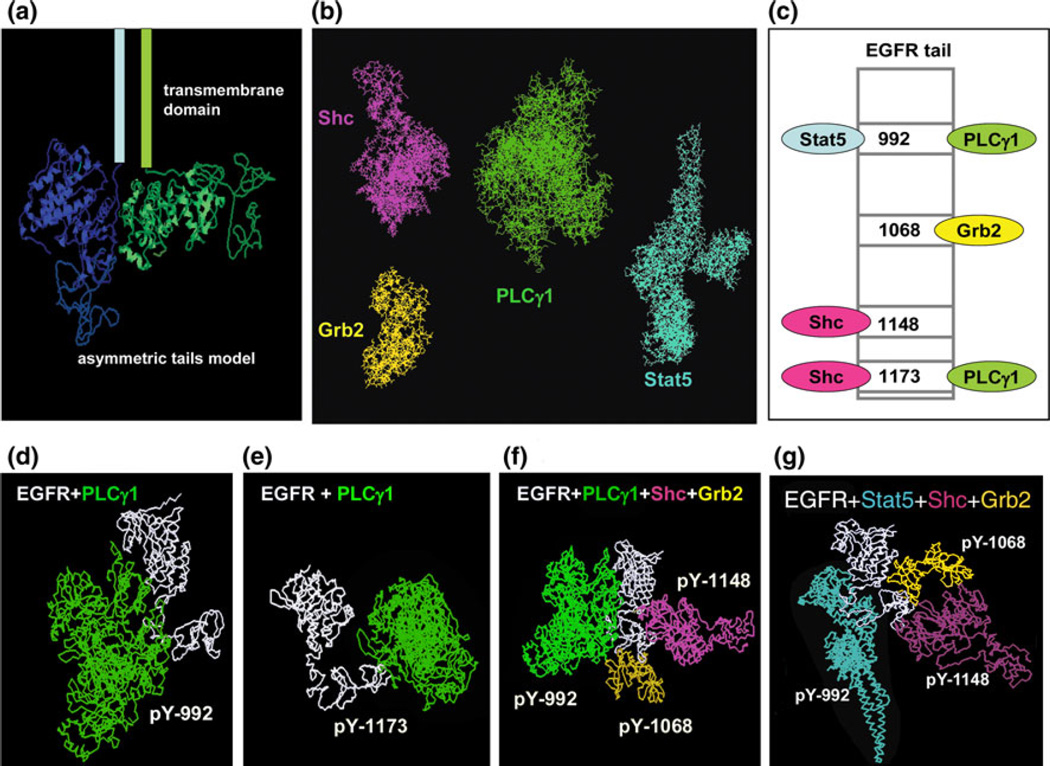
Hsieh et al.21 implemented an agent-based, CG model to investigate the kinetics of EGFR adaptor protein recruitment to the plasma membrane during EGFR signal initiation (see Fig. 2). Specifically, the model explored the combinatorial complexity associated with the simultaneous binding of adaptor proteins to the multiple phosphorylation sites positioned in the C-terminal tail of the EGFR kinase. The authors applied a set of rules, or constraints on the steric and diffusive properties of EGFR-adaptor protein binding, to govern the various possibilities for simultaneous docking of multiple proteins to the EGFR tail. In particular, the rules establish whether competitive or simultaneous adaptor protein recruitment is likely to occur. A multiscale approach was applied in that the model rules, which apply to the scale of receptor signaling and diffusion, were derived from CG molecular docking simulations, which consider the 3D structure of the proteins (Fig. 2). The model demonstrated that receptor clustering promotes efficient adaptor retainment (and hence, efficient signal transduction), and that binding of multiple adaptor proteins depends upon receptor density, binding kinetics, and membrane spatial organization. This study represents a novel approach to addressing the challenge of combinatorial complexity, as previous models of ErbB signaling3,25 rely on key assumptions in order to simplify the problem. One such assumption is disallowing the simultaneous recruitment of multiple adaptor proteins to the same phosphorylated EGFR tail based on competitive binding. Thus, by using information from one scale of representation (in this case, atomic-level modeling of receptor-adaptor binding) to parameterize a lower resolution model (i.e., assigning rules to govern the possibilities for combinations of adaptors docking to EGFR), the authors were able to address the combinatorial complexity in a unique way. Still, many challenges remain in our structural and mechanistic understanding of the protein–protein interactions in the ErbB pathway, including how the signals are regulated even within the receptor dimer system.6
FIGURE 2.
Coarse-grained molecular modeling of adaptor protein recruitment to the EGFR tail (reproduced with permission from Hsieh et al.21). In this model, EGFR has four cytoplasmic interaction partners. Some possible docking modes between the four adaptors and EGFR are shown.
