Abstract
The non-bilaterian animals comprise organisms in the phyla Porifera, Cnidaria, Ctenophora and Placozoa. These early-diverging phyla are pivotal to understanding the evolution of bilaterian animals. After the exponential increase in research in evolutionary development (evo-devo) in the last two decades, these organisms are again in the spotlight of evolutionary biology. In this work, I briefly review some aspects of the developmental biology of nonbilaterians that contribute to understanding the evolution of development and of the metazoans. The evolution of the developmental genetic toolkit, embryonic polarization, the origin of gastrulation and mesodermal cells, and the origin of neural cells are discussed. The possibility that germline and stem cell lineages have the same origin is also examined. Although a considerable number of non-bilaterian species are already being investigated, the use of species belonging to different branches of non-bilaterian lineages and functional experimentation with gene manipulation in the majority of the non-bilaterian lineages will be necessary for further progress in this field.
Keywords: early-diverging metazoans, BMP, FGF, Wnt, stem cells, evolution, nervous system
Introduction
The field of evolutionary developmental biology (evo-devo) has gained momentum since its foundation in the 1980s, when HOX genes were discovered in both fly and mouse (Carroll et al., 2005). In its infancy, evo-devo studies were mainly carried out on classical developmental biology models, capitalizing on previously established techniques and a large body of knowledge on the developmental molecules and genetics of these organisms (Sommer, 2009; Giles and Averof, 2014). As developmental biology is traditionally linked to medical sciences, the animal models utilized in these former studies were mainly vertebrates and the fly. However, animal diversity greatly surpasses the variety represented by these organisms. If we are to understand the evolution of developmental processes, and how these processes are related to the evolution of the animals, more models are needed (Jenner and Wills, 2007; but see Sommer, 2009 for a different perspective). Thus, we are now experiencing an explosion of new model systems (Sommer, 2009), especially due to lower costs for sequencing genomes and transcriptomes and other technological improvements (Giles and Averof, 2014).
To understand how the diversity of forms and body plans of multicellular animals arose it is necessary to investigate the five major lineages of metazoans living today. Vertebrates, Caenorhabditis elegans (a nematode) and the fly belong to the Bilateria lineage. The other four lineages (the phyla Porifera, Cnidaria, Ctenophora and Placozoa) consist of comparatively simple animals that generally lack bilateral symmetry in their body plans (however bilateral symmetry is present in the body plan of some cnidarians). These lineages are artificially grouped as non-bilaterian animals (also known as ‘early-branching’ or ‘basal’ metazoans) (Dohrmann and Wörheide, 2013). Non-bilaterians range from benthic sessile filter feeders to gelatinous planktonic carnivorous animals. One of these phyla, Porifera (sponges), has been traditionally described as organisms devoid of true epithelia and lacking the gastrulation stage during development (Leys, 2004; Leys and Hill, 2012; see below). Sponges lack a clear anterior-posterior (AP) polarity as adults (but the larvae do swim directionally), have no organs, are filter feeders, and have a body plan adapted to process as much water as needed for feeding and respiration (Leys and Hill, 2012).
The other three non-bilaterian phyla are diploblastic (adult tissues are derivatives of two germ layers: ectoderm and endoderm [also known as endomesoderm]; Martindale, 2005). Placozoa lack AP polarity, nerve and muscle cells and a proper gut. The body plan of organisms in this phylum is very simple compared to other metazoans. They have a disc-shaped, double-layered body comprising only six cell types (Smith et al., 2014). Representatives of this phylum absorb nutrients through contact of the lower epithelium with the substratum (Brusca and Brusca, 2003; Srivastava et al., 2008). Very little is known about their development (Eitel et al., 2008). Cnidaria (anemones, jellyfish, and their kin) and Ctenophora (comb jellies) pass through the gastrulation stage during embryogenesis, producing a gut that persists throughout the life cycle. The gut is blind ending in cnidarians, while ctenophores have an anal pore. Anal pores may serve as a primitive anus, assisting the mouth to egest undigested remains of the prey, and as an exit for metabolic wastes (Brusca and Brusca, 2003). Adults of both phyla have tentacles to capture prey and oral-aboral polarity is seen throughout their life cycle (Martindale, 2005).
Although the phylogenetic relationships among nonbilaterians and also between non-bilaterians and Bilateria are still contentious (see below), it has been assumed that the developmental biology of these four early-branching phyla may play a pivotal role in interpreting the evolution of metazoans. Although genomic and transcriptomic data indicate that these phyla are similar to other metazoans, their body plans are quite different from those of Bilateria and their development may be key to understanding many questions about animal evolution. As many of the events in development may be obscured due to evolutionary divergence, use of molecular markers can facilitate our understanding about deep homologies and the evolution of animals (Gold and Jacobs, 2003). Here, I provide a brief review of how studies on the development of non-bilaterians are aiding (or not) understanding of the evolution of animals. I present the findings that contributed to this knowledge and the gaps that still require research. My objective is to compile information about different topics related to non-bilaterian evo-devo in a single work. Consequently, I do not provide a thorough review of the literature on these topics, as others have comprehensively discussed each issue, usually for each specific taxon (e.g. Technau and Scholz, 2003; Lee et al., 2006; Burton, 2008; Galliot et al., 2009; Watanabe et al., 2009; Bosch et al., 2010; Technau and Steele, 2012; Ereskovsky et al., 2013; Funayama et al., 2013; Gold and Jacobs, 2013).