Predictive Multiscale Modeling of the ErbB Mutational Landscape
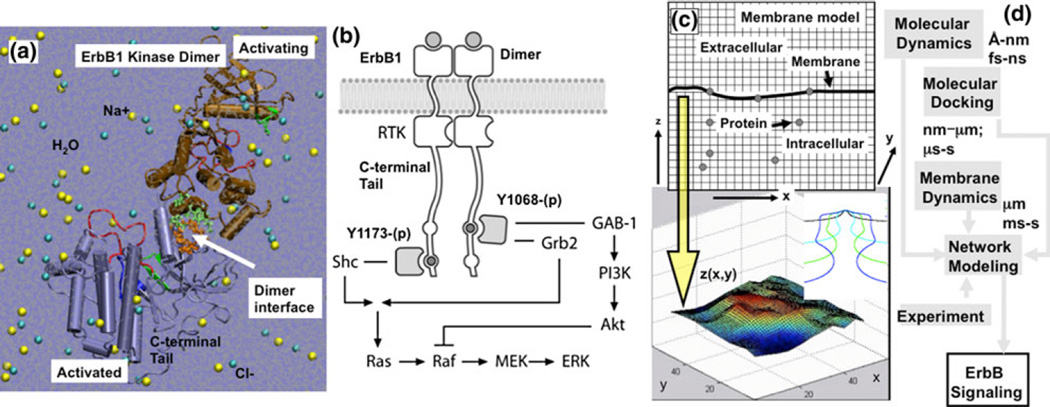
The previous examples (FGFR and EGFR) illustrate how known structures, when combined with modeling, can provide insights into the interacting components of a well-studied pathway. However, a more intriguing possibility is to apply modeling in a predictive mode as a means to propose new pathway elements. A combination of protein activity and interaction data can provide insights into the molecular details of the interactions, as well as the cellular and biochemical significance. More specifically, it is of great interest to investigate the molecular mechanisms that lead to the misregulation of ErbB signaling in pathologies such as cancer.45,47 Liu et al. and Shih et al. describe the application of a hierarchical multiscale modeling procedure28,45 to elucidate how point mutations in the ErbB1 receptor can profoundly alter signaling characteristics, leading to the onset of oncogenic transformation (Fig. 3). The flow of information between the models is depicted in the Fig. 3d. The researchers model the dimer-mediated receptor activation characteristics of the ErbB1 RTK using MD simulations. Figure 3a depicts the atomistic model of the explicitly solvated ErbB1 kinase dimer employed in the MD: trajectories of fully atomistic, explicitly solvated systems of wildtype and mutant ErbB1 kinase monomers and dimers are simulated and analyzed for specific stabilizing interactions such as hydrogen bonds and salt-bridges.46,48 They and others have also extended the molecular modeling to other ErbB family kinases, namely ErbB2/Her2,54,55 ErbB3/Her3,44,56 and ErbB4/Her4,46,48 and to binding of ErbB1 phosphotyrosines to cytosolic adaptor proteins such as Shc.51 Authors of these works treat receptor-mediated signaling using a deterministic network-based kinetic model,28,38,39,45,47 see Fig. 3b: the figure depicts the branched signaling network model for ErbB1-mediated signaling employed in this study. In the branched kinetic model, signaling through ErbB1 is modeled by combining three published models and augmented by an additional set of reactions to include the MAPK pathway, Akt, and PI3K activation. Shih et al. resolve phosphorylation of the active ErbB1 dimer at tyrosine site Y1068, a residue which, when phosphorylated, can bind to growth-factor-receptor bound-2 (Grb2) or Grb2-associated binding protein (GAB-1) proteins, and at tyrosine site Y1173, whose phosphorylated form can bind to the Src-homology-2-containing (Shc) adaptor protein. Phosphorylation of the signaling mediators Akt and ERK are used as indicators of downstream activation. Differences in ERK and Akt phosphorylation levels due to mutations in the ErbB1 receptor are implemented through changes to the kinetic parameters of the deterministic model. The altered parameters are computed through a combination of molecular dynamics and molecular docking simulations as well as through experiments published in the literature.45 Altogether, the network model comprises 74 reactions and 67 species. 17 of these reactions are novel to this work and represent enhanced molecular resolution to the ErbB1 activation, phosphorylation, and docking reactions, and enable separate parameterization of ErbB1 wildtype and mutant systems. Recently, Agrawal et al. and Ramanan et al. have extended the signaling module to include ErbB1 receptor internalization, which is treated using a hybrid discrete/continuum stochastic dynamics protocol.1,40 Figure 3c depicts the hybrid stochastic model for ErbB1 internalization: (top) grids in finite difference scheme for solving membrane dynamics (based on a continuum field model) and for modeling protein diffusion via lattice hopping; (bottom) snapshot of vesicle bud on the membrane in response to a specific spatial ordering of the curvature-inducing protein, epsin, on the membrane.
FIGURE 3.
(Reproduced with permission from Shih et al.45). Hierarchical multiscale modeling scheme for ErbB signaling. The dimer-mediated receptor activation characteristics of ErbB1 receptor tyrosine kinase are studied using molecular dynamics simulations. The interactions of substrate tyrosine containing peptides derived from the C-terminal tail with the ErbB1 kinase are studied using molecular docking simulations. Shih et al. have employed a deterministic network-based kinetic modeling scheme to study ErbB1-mediated signaling, and a hybrid discrete/continuum stochastic dynamics protocol to study the initiation of ErbB1 receptor internalization. (a) Atomistic model for ErbB1 dimer employed in the molecular dynamics and molecular docking calculations. (b) Branched network model for ErbB1-mediated signaling in which phosphorylation of the ErbB1 dimer occurs at either tyrosine Y1068, which can bind GAB-1 or Grb2, or at tyrosine Y1173, which binds Shc. Phosphorylation of the factors Akt and ERK were used as indicators of downstream activation. (c) Hybrid stochastic model for ErbB1 internalization. (Top) grids in finite difference scheme for membrane dynamics and the lattice in the kinetic Monte Carlo scheme for protein diffusion; (bottom) snapshot of vesicle bud on the membrane in response to a specific spatial ordering of the curvature-inducing protein, epsin, on the membrane; inset depicts a stabilized vesicle neck. (d) Flow of information between different simulation methods.