Phylogenetic Relationship of Non-Bilaterian Animals
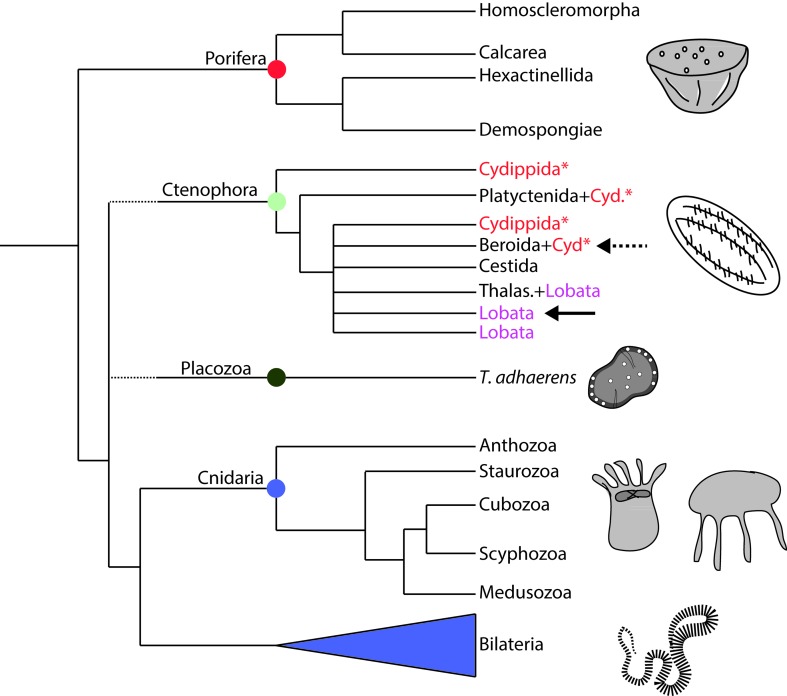
One of the most disputed issues on the phylogeny of animals is the topology of the five major extant lineages of the metazoan tree (Porifera, Cnidaria, Ctenophora, Placozoa, and Bilateria) (Edgecomb et al., 2011; Dohrmann and Wörheide, 2013). Understanding the relationships forming the basis of the metazoan tree of life is important for answering questions about how metazoans traits (including development) arose and evolved over the last 700 million years (Dohrmann and Wörheide, 2013). In the last two decades a forest of trees with very conflicting scenarios has been published. From paraphyletic Porifera (the ‘we are all derived from sponge larvae’ scenario; Nielsen, 2008), to ctenophores branching out of the metazoan tree earlier than any other phyla (the ‘Ctenophora-first’ scenario; Ryan et al., 2013; Moroz et al., 2014;), a wide variety in the phylogeny of basal metazoans has been proposed (reviewed in Dohrmann and Wörheide, 2013). These inconsistencies may be due to insufficient molecular sampling and/or inadequate sampling of taxa (Phillipe et al., 2009). However, addition of more sequences is not sufficient to provide a more reliable phylogeny of metazoans, and more integrative approaches combining morphology, development, cytology and genome architecture are fundamental to comprehend animal phylogeny (Philippe et al., 2011; Dohrmann and Wörheide, 2013). Dohrmann and Wörheide (2013) suggest that it is better to rely on solid evidence (such as it is) instead of proposing new scenarios with every newly published tree. I agree with this view and use the ‘conservative’ evolutionary scenario proposed by these authors in this current work (Figure 1). Nonetheless, when necessary I discuss different interpretations of findings from the perspective of alternative phylogenetic scenarios.
Figure 1. Phylogenetic relationship among the non-bilaterians phyla and their respective subclasses. Dashed lines indicate uncertain locations of the branches. Many of the orders of Ctenophora are not monophyletic (Cyd* = Cydippida). Solid arrow points to phylogenetic position of M. leidyi and dashed arrow points to the phylogenetic position of P. pileus. See ‘Phylogenetic relationship of non-bilaterian animals’ for references.
Even though the relationship among phyla is still not agreed, the relationships of groups within phyla appear robust (Figure 1), at least in sponges and cnidarians. Sponges have four monophyletic classes (Calcarea, Homoscleromorpha, Demospongiae, and Hexactinellida) divided into two groups: Silicea (Demospongiae and Hexactinellida) and another group containing Homoscleromorpha and Calcarea, for which no morphological synapomorphy has been found (Philippe et al., 2009; Wörheide et al., 2012).
Placozoa is an enigmatic phylum that historically been classified as monospecific, with Trichoplax adhaerens the single species of the group. However, Voigt et al.(2004)showed that the high genetic divergence among placozoan lineages is similar to that observed, in other metazoans, for different families.
Cnidaria has five monophyletic classes: Anthozoa, Staurozoa, Cubozoa, Scyphozoa, and Hydrozoa; Anthozoa is a sister group of Medusozoa, which contains the other the four classes (Figure 1; Collins et al., 2006). Ctenophora is considered the most problematic branch among the nonbilaterians (Simion et al., 2015). The classical taxonomy of this group splitted the phylum in two classes Tentaculata (with six orders) and Nuda (one order). Even before the use of molecular markers for phylogenetic studies, Harbison (1985)suggested that these classes were not valid, abandoning the use of this hierarchical group in ctenophores. More recently, Simion et al. (2015) have shown that many of the orders, families and even genera are not monophyletic (Figure 1). Analysis of 18S RNA and ITS1 + 5.8S + ITS2 sequences of several species of the phylum suggests that present-day ctenophores have passed through a recent radiation followed by a bottleneck (Podar et al., 2001; Simion et al., 2015). Even though there are still large gaps in understanding of the phylogeny of the non-bilaterians, current knowledge is already helping us to understand how the development of these animals evolved and promoted the diversification of the metazoans.
Brief Overview of the Embryogenesis of Non-Bilaterian Animals
Porifera
It is very difficult to generalize the development of sponges. There is large variation in developmental types even within a single class (Ereskovsky, 2010; Wörheide et al., 2012). Gametogenesis starts from somatic cells (choanocytes or archaeocytes) that transdifferentiate either into spermatogonia or oogonia. The gametogenic steps, however, are similar to other metazoans. There are four main types of cleavage: chaotic in Homoscleromorpha and some oviparous Demospongiae; radial in oviparous Demo-spongiae and Hexactinellida; polyaxial in a few Demo-spongiae and one of the subclasses of Calcarea (Calcinea); and incurvational (palyntomic cleavage) in the other subclass of Calcarea (Calcaronea) (Ereskovsky et al., 2013). The boundaries of subsequent stages of development (blastulation, gastrulation, histogenesis and organogenesis) are fuzzy in all groups of sponges. Blastulation is hard to delineate because by the end of cleavage the cells of the embryo have already started to differentiate (Ereskovsky, 2010). Gastrulation in Porifera is also controversial, but there is extensive morphogenesis and cell differentiation following cleavage. Finally, the embryo leads to a free-living larva (there are eight types of larvae; see Maldonado and Bergquist, 2002; Ereskovsky, 2010) that swims unidirectionally using cilia until it settles and starts metamorphosis. In demosponges and homoscleromorphs a series of cell rearrangements and differentiation takes place to form the rhagon, a juvenile with a single osculum and several choanocyte chambers, while in Calcarea, the olynthus, a juvenile with a single, sac-like, choanocyte chamber that opens in a apical osculum, is formed (Ereskovsky, 2010).
One of the main characteristics of sponges is their high potential for asexual reproduction by fragmentation, budding or gemmulation (Funayama, 2010). Gemmules are mainly found in freshwater sponges as bodies resistant to the harsh environment. This characteristic allows them to be kept in laboratory conditions and, subsequently, be used as a model to study cell differentiation and aquiferous system development (e.g. Funayama, 2010; Windsor and Leys, 2010; Leys, 2015).