Predicting the Effects of Molecular Perturbations on Receptor Kinase Activation
Small molecule tyrosine kinase inhibitors of ErbB1/ 2 tyrosine kinase such as gefitinib, erlotinib, and lapatinib, which are ATP analogues, are of significant interest as cancer drugs. While the RTK inhibition approach has shown promise in some clinical trials, success has been mixed. In particular, the occurrence of somatic mutations in the ErbB1/HER1/EGFR kinase domain (L834R: where the leucine residue in position 834 is replaced by an arginine, L837Q: leucine at 837 replaced by a glutamine, G685S, del L723-P729 ins S: deletion of residues 723–729 and an insertion of a serine) as seen in non-small cell lung cancer patients renders the cell lines harboring such mutations more sensitive to treatment.29,36
To determine how such molecular perturbations can shape cellular fate, there is a need to characterize how the mutations affect the regulatory mechanisms within the kinase domains of ErbB1, ErbB2, and ErbB4.22 A recent structural and biochemical study of ErbB1 by Zhang et al.62 proposed a new dimer-mediated allosteric activation mechanism through which the ErbB1 RTK dimerizes in an asymmetric head-to-tail configuration. While the monomer ErbB1 kinase is stable in an inactive conformation, which interferes with ATP binding, in the asymmetric dimer configuration, one ErbB1 kinase domain serves as an activating protein and activates the other ErbB1 kinase in the dimer through allosteric contacts. The kinase–kinase contact at the asymmetric dimer interface allosterically stabilizes the active conformation. Shih et al. hypothesized that an underlying network of stabilizing hydrogen bonds dominates the relative stabilities of the inactive and active conformations and governs the kinase activation. The authors then performed a hydrogen bond analysis of the MD data, focusing on the interactions surrounding the activation-loop and the αC-helix sub-domains in the active and inactive conformations, and identified this stabilizing bonding network.46,48
To consider the effect of ErbB1 kinase dimerization on the network of stabilizing interactions, Shih et al. identified the protein residues participating in stabilizing bonds that are also proximal to the residues involved in the formation of the asymmetric dimer interface. The rationale for the proximity analysis is that the interactions in the stabilizing network could be compromised due to the molecular-level reorganization upon kinase dimerization. The proximity analysis suggested that 3 out of 13 stabilizing residues in the inactive state might be perturbed or lost upon dimerization, which suggests a potential molecular relay mechanism by which the kinase dimerization event activates the kinase domain. Indeed, when the authors performed MD simulations of an ErbB1 kinase dimer system (shown in Fig. 3a), they observed a significant rearrangement (change in the root-mean-squared deviation or RMSD of 3 Å) of the αC-helix position. The shift in the αC-helix position was accompanied by several changes in the stabilizing network consistent with the predicted bonding patterns. The dimer simulations reaffirm their hypothesis that the stabilizing network is susceptible to perturbation in the inactive conformation of the kinase, and that formation of the asymmetric dimer directly disrupts the network of interactions surrounding the αC-helix, thereby destabilizing the inactive state. The loss of these interactions and the shift of the αC-helix conformation towards the active state provide the impetus for kinase domain activation. Intriguingly, several of the clinically identified mutations that have been reported to constitutively activate the kinase also directly perturb the stabilizing network by breaking key stabilizing bonds. Thus, the delineation of the stabilizing hydrogen bond network provides molecular-level insight into the possible mechanisms by which activating mutations of ErbB1 kinase, such as L834R and del L723-P729 ins S, destabilize the inactive conformation. This preferential destabilization of the inactive conformation renders the receptor kinase constitutively active even as a monomer, producing high basal activation levels in the absence of growth factor-induced dimerization. Considering that there is an excellent correlation between the stabilizing network of interactions and the clinically identified activating mutations in ErbB1,38 the structural studies of kinase activation are well poised to forecast the mutation landscape associated with other ErbB family members. Shih et al. have extended their analysis for ErbB1 to the ErbB2 and ErbB4 kinases, in which they have identified similar networks of stabilizing interactions. Based on the similarities between the stabilizing interactions in the ErbB1 and ErbB4 kinase domains, the authors can predict the effect of analogous mutations in ErbB4 on kinase activation.46 With experimental validation of these predictions, we believe that such approaches are valuable for evaluating the likely effect of mutations on kinase activation and inhibition efficacies in cancer.