Some species of Porifera are currently used as evodevo model species (Table 1). The main models are the demosponge Amphimedon queenslandica and the calcarean Sycon ciliatum, in which several steps of development have been investigated, and genomes and transcriptomes sequenced. In addition, many studies investigating the pattern of gene expression during development are available (e.g. Adamska et al., 2007; Srivastava et al., 2010a; Fortunato et al., 2014; Leininger et al., 2014). There are also other species that have been used in experimental manipulations. Borojevic and Levi (1964) and Borojevic (1966) investigated cell differentiation during metamorphosis of the larvae of the demosponge Mycale contarenii. In addition, in the last twenty years, studies have investigated the pattern of gene expression in the demosponges Ephydatia spp. and Suberites massa and in the homoscleromorphs Oscarela spp. (Table 1) (Adell et al., 2003; Perovik-Ottstadt et al., 2004; Lapébie et al., 2009; Windsor and Leys, 2010; Okamoto et al., 2012; Funayama, 2013). More recently, gene expression has been manipulated using drug treatments and RNA interference. Although drug treatments to disturb gene function are not specific, there have been some attempts to manipulate development using different protocols that have provided promising results (Lapébie et al., 2009; Windsor and Leys, 2010). Rivera et al. (2011) demonstrated that, by soaking sponges with double-stranded RNA (RNAi) or feeding them with bacteria expressing RNAi, it was possible to efficiently affect target genes. However, little progress has been achieved since this publication.
Table 1. Summary list of non-bilaterian models being used in evo-devo research. (Modified from Technau and Steele, 2012).
| Phylum | Class * | Model | Genome | Transcriptome | T/M/KDM * | Font |
|---|---|---|---|---|---|---|
| Porifera | Demospongiae | Amphimedon queenslandica | YES | YES | NO | Srivastava et al., 2010a |
| Ephydatia muelleri | NO | NO | NO | Windsor and Leys, 2010 | ||
| Ephydatia fluviatilis | NO | NO | NO | Funayama, 2010 | ||
| Calcarea | Sycon ciliatum | YES | YES | NO | Leininger et al., 2014 | |
| Sycon coactum | NO | YES | NO | Riesgo et al., 2014 | ||
| Leucosolenia complicata | YES | YES | NO | Leininger et al., 2014 | ||
| Homoscleromorpha | Oscarella lobularis | NO | YES | NO | Ereskovsky et al., 2009 | |
| Oscarella carmela | YES | YES | NO | Nichols et al., 2006 | ||
| Cnidaria | Hydrozoa | Hydra magnipapilata | YES | YES | Yes, No, RNAi | Chapman et al., 2010 |
| Clytia hemispherica | YES | YES | No, No, Mo ** | Houliston et al., 2011 | ||
| Hydractinia echinata | NO | NO | Yes, No, RNAi | Kraus et al., 2014 | ||
| Podocoryne carnea | NO | NO | NO | Spring et al., 2002 | ||
| Scyphozoa | Aurelia aurita | NO | YES | NO | Fuchs et al., 2014 | |
| Anthozoa | Nematostella vectensis | YES | YES | Yes, No, Mo/RNAi | Putnam et al., 2007 | |
| Acropora digitifera | YES | YES | NO | Shinzato et al., 2011 | ||
| Cubozoa | Tripedalia cystophora | NO | NO | NO | Piatgorsky and Kozmik, 2004 | |
| Ctenophora | Lobata | Mnemiopsis leidyi | YES | YES | NO | Ryan et al., 2013 |
| Pleurobrachiidae | Pleurobrachia bachei | YES | YES | NO | Moroz et al. | |
| Pleurobrachia pileus | NO | YES | NO | Alié et al., 2011 | ||
| Placozoa | - | Trichoplax adhaerens | YES | YES | Yes, No, No | Schierwater et al., 2009 |
T/M/KDM = Transgenics, Mutants and knockdown methods.
Mo = morpholino.
Placozoa
Placozoans present the simplest morphology among all metazoans: they have a clear baso-apical polarity (upper vs. lower epithelia), no mesenchymal tissues and are formed by only six somatic cell types (Smith et al., 2014). The T. adhaerens genome was sequenced even before the basic biology (e.g. life cycle and development) was unraveled, probably because of the difficulties in culturing placozoan embryos as they usually die around the 128-cell stage (Srivastava et al., 2008; Eitel et al., 2011). Thus, knowledge of the development of Placozoa is more complete for investigation of the molecular toolkit than for embryological studies. For instance, in the last decade some studies using in situ hybridization (ISH) showed that developmental molecules are not only present, but also are expressed in adults (Jakob et al., 2004; Hadrys et al., 2005; Schierwater et al., 2009). Morpholinos and RNAi have been used to investigate gene function in these animals (Jakob et al., 2004).
Cnidaria
Numerous studies during the last two centuries have investigated embryogenesis and the developmental biology of cnidarians. Cnidarians are diploblastic, radially symmetrical animals that include species of jellyfish, corals, and sea anemones (Brusca and Brusca, 2003). Cnidarians have a remarkably complex life cycle. To some extent, all groups (except Anthozoa) present metagenesis, i.e. an alternation between polyp and medusa forms (Brusca and Brusca, 2003). Gametogenesis takes place in gonads growing in contact with the coelenteron (a cavity inside the organism). Eggs and sperm cells derive from mesenchymal cells (Extavour et al., 2005). Fertilization occurs either in the water column or inside the parental organism bearing the eggs. The eggs usually lack a primary animal-vegetal axis, which is achieved with sperm entry, setting the body axis just before the first cleavages (Byrun and Martindale, 2004; see Momose et al., 2008 for a different perspective in the hydrozoan Clytia). Cleavages in this phylum are usually random, but some species of direct-developing medusozoans present a more defined pattern during the first three divisions (Freeman, 1983).
After cleavage, the embryo becomes a blastula (either solid or hollow) that gastrulates to form a ciliated planula larva. Gastrulation in this phylum takes nine different forms. There are two types of ingression (unipolar and multipolar), three forms of delamination (blastula, morula, and syncytial), invagination, epiboly, and at least two forms of mixed gastrulation (involving more than one form of morphogenetic movements: (i) mixed delamination and (ii) forms combining ingression and invagination) (Byrun and Martindale, 2004). These different forms of gastrulation are not exclusive or specific for the different evolutionary lineages of the group, although invagination seems to be the ancestral form (Byrun and Martindale, 2004). Gastrulation leads to a diploblastic embryo, with an archenteron and a blastopore. The archenteron becomes the endoderm, from which the gut develops, and the ectoderm becomes the epidermis (including cnidocytes) and the nervous cells (however, in hydrozoans, the nervous cells are derived from interstitial cells, which derive from endodermal cells) (Byrun and Martindale, 2004; Technau and Steele, 2012). During metamorphosis, cell proliferation starts at the bud primordia located at the apical region of the embryo, which eventually form the tentacles.