Transcribing the Effects of Molecular Alterations into Downstream Signal Activation
The preferential binding characteristics of different cytosolic substrates to specific phosphotyrosine sites of ErbB family kinases were recently reported.43 Differences in the phosphorylation kinetics associated with specific tyrosine sites of the cytoplasmic C-terminal tail of the ErbB1 kinase can induce differential patterns of downstream signaling and activation of key transcription factors. This leads to the hypothesis that the clinically identified activating mutations of ErbB1 kinase can potentially influence cellular homeostasis by directly altering the phosphorylation kinetics of ErbB1 substrate tyrosines. Indeed, the identity-specific phosphotyrosine kinetics for Y1068 and Y1173 (as well as other phosphotyrosine sites in ErbB1) for wildtype, L834R, and del L723-P729 ins S mutant systems are supported by the kinetic experiments of Mulloy et al.34 In particular, the relative catalytic turnover rates (λ = kcat/KM, where kcat represents the rate of tyrosine phosphorylation in the bound complex, and KM represents the affinity between the tyrosine substrate and the ErbB1 kinase) of Y1068 phosphorylation and Y1173 phosphorylation were measured in the experiments. The structural basis for the context-specific kinetics of the C-terminal tyrosine substrates is provided by computational docking calculations: substrate peptides derived from tyrosine sites of the ErbB1 C-terminal tail—Y1068 (VPEYINQ) and Y1173 (NAE-YLRV)—bound to the wildtype and the L834R mutant ErbB1 kinase revealed how the structure of the bound peptide/protein complex is altered at the catalytic site due to the arginine substitution of leucine in L834R.28,38
To translate differences in substrate specificity into tangible differences in the downstream response (ERK and Akt activation), Purvis et al. utilized a branched signaling model for ErbB1, see Fig. 3b, which features two parallel phosphorylation pathways corresponding to Y1068 and Y1173.38,45 Based on the results of docking simulations, they developed a molecularly resolved systems model in which phosphorylated Y1068 binds only to Gab-1 and Grb2 and not Shc, and phosphorylated Y1173 binds only to Shc and not to Gab-1 or Grb2, as depicted in Fig. 3b. The authors were then able to re-parameterize the model based on the identity-specific phosphotyrosine kinetics of Y1068 and Y1173 for wildtype and mutant (L834R and del L723-P729 ins S) ErbB1 and on the relative catalytic turnover (λ) rates. They also extended this model to ErbB1 kinase inhibition upon treatment with the small molecule inhibitor erlotinib in wildtype and mutant systems, again based on experimentally available inhibitor/ATP affinity data.
Using the different parameter values corresponding to wildtype, L834R, and del L723-P729 ins S mutant systems, they ran network simulations for different levels of EGF stimulation and ErbB1 expression levels: normal receptor expression and over-expression of ErbB1, with or without EGF stimulation. Based on the altered λ, the L834R mutant was shown to have a stronger preference for both Y1068 and Y1173 phosphorylation compared to the wildtype receptor, while the del L723-P729 ins S mutant exhibited increased Y1068 and decreased Y1173 phosphorylation. To gauge the downstream effects of differential signaling through Y1068 and Y1173 phosphorylation sites of ErbB1, they calculated the levels of ERK-(p) (phosphorylated ERK) and Akt-(p) (phosphorylated Akt) in the system simulations in response to changes in the phosphotyrosine kinetics (λ values) of Y1068 and Y1173. The effect of altered affinities of the Y1068 and Y1173 sites to the catalytic domain of ErbB1 is that the L834R mutant (for normal ErbB1 expression levels) exhibits differential downstream response, i.e., a pronounced decrease in ERK activation and relatively smaller decrease in Akt activation. The del L723-P729 ins S mutant, however, demonstrates sustained ERK as well as Akt activation relative to wildtype. For ErbB1 over-expressed cells, both ERK and Akt activation characteristics show relative insensitivity to ErbB1 as a result of signal saturation. The trends also reveal that the mutants can continue to signal even in the absence of the growth factor. In addition, the mutant signaling can differ due to changes in the ATP affinity. However, neither of these factors introduces any differential characteristics (in terms of preferring Y1068 to Y1173); each factor impacts the overall activation levels of ERK and Akt uniformly.38,45
Clinical Implications of Multiscale Modeling of ErbB Receptor Signaling
Using a model of the effect of Akt activation on cell response, Purvis et al. showed that preferential Akt activation is conducive for the cell to rely on ErbB1-mediated Akt activation for generation of pro-survival signals while requiring the initiation of death-inducing signals from other pathways. Their simplified model illustrates a mechanism by which inhibiting the dominant source of the pro-survival signals shifts the cellular homeostasis to a cellular state devoid of pro-survival signals, providing grounds for a remarkable inhibitor sensitivity.38,45 Purvis et al. hypothesized that the mechanisms, which lead to inhibitor hypersensitivity (as well as resistance) target points of network hypersensitivity and fragility. Since preferential Akt activation is a hallmark of the hyper-sensitive mutants and the efficacy of the inhibitors, they determined through a global sensitivity analysis38,45 the combinations of model parameter perturbations that drive enhanced production of Akt-(p) and ERK-(p). Interestingly, they noted a striking correlation between the components that drive enhanced production of Akt-(p) and ERK-(p) resulting from the model sensitivity analysis and patterns of oncogenic mutations and mechanisms of drug resistance found in clinical studies. For example, high frequency of mutations of PI3K, Ras (a GTPase named as an acronym for rat sarcoma), Gab-1, MEK (mitogen activated protein kinase), Raf (a serine/threonine kinase which activates MEK) have all been observed in several human cancers.46,57 Moreover, it has been established in screened breast and colorectal cancer patients that the GAB-1, MEK, and Ras mutations are non-random and likely arise from selective evolutionary pressures that give the cancer cells a survival advantage.46 With reference to the hypersensitive ErbB1 mutants found in non-small cell lung cancer patients, the perturbation of the phosphotyrosine kinetics of Y1068 and Y1173 through mutations (L834R and del L723-P729 ins S) is directly responsible for the differential signaling leading to preferential Akt activation. The restoration of signaling via reduction of Ki of the inhibitor and the simultaneous enhancement of KM associated with ATP binding has also been reported through a double mutation of L834R/T766M in ErbB1 kinase. This double mutant increases receptor phosphorylation (Y1068 and Y1173) kinetics 100-fold while simultaneously decreasing inhibitor affinity. Another drug resistance mechanism related to Y1068 kinetics (i.e., by circumventing Y1068 involvement and restoring downstream signaling through an alternative branch) has been identified: in the presence of ErbB3, a branch of signaling analogous to that through Y1068 becomes available through ErbB1-3 heterodimerization, directly resulting in PI3K recruitment to ErbB3 and subsequent Akt activation.