Similar to sponges, cnidarians are well-known for their ability to regenerate lost parts. Hydra has been used for centuries to investigate the regeneration process in metazoans. Regeneration in these organisms occurs by a mechanism that does not require proliferation and growth (morphalaxis) (Cummings and Bode, 1984; Technau and Steele, 2012). It seems that the WNT pathway and transcription factor Brachyury play an important role in patterning and differentiation of cells during this process in Hydra (Technau and Bode, 1999; Technau and Steele, 2012). Investigation of the regeneration process of Nematostella vectensis indicates that an alternate developmental trajectory is taken to reconstruct the body of the animal. Anthozoan regeneration uses molecular mechanisms different to both anthozoan embryonic development and regeneration in Hydra (Burton and Finnerty, 2009; Technau and Steele, 2012).
The developmental biology of many species of Cnidaria is currently being investigated. The genomes of a few species are already available (Table 1). In addition, there are many other species that are routinely investigated in other lineages of the phylum (Table 1). There are many studies dealing with gene expression and experimental embryology of Hydra, Clytia, Acropora, Nematostella and Aurelia (Piatgorsky and Kozmik, 2004; Houliston et al., 2010; Fuchs et al., 2014; and other studies reviewed in Technau and Steele, 2012). Transgenic lineages and mutants are available for Hydra, while gene knockdown through RNAi and morpholinos has already been used in some models (Wittlieb et al., 2006; Kunzel et al., 2010; Renfer et al., 2010; Technau and Steele, 2012).
Ctenophora
Ctenophore development has been studied for a long time but in a limited number of species. Most ctenophores are hermaphroditic. Gametogenesis takes place in gonads located in the walls of the meridional canals, with sperm and eggs produced at the same time (Byrun and Martindale, 2004; Alié et al., 2011). Gametes are shed into the water through the mouth or, in some cases, through a gonadal duct (Brusca and Brusca, 2003). After spawning, the gametes meet in the water column and polyspermy is common. The embryogenesis of ctenophores is unique and stereotyped. The first cleavage occurs in the sagittal plane, while the second occurs in the tentacular plane. The following cleavage generates E and M blastomeres that subsequently divide to form a morula with large and small blastomeres (see Martindale and Henry, 1997). Later, micromeres cover the macromeres through epiboly generating the ectoderm (micromere-derived) and endoderm (macromere-derived). Mesodermal cells appear later in development, deriving from micromeres located in the oral region (oral micromeres), with the fate of these cells established early in development. The oral micromeres invaginate to form the pharynx. Thereafter, the embryo reaches the cydippid larval stage which shares the same symmetry and body plan of the adults, and grows until the adult forms (some groups do not pass through this stage). Therefore, ctenophores can be considered direct developers (Byrun and Martindale, 2004).
Comb jellies can regenerate virtually any lost part of their body, including the complex apical system (Brusca and Brusca, 2003). However, the molecular mechanisms of regeneration and the repatterning of tissues and cells are still poorly investigated (Martindale and Henry, 1996). The main ctenophore models used in evo-devo are the invasive species Mnemiopsis leidyi, for which data on fate maps, gene expression, functional experimentation and genomics are available (e.g. Martindale and Henry, 1997; Ryan et al., 2013), and Pleurobrachia pileus, which has attracted increased attention in recent years (e.g. Alié et al., 2011; Jager et al., 2011, 2013; Dayraud et al., 2012).
Investigations of the developmental biology of cnidarians have led to the establishment of new non-bilaterian models. As Technau and Steele (2012) highlighted, when investigations on cnidarians started the "major goal was to determine whether the genetic toolkit used to construct the bilaterian embryo [...] was in place in the ancestor of cnidarians and bilaterians." Recently, other models have been added to answer other specific and more general questions. Below, I review some of the evolutionary questions that understanding of the development of non-bilaterians is helping to address.
Was the Basic Developmental Genetic Toolkit Present in the Animal Ancestor?
In their famous evo-devo book, Carroll et al. (2005) stated: "Basal animal lineages, including the diploblast phyla (Cnidaria, Ctenophora) and the Porifera (sponges) have much less developmental and morphological complexity than do bilaterians." At that time, little was known about the genome and developmental genetics of the nonbilaterians, leading them to ask: "Does their simple body organization reflect a smaller complement of toolkit genes? Or did the bilaterian toolkit predate the origin and radiation of animals altogether?" (Carroll et al., 2005). With more than 10 genomes of non-bilaterians currently available and many other transcriptomes, we are now closer to answer these questions.
We now know that much of the developmental signaling and transcription factors repertoire was already present in the metazoan stem (Degnan et al., 2009; Srivastava et al., 2010a; Riesgo et al., 2014). Consequently, the comparatively morphological simplicity of non-bilaterian animals and the great differences in morphology between sponges and arthropods or vertebrates cannot be due to the absence of these protein-coding gene families. The increase in the complexity of the body plan observed in bilaterians must involve the further expansion of transcription factor families, as well as differences in the temporal and spatial expression of these genes and their regulation (Degnan et al., 2009; Srivastava et al., 2010a; Erwin et al., 2011). Srivastava et al.(2010a)suggested a correlation between the appearance or expansion of gene families and animal complexity. For instance, they argued that the enrichment of homeobox transcription factors and gap-junction proteins were necessary for the increase in complexity observed in bilaterians (Srivastava et al., 2010a). The developmental toolkit genes are not only present in the genome of the non-bilaterians, as are also expressed during their development. However, there are striking absences in the genomes of each lineage, such as the antagonists of some signaling pathways (such as WNT and TGF-BETA) in sponges and ctenophores, some genes related to the deployment of mesoderm in sponges, and some downstream genes of genetic developmental pathways (e.g. in placozoans, Notch and JAK/STAT signaling pathways are present, but molecules involved in the transduction of the signal, such as Janus kinase in the JNK pathway, or Notch-like gene with true Notch domain are absent) (Srivastava et al., 2008, 2010a; Ryan et al., 2013).