Multiscale Modeling of the ErbB3/HER3 Signaling Network
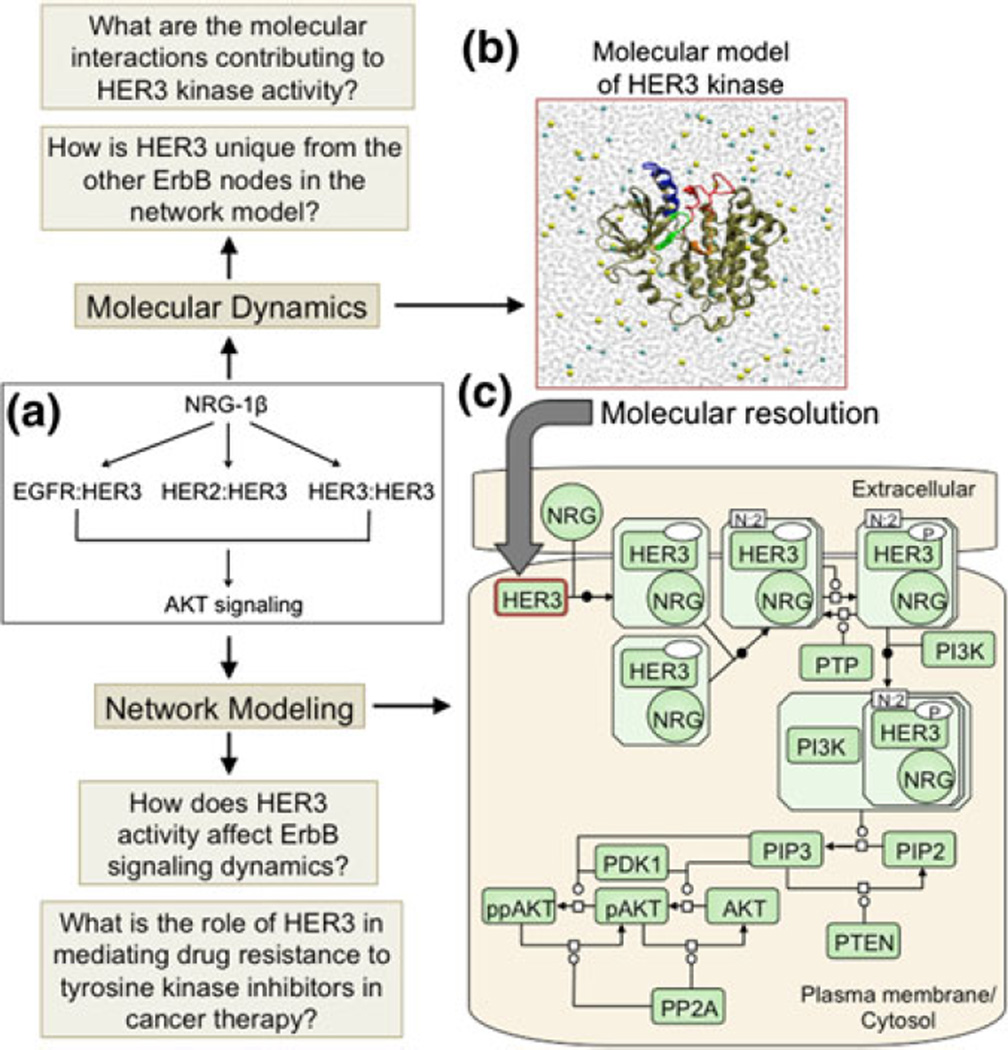
Recently, Telesco et al. described the application of a multiscale modeling scheme to the HER3/ErbB3 RTK signaling network (Fig. 4),56 which regulates critical cellular processes including proliferation, migration, and differentiation.16 Through a multiscale model, the authors investigated the most significant molecular interactions that contribute to potential mechanisms of HER3 activity and the physiological relevance of this activity to mechanisms of drug resistance in an ErbB-driven tumor cell in silico. The HER3 kinase is a topic of current interest and investigation,9 as it has been implicated in mechanisms of resistance to tyrosine kinase inhibition (TKI) of EGFR and HER2 in the treatment of many human malignancies.16 Moreover, the commonly regarded status of HER3 as a catalytically inactive ‘pseudokinase’ has recently been challenged by a combined modeling and crystallographic study,44 which demonstrated robust in vitro residual kinase activity for HER3 and raised the possibility that HER3 may play an active role in ErbB signaling dynamics. To investigate the relevance of HER3 activity to ErbB signaling in a cellular context, Telesco et al. proposed a pathway model of the HER3 signaling network (Fig. 4c), in which ligand stimulation of the HER3 RTK (as well as the EGFR and HER2 RTKs) results in induction of the PI3K-AKT cascade.56 The ligand-induced coupling of the EGFR, HER2, and HER3 nodes to the PI3K-AKT pathway has been extensively validated computationally and experimentally, although thus far HER3 has been postulated to play a passive role in the ErbB-AKT signaling network, in that its phosphorylation (and hence, recruitment of PI3K/AKT) depends upon the catalytic activities of the EGFR and HER2 RTKs. Their model proposed that HER3 can activate independently of its ErbB family members, a hypothesis which is reflected in the topology of the HER3 signaling pathway (Fig. 4c).56
FIGURE 4.
(Reproduced with permission from Telesco et al.56). Representation of the multiscale model of HER3 activity. (a) Schematic of the HER3 network model topology, which focuses on ligand stimulation of EGFR:HER3, HER2: HER3, and HER3:HER3 dimers, inducing the AKT cascade. (b) The HER3 node in (a) is examined at molecular resolution. The molecular model comprises two parts: homology modeling to refine the HER3 kinase crystal structure, and molecular dynamics simulations of the refined HER3 structure to identify the molecular features which distinguish HER3’s unique mechanism of activity from that of the EGFR and HER2 nodes in the HER3 network model. (c) Process diagram of the HER3 network model in SBGN notation. The aim of the HER3 network model is to investigate the implications of HER3 activity for ErbB signaling dynamics and mechanisms of HER3-mediated drug resistance in an ErbB-driven tumor cell in silico. Note that, for clarity, only HER3 dimers are illustrated in (c), although EGFR:HER3 and HER2:HER3 dimers are also present in the network model.
The multiscale model of HER3 activity was informed by atomic-level simulations of the HER3 kinase crystal structure (Fig. 4), in order to identify the molecular features that distinguish HER3 from the other nodes (EGFR, HER2) in the proposed ErbB signaling network. The results of the molecular-scale simulations supported the characterization of HER3 as a weakly active kinase44 that, in contrast to its fullyactive ErbB family members, depends upon a unique hydrophobic interface to coordinate the alignment of specific catalytic residues required for its activity.56 Translating the molecular simulation results of the uniquely active behavior of the HER3 kinase into a physiologically relevant environment, the HER3 signaling model demonstrated that even a weak level of HER3 activity may be sufficient to induce AKT signaling and tyrosine kinase inhibitor resistance in the context of an ErbB signaling-dependent tumor cell, and therefore therapeutic targeting of HER3 may represent a superior treatment strategy for specific ErbB-driven cancers.56
CONCLUDING REMARKS
Protein interaction networks provide an abstract view of macromolecular association and signaling, which can be useful for deducing the global features of a cellular network. However, such networks have a limited relationship with physical reality. A more realistic picture of the cell will ultimately develop when pathways and high-resolution structures can be integrated by a near complete repertoire of the 3D structures of protein complexes. This places experimental and computational structural biology in a crucial partnership and on equal footing with systems biology.3 Structural information describing interacting cellular components will produce a more complete whole-cell framework at atomic-level detail, which will be of immense benefit to modeling biological systems.
ACKNOWLEDGMENTS
We acknowledge financial support from NSF grants CBET-0853389 and CBET-0853539. Computational resources were provided in part by the National Partnership for Advanced Computational Infrastructure (NPACI) under the allocation grant MRAC MCB060006. S.E.T. was supported by the National Institutes of Health under Ruth L. Kirschstein National Research Service Award 2T32HL007954 from the NIH-NHLBI, a National Science Foundation Graduate Research Fellowship, and a Graduate Assistantship in Areas of National Need (GAANN) from the Department of Education.
Footnotes
CONFLICTS OF INTEREST
No conflicts of interest apply to this manuscript and related work.