The Antennapedia (ANTP)-superclass of homeobox-containing transcription factors counts for an interesting story. In bilaterians, these genes are grouped in three clusters: Hox, Para-Hox and NKL (Holland, 2013). Multiple members of these classes were initially found in anthozoans, indicating that they preceded the cnidarianbilaterian divergence (Ryan et al., 2006). Later, members of the ANTP-superclass were also found in the genome of sponges, placozoans, ctenophores and other cnidarians (e.g. Monteiro et al., 2006; Derelle and Manuel, 2007; Larroux et al., 2007; Chapman et al., 2010; Fortunato et al., 2014). Larroux et al. (2007) found different genes belonging to the NKL cluster in Amphimedon, but no Hox or ParaHox genes, which made them suggest that the NKL cluster pre-dated the appearance of the other ANTP clusters in metazoans. Furthermore, as non-bilaterians lack a distinct mesoderm (see below) the maintenance of the NKL cluster in these organisms could be the result of fundamental regulatory constraints inherent to the cluster’s organization (Derelle and Manuel, 2007; Larroux et al., 2007). In anthozoans, Hox genes are present collinearly and spatially expressed, as observed in modern bilaterians (Finnerty et al., 2004; Putnam et al., 2007). Placozoans present a single gene with sequence similar to a Hox-gene (TROX-2), which is currently considered as a ParaHox gene (Mendivil Ramos et al., 2012). However, Hox and ParaHox genes were not recovered in the genome of Amphimedon and Mnemiopsis, raising the hypothesis that all animals except sponges and ctenophores form a group called ‘ParaHoxozoa’ (Ryan et al., 2010; Fortunato et al., 2014). Ctenophores appear to lack Hox and ParaHox genes, as these genes were not recovered in 10 species belonging to different evolutionary lineages of the phylum (Moroz et al., 2014). However, a recent analysis of transcriptomes and genome assemblies of calcareous sponges Sycon and Leucosolenia indicated the presence of CDX ParaHox genes in Porifera, indicating that Amphimedon may have lost these genes (Fortunato et al., 2014). Fortunato et al. (2014) further found that ParaHox genes were already present in pre-poriferan lineages corroborating the ‘ghost locus’ hypothesis for the origin of Hox/ParaHox genes in animals (Mendivil Ramos et al., 2012). These findings highlight the importance of sampling a larger pool of species belonging to different evolutionary lineages to understand the evolution of the developmental genetic toolkit.
Could the Establishment of the Axial Properties be Conserved Throughout Metazoan Evolution?
The body plans of animals can be defined in terms of symmetry and axes of polarity. Bilaterians have well defined anteroposterior and dorsoventral axes, while the majority of sponges, cnidarians and ctenophores (at least during the larval stage) present a single axis: the apicobasal (or oral-aboral) (Manuel, 2009; Houliston et al., 2010). Hox genes define the anteroposterior axis in many bilaterians (Ryan and Baxevanis, 2007; Petersen and Reddien, 2009). However, Hox genes are mostly absent in sponges and ctenophores (Adamska et al., 2011; Ryan et al., 2013; Fortunato et al., 2014). In cnidarians the evolutionary history of Hox is complicated and these genes seem to play no role in the establishment of the primary axis of these animals (Houliston et al., 2010). So, what sets the primary axis of non-bilaterians? And is the non-bilaterian embryonic polarity homologous to that observed in bilaterians?
There is mounting evidence that the WNT pathway establishes the polarity of the main axis in sponges and cnidarians (Lee et al., 2006; Adamska et al., 2007; Leininger et al., 2014). WNT ligands and downstream genes are expressed in the posterior region of the larvae and the apical region of the juvenile in both Amphimedon and Sycon (Adamska et al., 2007; Leininger et al., 2014). The same pattern can be observed in Hydra, Nematostella, Clytiaand other cnidarians (reviewed in Lee et al., 2006; Momose et al., 2008). Functional investigations with both repression and over-expression of genes of this signaling pathway indicate that the WNT gradient (formed by WNT ligands and their repressors) is responsible for setting the oral-aboral axis of cnidarians (Lee et al., 2006; Plickert et al., 2006; Momose et al., 2008; Petersen and Reddien, 2009; Marlow et al., 2013). Treatment with drugs that mimic WNT signaling (alsterpaullonne, lithium chloride, and a highly specific inhibitor called BIO) in both gemmule-derived juveniles and adults also indicated that this pathway is involved in establishing the polarity of the aquiferous system of sponges (Lapébie et al., 2009; Windsor and Leys, 2010) and growth modules in arborescent demosponges (Wiens et al., 2008). In fact, WNT/Beta-catenin pathways are widely used in patterning the anteroposterior axis in metazoans. Besides sponges and cnidarians, WNT ligands are related to the patterning of the posterior region of deuterostomes, lophotrocozoans (e.g. planarians, annelids, and mollusks) and ecdysozoans (e.g. C. elegans and arthropods) (reviewed in Petersen and Reddien, 2009). These findings may indicate that the ‘WNT code’ predated the ‘Hox code’ during animal evolution and suggest that the primary axis of non-bilaterians and bilaterians could have a common origin (Ryan and Baxevanis, 2007; Adamska et al., 2011).
Surprisingly, however, WNT ligands and other components are expressed only after gastrulation in the development of comb jellies (Pang et al., 2010), suggesting that the patterning of the primary axis in Ctenophora is set by another mechanism (Ryan and Baxevanis, 2007; Adamska et al., 2011). The development of ctenophores is stereotypical, with the oral-aboral axis established after the third cleavage, without expression of zygotic genes. These observations suggest that the primary axis polarity is provided by maternal products (possibly proteins) located in the cytoplasm of the fertilized egg (Pang et al., 2010). Nonetheless, the WNT signaling pathway appears to be important in the establishment of polarity later in the development of ctenophores, as indicated by the expression of many components around the mouth and other areas of Pleurobranchia pileus adults (Jager et al., 2013). These findings led Jager et al. (2013) to suggest that WNTs are a universal metazoan feature for axial polarization that is secondarily lost during early development but retained in adult polarization of ctenophores. Alternatively, if the ‘ctenophora-first’ hypothesis is accepted, the use of WNTs in establishing primary axis polarity could be a novelty in the lineage that led to all animals except ctenophores. New research on the expression of these genes during embryonic development of other comb jellies will help to understand the early evolution of the establishment of the primary axis.
Bilaterians are also characterized by a secondary axis (dorsoventral) perpendicular to the primary axis. This axis is patterned by a complex interplay of BMP (a member of the TGF-BETA superfamily) signaling regulators (De Robertis, 2008; Araújo et al., 2011; Leclère and Rentzsch, 2014). The interaction of BMP (or DPP in insects) and Chordin (short gastrulation in insects), in addition to other BMPextracellular modulators, is used in the establishment of the dorsoventral polarity of bilaterians (Araújo et al., 2011). In sponges, TGF-BETAgenes are expressed along the main axis of the larvae, suggesting that, in conjunction with WNT, these signaling pathways were already setting the radial symmetry of the ancestor of all metazoans (Adamska et al., 2007; Leininger et al., 2014). In situhybridization of TGF-BETA signaling pathway genes in the ctenophore Mnemiopsis embryos indicated that this pathway is probably not related to axial specification in ctenophores (Pang et al., 2011). Both radial and bilateral forms can be found among cnidarians (Brusca and Brusca, 2003; Leclère and Rentzsch, 2014). The majority of medusozoans show only radial symmetry, while anthozoans have a secondary body axis perpendicular to the primary axis. The secondary axis is observed in adult anthozoans as anatomic detail in the retractor muscles of the mesenteries (longitudinal ridge-like expansions in the gastrovascular cavity) (Brusca and Brusca, 2003; Leclère and Rentzsch, 2014). In addition, the expression pattern of BMPs and associated genes (e.g. CHD, RGM) is asymmetrical in relation to the main axis. Together, these findings support the existence of a second body axis, the directive axis, in anthozoans (Saina et al., 2009; reviewed in Technau and Steele, 2012; Leclère and Rentzsch, 2014). As an asymmetric expression of the BMPsignaling pathway components is not observed in Hydra (Rentzsch et al., 2007), it was suggested that the break in symmetry caused by BMP signaling was either lost during the evolution of cnidarians or reverted to a radial pattern in the polyp stage of medusozoans, leading to a secondary radialization of the body plan (Rentzsch et al., 2007; Technau and Steele, 2012). Although the same molecular machinery seems to be used by anthozoans and bilaterians to break the symmetry of the secondary axis, there is mounting evidence that BMP/Chordin mechanisms are used in a significantly different manner in cnidarians and bilaterians (Rentzsch et al., 2007; Saina et al., 2009).
An interesting issue related to body plan organization is the presence of ‘organizers’. Organizers are localized signaling centers in the embryo that are able to generate polarity in the surrounding tissues and induce differentiation in cell fate and behavior (Rentzsch et al., 2007). In amphibians, the transplantation of the dorsal blastopore lip to an ectopic region induces the formation of a second body axis in the host embryo. Thus, the blastopore lip was labeled the ‘organizer’ of amphibian embryos (Spemann and Mangold, 1924). Later on, similar organizer fields were discovered in other vertebrates, such as the embryonic shield in fishes, Hensen’s node in birds and the node in mammals (Gilbert, 2010). Interestingly, non-bilaterians also seem to have organizers. Windsor and Leys (2010) demonstrated, in an elegant experiment, that the oscula of Ephydatia act as organizers of the aquiferous system, probably through the action of a diffusive molecule (WNT?). Further investigations to identify this diffusive molecule, followed by loss-of-function studies, are necessary to confirm whether the sponge osculum could be considered a bona fide organizer. On the other hand, transplantation of the hypostome (which expresses Chordin and WNT)of Hydra polyps to lateral regions of hosts also induces the development of additional body axes in hydrozoans (Browne, 1909; Broun et al., 2005; Rentzsch et al., 2007). In anthozoans, BMP, Chordin and WNT are expressed around the blastopore of the embryos and the transplantation of part of the blastopore lip to other parts of the embryo induces the formation of a second oral-aboral axis in the organisms (Kraus et al., 2007). These findings support the proposition that cnidarian organizers (head organizer in hydrozoans and blastoporal lip in anthozoans) are homologous to the organizers of vertebrates (Kraus et al., 2007; Technau and Steele, 2012). The establishment of a proper phylogeny of non-bilaterians and further investigation of organizing fields in sponges and ctenophores are necessary to understand whether organizers are a characteristic of metazoan development.
The Origin of Gastrulation and the Origin of Germ Layers Are Still Controversial
In an important review about the evolution of meta-zoan axial properties Martindale (2005) concluded that "gastrulation is perhaps the most important event in meta-zoan evolution: it is the reason that animals are not all a hollow ball of cells, and provides the opportunity for the interaction of different tissue layers to give rise to complex structures and organ systems." There is no doubt that cnidarians and ctenophores gastrulate, but there is a longstanding debate whether sponges gastrulate or not. If we consider sponges as the basal-most branch of the metazoan tree, it is in these animals that we should focus to understand the origin of gastrulation.
Historically, there are three schools of thought about gastrulation in Porifera: (i) one that denies gastrulation entirely; (ii) one that affirms that sponges gastrulate during embryogenesis; and (iii) one that affirms that sponges gastrulate during metamorphosis (reviewed in Leys, 2004). Much of the debate took place before the advent of molecular techniques now available and this is thoroughly summarized in Leys (2004)and Ereskovsky (2010). More recently, Nakanishi et al.(2014) and Leininger et al. (2014)provided conflicting results about the gastrulation of sponges that increase the current debate. Cell labeling techniques indicated that the cell layers established during embryogenesis were not restricted to juvenile cell layers in the demosponge Amphimedon and that the epithelial cells of the juvenile could transdifferentiate into other cell types of the sponge (Nakanishi et al., 2014). These results suggest that the progressive cell differentiation expected for germ cell layers is not present in this demosponge. In contrast, the endomesodermal marker GATA was expressed only (and constantly) in choanocytes. Nakanishi et al.(2014) thus suggested that the conserved expression of GATA in the internal cells of the body of animals, together with the conserved differential expression of TGF-BETA and WNT along the metazoan primary axes, were necessary to provide positional information to cells in the metazoan ancestor, not to specify germ layers.
Applying in situ hybridization techniques to the calcareous sponge Sycon, Leininger et al. (2014) demonstrated that several genes related in cnidarians and bilaterians to endomesoderm specification (e.g.β-catenin, Brachyury and GATA) were co-expressed in both embryo/larva ciliated cells (micromeres, which are precursors of internal cells, choanocytes included) and adult choanocytes. These findings suggest that the sponge choanoderm and bilaterian endoderm are homologous structures (Fortunato et al., 2014; Leininger et al., 2014) and ciliated cells/choanocytes are germ layers. The development of the heterocoelic aquiferous system of the juvenile of calcareous sponges relies on epithelial morphogenetic movements (E. Lanna, personal observation), while in demosponges it is based on mesenchymal movements of the cells (Ereskovsky, 2010). Conservation of GATA expression in choanocytes of both calcareous sponges and demosponges (Leininger et al., 2014; Nakanishi et al., 2014) raises even more questions on whether deployment of germ layers (gastrulation) occurs in poriferans or not. One may hypothesize that the maintenance of germ layers was lost in demosponges or that its maintenance in calcareous sponges is convergent in Calcarea and other metazoans. ‘Epithelial’ development is also seen in homoscleromorphs, which could putatively have more stable germ layers than the calcareous sponges (Ereskovsky, 2010). The investigation of these and expression of other endomesodermal genes in homoscleromorphs will be important to answer the conundrum about the homology of gastrulation and germ layers in animals.
The Metazoan Ancestor Had an Endomesoderm That Later Gave Rise to the Bilaterian Mesoderm
Non-bilaterians are diploblastic animals. Although homology of germ layers is contested in sponges, the other three phyla present endoderm and ectoderm sandwiching an amorphous non-cellular layer. Some members of the Medusozoa present another layer that is linked neither to endoderm nor to ectoderm: the entocodon (a mesoderm-like structure that contributes to the formation of epidermal muscles in some developmental stages of some hydrozoans; Spring et al., 2002; Seipel and Schmid, 2006). Thus, it is surprising that most of the genetic toolkit related to the development of mesoderm is found in all four phyla.