REFERENCES
- 1.Agrawal NJ, Nukpezah J, Radhakrishnan R. Minimal mesoscale model for protein-mediated vesiculation in clathrin-dependent endocytosis. PLoS Comput. Biol. 2010;6(9):e1000926. doi: 10.1371/journal.pcbi.1000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alber F, et al. Determining the architectures of macro-molecular assemblies. Nature. 2007;450(7170):683–701. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- 3.Aloy P, Russell RB. Structural systems biology: modelling protein interactions. Nat. Rev. Mol. Cell Biol. 2006;7(3):188–197. doi: 10.1038/nrm1859. [DOI] [PubMed] [Google Scholar]
- 4.Ayton GS, Voth GA. Multiscale simulation of protein mediated membrane remodeling. Semin. Cell Dev. Biol. 2010;21(4):357–362. doi: 10.1016/j.semcdb.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayton GS, Voth GA. Multiscale computer simulation of the immature HIV-1 virion. Biophys. J. 2010;99(9):2757–2765. doi: 10.1016/j.bpj.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bessman NJ, Lemmon MA. Finding the missing links in EGFR. Nat. Struct. Mol. Biol. 2012;19(1):1–3. doi: 10.1038/nsmb.2221. [DOI] [PubMed] [Google Scholar]
- 7.Bhalla US, Iyengar R. Emergent properties of biological networks. Science. 1999;283:381–387. doi: 10.1126/science.283.5400.381. [DOI] [PubMed] [Google Scholar]
- 8.Birtwistle MR, et al. Ligand-dependent responses of the ErbB signaling network: experimental and modeling analyses. Mol. Syst. Biol. 2007;3:144. doi: 10.1038/msb4100188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boudeau J, et al. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16(9):443–452. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Bursulaya BD, et al. Comparative study of several algorithms for flexible ligand docking. J. Comput. Aided Mol. Des. 2003;17(11):755–763. doi: 10.1023/b:jcam.0000017496.76572.6f. [DOI] [PubMed] [Google Scholar]
- 11.Camacho JC, et al. Scoring dockied conformations generated by rigid body protein-protein docking. Proteins. 2000;40:525–537. doi: 10.1002/1097-0134(20000815)40:3<525::aid-prot190>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 12.Chen B, Tycko R. Simulated self-assembly of the HIV-1 capsid: protein shape and native contacts are sufficient for two-dimensional lattice formation. Biophys. J. 2011;100(12):3035–3044. doi: 10.1016/j.bpj.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C, Saxena R, Wei A. A multiscale model for virus capsid dynamics. Int. J. Biomed. Imaging. 2010;2010:308627. doi: 10.1155/2010/308627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen HM, et al. Evaluating molecular-docking methods for pose prediction and enrichment factors. J. Chem. Inf. Model. 2006;46(1):401–415. doi: 10.1021/ci0503255. [DOI] [PubMed] [Google Scholar]
- 15.Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat. Rev. Mol. Cell Biol. 2006;7(7):505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 16.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Martinez J, et al. Structure-function mapping of a heptameric module in the nuclear pore complex. J. Cell Biol. 2012;196:419–434. doi: 10.1083/jcb.201109008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 19.Gebremichael Y, Chu JW, Voth GA. Intrinsic bending and structural rearrangement of tubulin dimer: molecular dynamics simulations and coarse-grained analysis. Biophys. J. 2008;95(5):2487–2499. doi: 10.1529/biophysj.108.129072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hicks SD, Henley CL. Coarse-grained protein– protein stiffnesses and dynamics from all-atom simulations. Phys. Rev. E. 2010;81(31):030903. doi: 10.1103/PhysRevE.81.030903. [DOI] [PubMed] [Google Scholar]
- 21.Hsieh MY, et al. Spatio-temporal modeling of signaling protein recruitment to EGFR. BMC Syst. Biol. 2010;4:57. doi: 10.1186/1752-0509-4-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 23.Johnston IG, Louis AA, Doye JP. Modelling the self-assembly of virus capsids. J. Phys. Condens. Matter. 2010;22(10):104101. doi: 10.1088/0953-8984/22/10/104101. [DOI] [PubMed] [Google Scholar]
- 24.Karplus M, Kuriyan J. Molecular dynamics and protein function. Proc. Natl. Acad. Sci. USA. 2005;102(19):6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kholodenko BN. Cell-signalling dynamics in time and space. Nat. Rev. Mol. Cell Biol. 2006;7(3):165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirchhausen T. Three ways to make a vesicle. Nat. Rev. Mol. Cell Biol. 2000;1(3):187–198. doi: 10.1038/35043117. [DOI] [PubMed] [Google Scholar]
- 27.Kloth MT, et al. STAT5b, a mediator of synergism between c-Src and the epidermal growth factor receptor. J. Biol. Chem. 2003;278:1671–1679. doi: 10.1074/jbc.M207289200. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, et al. A multiscale computational approach to dissect early events in the Erb family receptor mediated activation, differential signaling, and relevance to oncogenic transformations. Ann. Biomed. Eng. 2007;35(6):1012–1025. doi: 10.1007/s10439-006-9251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 30.Mohammadi M, Schlessinger J, Hubbard SR. Structure of the FGF receptor tyrosine kinase domain reveals a novel autoinhibitory mechanism. Cell. 1996;86(4):577–587. doi: 10.1016/s0092-8674(00)80131-2. [DOI] [PubMed] [Google Scholar]
- 31.Mohammadi M, et al. Structures of tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276:955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- 32.Mohammadi M, et al. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. EMBO J. 1998;17:5896–5904. doi: 10.1093/emboj/17.20.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mosesson Y, Mills GB, Yarden Y. Derailed endocytosis: an emerging feature of cancer. Nat. Rev. Cancer. 2008;8(11):835–850. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- 34.Mulloy R, et al. Epidermal growth factor receptor mutants from human lung cancers exhibit enhanced catalytic activity and increased sensitivity to gefitinib. Cancer Res. 2007;67(5):2325–2330. doi: 10.1158/0008-5472.CAN-06-4293. [DOI] [PubMed] [Google Scholar]