Cnidarians present an enriched set of genes involved in mesoderm development in bilaterians that is expressed during the development of the cnidarian embryo (Martindale et al., 2004; Putnam et al., 2007; Chapman et al., 2010; Houliston et al., 2010). These findings raised three hypotheses about the origin of mesoderm (Martindale et al., 2004; Houliston et al., 2010). The first hypothesis is that mesoderm may have had a more ancient origin during early animal evolution, with subsequent reduction or loss in some cnidarian lineages. The second hypothesis is that these genes could have been engaged in regulation of cell movement during gastrulation and/or in the specification of an endodermal territory in the metazoan ancestor and were later co-opted to specify the bilaterian mesoderm (Martindale et al., 2004; Houliston et al., 2010). Finally, the third hypothesis is that the ancestor of cnidarians and bilaterians was a triploblastic animal, which employed the same genes involved in the mesoderm of current bilaterians in the development of its mesoderm. In this scenario, diploblasty would be a derived characteristic of cnidarians generated by the fusion of the endo- and mesoderm (Martindale et al., 2004).
This last hypothesis can be considered in light of the idea that the entocodon of some hydrozoans and the mesoderm of bilaterians as homologous structures (Seipel and Schmid, 2006; reviewed in Burton, 2008). The entocodon arises from the polyp ectoderm and gives rise to the striated muscles of medusae (Martindale et al., 2004; Burton, 2008). If this homology is accepted, the hypothesis suggests that the third germ layer (entocodon/mesoderm) was present in the ancestor of the Cnidaria and Bilateria and was lost in all classes of Cnidaria except Hydrozoa. However, mesoderm ‘markers’, such as Twist, MEF2and Brachyury, are not specifically expressed in the entocodon of Podocoryne carnea (Spring et al., 2002; Martindale et al., 2004). Furthermore, since the mesoderm appears to be derived from endoderm (based on both developmental and molecular data, reviewed in Martindale et al., 2004) and the entocodon is derived from ectoderm, it is most plausible that mesoderm and entocodon are not homologous germ layers (however, see Seipel and Schmid, 2006 for an alternative view).
Therefore, the most plausible hypothesis for the evolution of the bilaterian mesoderm is the second presented above (Martindale et al., 2004). This hypothesis implies that mesoderm is derived from endoderm, followed by the restriction of the expression of the mesodermal marker genes in the new germ layer (Martindale et al., 2004). Interestingly, many of the endomesodermal markers are expressed in the embryonic precursors of choanocytes and in the choanoderm of Sycon (Leininger et al., 2014), implying that the origin of endomesoderm could have occurred in the ancestor of all metazoans. Further investigation of expression pattern through functional approaches in homoscleromorphs, ctenophores and different cnidarian classes are necessary for a better comprehension of the evolution of the third germ layer.
The Origin of Two Important Animal Traits: Muscle and Nerve Tissues
One of the most intriguing issues in the evolution of animals is the origin of complex cell types such as muscles and neurons (Arendt, 2008). Muscles and neurons are typical cell types of bilaterians. Although some sponges have contractile cells and others are capable of transmitting information through electrical signaling (Leys and Hill, 2012), they lack typical muscles and neurons. Yet cnidarians, ctenophores and bilaterians share smooth and striated muscles (Burton, 2008; Steinmetz et al., 2012). While the majority of bilaterians present cord-like neural tissue, cnidarians and ctenophores possess a neural net, although some hydrozoan medusae and some ctenophores also possess nerve cords and nerve plexuses (Koizumi, 2006; Jager et al., 2011).
In metazoans, there are two types of contractile cells: epitheliomuscular cells and muscle fiber cells. Both types of cells have thick (myosin) and thin (actin) filaments, but muscle fiber cells are connected to the body through connective tissue, while epitheliomuscular cells, as the name suggests, are restricted to the epithelium (such as the epidermis and gastrodermis of cnidarians) (Burton, 2008). The former derive from endomesodermal precursors and the latter from either endo- or ectoderm (Burton, 2008). It is interesting that the core of the molecular machinery necessary for muscle functioning was present earlier than the origin of the muscle cell sensu stricto, antedating the origin of animals (Steinmetz et al., 2012). The myosin heavy chain (MyHC) seems to be the most ancient molecule related to the regulation of muscle contraction. Phylogenetic approaches indicated that the MyHC gene was duplicated in the metazoan ancestor, giving rise to a non-muscle paralog (functioning in common cellular behavior, e.g. cell division and migration, and smooth muscle contraction) and a muscle paralog (functioning in vertebrate striated muscle contraction) (Dayraud et al., 2012; Steinmetz et al., 2012). All four non-bilaterian phyla have both non-muscle and muscle MyHC paralogs and the expression pattern of these genes in demosponges suggests that the segregation of these paralogs occurred before the split of sponges and eumetazoans (Steinmetz et al., 2012). In cnidarians, the muscle paralog is expressed in both smooth and striated muscles, suggesting that the presence of this gene is not sufficient for striation. However, genes coding other molecules related to the striation of striated muscles in bilaterians are absent in the cnidarian genome. Together, these findings indicate that striated muscles of cnidarians and bilaterians (and most probably those of the ctenophore Euplokamissp., as the genome of ctenophores lacks the bilaterian machinery for striated muscles; Ryan et al., 2013) are convergent structures that evolved from a common contractile cell (Dayraud et al., 2012; Steinmetz et al., 2012).
The function of the nervous system is to sense and rapidly relay information about the environment (Watanabe et al., 2009). Nervous systems rely on neurons that transmit rapid signals directly through long distances between the sensory cells and also with other unities, such as muscles. The appearance of neurons, then, was a crucial event during animal evolution (Arendt, 2008; Anderson, 2015; Leys, 2015). The success of metazoans can be attributed, in large part, to the origin and evolution of nerve cells. Thus, it is not surprisingly that the development of nerve cells is of great interest to the scientific community (Galliot et al., 2009; Moroz, 2009; Watanabe et al., 2009; Ryan, 2014; see also Anderson, 2015 and the special edition of The Journal of Experimental Biology published in February 2015).
Non-bilaterian animals are central to understanding how nerve cells, and consequently nervous systems, evolved in Metazoa. Cnidarians and ctenophores have undisputable nerve cells, organized in a myriad of patterns of nerve net, although nerve cords and plexuses are found in some species (Koizumi, 2006; Jager et al., 2011). However, sponges and placozoans lack any recognizable cell type (both at morphological and gene expression levels) exclusively for transmission and reception of electric signals (Schierwater et al., 2009; Leys and Hill, 2012).