- 35.Oved S, Yarden Y. Molecular ticket to enter cells. Nature. 2002;416:133–136. doi: 10.1038/416133a. [DOI] [PubMed] [Google Scholar]
- 36.Paez JG, et al. EGFR mutations in lung, cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 37.Periole X, et al. Combining an elastic network with a coarse-grained molecular force field: structure, dynamics, and intermolecular recognition. J. Chem. Theory Comput. 2009;5(9):2531–2543. doi: 10.1021/ct9002114. [DOI] [PubMed] [Google Scholar]
- 38.Purvis J, Ilango V, Radhakrishnan R. Role of network branching in eliciting differential short-term signaling responses in the hyper-sensitive epidermal growth factor receptor mutants implicated in lung cancer. Biotechnol. Prog. 2008;24(3):540–553. doi: 10.1021/bp070405o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Purvis J, et al. Proceedings of the Foundations in Systems Biology II. Stuttgart: IRB Verlag; 2007. Efficacy of tyrosine kinase inhibitors in the mutants of the epidermal growth factor receptor: a multiscale molecular/systems model for phosphorylation and inhibition; pp. 289–294. [Google Scholar]
- 40.Ramanan V, et al. Systems biology and physical biology of clathrin-mediated endocytosis. Integr. Biol. 2011;3(8):803–815. doi: 10.1039/c1ib00036e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saunders MG, Voth GA. Coarse-graining of multiprotein assemblies. Curr. Opin. Struct. Biol. 2012;22:144–150. doi: 10.1016/j.sbi.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 43.Schulze WX, Deng L, Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol. Syst. Biol. 2005;1:E1–E13. doi: 10.1038/msb4100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi F, et al. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc. Natl. Acad. Sci. USA. 2010;107:7692–7697. doi: 10.1073/pnas.1002753107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shih AJ, Purvis J, Radhakrishnan R. Molecular systems biology of ErbB1 signaling: bridging the gap through multiscale modeling and high-performance computing. Mol. BioSyst. 2008;4(12):1151–1159. doi: 10.1039/b803806f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shih AJ, Telesco SE, Radhakrishnan R. Analysis of somatic mutations in cancer: molecular mechanisms of activation in the ErbB family of receptor tyrosine kinases. Cancers. 2011;3(1):1195–1231. doi: 10.3390/cancers3011195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shih A, et al. The role for molecular modeling in multiscale cancer models. In: Deisboeck TS, Stamatakos G, editors. Multiscale Cancer Modeling of Cancer. Mathematical and Computational Biology Series. Boca Raton: Chapman & Hall/CRC; 2010. pp. 31–43. [Google Scholar]
- 48.Shih A, et al. Molecular dynamics analysis of conserved hydrophobic and hydrophilic bond interaction networks in ErbB family kinases. Biochem. J. 2011;436:241–251. doi: 10.1042/BJ20101791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sorkin A, et al. Epidermal growth factor receptor interaction with clathrin adaptors is mediated by the Tyr974-containing internalization motif. J. Biol. Chem. 1996;271(23):13377–13384. doi: 10.1074/jbc.271.23.13377. [DOI] [PubMed] [Google Scholar]
- 50.Stark H, et al. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature. 2001;409(6819):539–542. doi: 10.1038/35054102. [DOI] [PubMed] [Google Scholar]
- 51.Suenaga A, et al. Molecular dynamics simulations reveal that Tyr-317 phosphorylation reduces Shc binding affinity for phosphotyrosyl residues of epidermal. Growth Factor Recept. 2009;96(6):2278–2288. doi: 10.1016/j.bpj.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tama F, Brooks CL. Symmetry, form, and shape: guiding principles for robustness in macromolecular machines. Annu. Rev. Biophys. Biomol. Struct. 2006;35:115–133. doi: 10.1146/annurev.biophys.35.040405.102010. [DOI] [PubMed] [Google Scholar]
- 53.Taylor WR, Katsimitsoulia Z. A coarse-grained molecular model for actin-myosin simulation. J. Mol. Graph. Model. 2010;29(2):266–279. doi: 10.1016/j.jmgm.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 54.Telesco SE, Radhakrishnan R. Atomistic insights into regulatory mechanisms of the HER2 tyrosine kinase domain: a molecular dynamics study. Biophys. J. 2009;96(6):2321–2334. doi: 10.1016/j.bpj.2008.12.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Telesco SE, Shih A, Liu Y, Radhakrishnan R. Investigating molecular mechanisms of activation and mutation of the HER2 receptor tyrosine kinase through computational modeling and simulation. Cancer Research Journal. 2011;4(4):1–35. [PMC free article] [PubMed] [Google Scholar]
- 56.Telesco SE, et al. A multiscale modeling approach to investigate molecular mechanisms of pseudokinase activation and drug resistance in the HER3/ErbB3 receptor tyrosine kinase signaling network. Mol. BioSyst. 2011;7(6):2066–2080. doi: 10.1039/c0mb00345j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang E, editor. Cancer Systems Biology. Mathematical and Computational Biology Series. London: CRC Press/Taylor and Francis; 2010. [Google Scholar]
- 58.E W, Engquist B. Multiscale modeling in computation. Notices AMS. 2003;50(9):1062–1070. [Google Scholar]
- 59.E W, Engquist B, Huang ZY. Heterogeneous multiscale method: a general methodology for multiscale modeling. Phys. Rev. B. 2003;67(9):092101. [Google Scholar]
- 60.Westerhoff HV, Palsson BO. The evolution of molecular biology into systems biology. Nat. Biotechnol. 2004;22(10):1249–1252. doi: 10.1038/nbt1020. [DOI] [PubMed] [Google Scholar]
- 61.Yarden Y, Sliwkowski MX. Untangling the ErbB signaling network. Nat. Rev. Mol. Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 62.Zhang X, et al. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125(6):1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]