The molecular machinery for a nervous system was already present in the last common animal ancestor. Sponges have orthologs of post-synaptic structural and proneural regulatory proteins in their genomes (Srivastava et al., 2010a; Riesgo et al., 2014). In fact, most of genes coding for post-synaptic proteins were already present in the genomes of the unicellular close relatives of metazoans (Alié and Manuel, 2010). Additionally, all the basic machinery for the synthesis, release, and uptake of neurotransmitters is also found in Trichoplax (Srivastava et al., 2008, in fact Smith et al., 2014recently described a neurosecretory cell in Trichoplax adhaerens). In fact, numerous genes known to be involved in neurogenesis, such as members of the homeobox and basic helix-loop-helix (bHLH) transcription factor families, can be traced to ancient eukaryotic genes with these signature domains, but many novelties appeared in the lineage of eumetazoans (Putnam et al., 2007; Richards et al., 2008).
Nevertheless, the current knowledge about neural development and specification in these organisms is still fragmentary. It has already been shown that some neural bHLH transcription factors are expressed in developing neurons in hydrozoans (Grens et al., 1995; Seipel et al., 2004; reviewed in Watanabe et al., 2009). The identification of neural inducing signals in cnidarians can also help us to understand the origin of neural induction and the evolution of a centralized nervous system in bilaterians. Little is known about this issue, but it seems that BMP signaling is probably not related to neural induction in Nematostella (Watanabe et al., 2009). This result was unexpected, as in the large majority of bilaterians early patterning of the neurogenesis is based on the behavior of BMPs and their antagonists (Watanabe et al., 2009). On the other hand, another widespread neural inducer, FGF signaling, was already observed to be important for the development of nervous cells of the apical tuft of Nematostella larvae (Rentzsch et al., 2008; Sinigaglia et al., 2015). Another important neural inducer (LHX, belonging to the LIM homeobox transcription factors) is not only present in sponge, placozoan, cnidarian and ctenophore genomes, but is also expressed in neural progenitor cells in cnidarians and ctenophores (Srivastava et al., 2010b; Simmons et al., 2012). In non-neural placozoans, the whole complement of LHX is expressed randomly throughout the body of Trichoplax, while in Amphimedon it is widely expressed during embryogenesis, including in the cells associated with the larval photosensory ring (Srivastava et al., 2010b). As indicated by Simmons et al. (2012), LHX may have had an ancient role in specifying sensory cells in the metazoan ancestor and was later co-opted to pattern more diversified complements of neural and non-neural cell types of eumetazoans.
The contentious placement of Ctenophora on the metazoan tree is of great significance for understanding the evolution of the nervous system (Ryan et al., 2013; Moroz et al., 2014; Ryan, 2014). If the ctenophore-first scenario is not considered, the sponges are the first metazoans to have a sensory system to protect their filtering apparatus. The nervous system of cnidarians, ctenophores and bilaterians was then later constructed based on the co-option of the sponge system and other more sophisticated regulatory networks (Leys, 2015). However, if the ‘ctenophore-first’ scenario is accepted, a different view of the evolution of the nervous system arises: either sponges and placozoans lost their nervous systems (single-origin hypothesis), or the nervous system in ctenophores and cnidarians+bilaterians evolved in parallel (Ryan et al., 2013; Moroz et al., 2014). Considering the single-origin hypothesis (all the components of the nervous system were present in the animal ancestor), ctenophores would have had to lose many of the genes involved in neuronal and signaling toolkits and to substitute them with novel signaling molecules and receptors (Moroz et al., 2014). We will have better understanding of how the neural cells arose in ctenophores, cnidarians and bilaterians only after we achieve a robust phylogeny of the early metaozans that enables the recognition of ancestral and derived traits and once functional experiments are carried out in ctenophores (Ryan, 2014).
What Are the Boundaries Between Germ and Somatic Cell Lineages?
Germ cell segregation is an important event during the development of multicellular organisms, as these cells carry the hereditary information for the next generation. This event is significant for evolutionary biology because it takes into account how the differences between somatic cells and germ cells are initiated and maintained during development while subject to different selective pressures (Extavour and Akam, 2003). The fact that many classic model organisms (e.g. fly, frogs) segregate their germ cell lineage early in development (i.e. preformation), led to the idea that there is a clear differentiation between germline and soma within the animal body (Extavour and Akam, 2003). However, there has been a history of disagreement about how germ cells are specified. It is known that the limits between germline and soma are fluid (especially in non-bilaterians) and that the germ cells in the majority of animals arise by an inductive process (i.e. epigenesis) (Extavour and Akam, 2003). Non-bilaterians played and are still playing an important role in contributing to understanding this important issue in the evolution of metazoans.
Sponges and placozoans lack gonads. In sponges, gametes are produced either from epithelial choanocytes or from mesenchymal archaeocytes (Maldonado and Riesgo, 2008; Lanna and Klautau, 2010). Gametes in cnidarians and ctenophores are formed in specialized tissues (game-togenic areas) located in the mesoglea in cnidarians, and the meridional canals in ctenophores (Eckelbarger and Larson, 1992; Brusca and Brusca, 2003). In cnidarians and ctenophores the gametes are formed from epithelial cells (endoderm in anthozoans, scyphozoans and ctenophores, and ectoderm in hydrozoans) that migrate to the mesoglea to start their development (Brusca and Brusca, 2003; Amiel and Houliston, 2009). Little is known about the gameto-genesis of placozoans. In fact, there are only few observations of egg-laying under harsh conditions in the laboratory (Eitel et al., 2011). Most importantly, non-bilaterians do not seem to have a resident population of cells that is clearly segregated during embryogenesis to generate their gametes. So, how can we recognize the cell populations that give rise to gametes in these lineages? In other words, how are the gametes of non-bilaterians specified?
There is a well-described germline molecular machinery in metazoans (Ewen-Campen et al., 2010; Riesgo et al., 2014). Among these genes, the markers NANOS and VASA are universally expressed in germ cells and have been used to localize them in different animals (Extavour and Akam, 2003). These genes are found in the genomes of all four non-bilaterian phyla (Putnam et al., 2007; Srivastava et al., 2008, 2010a; Ryan et al., 2013). These germline markers are expressed in oocytes and somatic cells of calcareous sponges (Leininger et al., 2014), in eggs and somatic tissues of cnidarians (Extavour et al., 2005; Leclère et al., 2012) and in the germline and somatic stem cells of adult ctenophores (Alié et al., 2011). The conserved expression pattern of these genes (in addition to the presence of many other genes of the germline machinery, see Riesgo et al., 2014) suggests that primordial germ cells may be homologous across all Metazoa (Extavour and Akam, 2003). Nonetheless, these genes are also observed in cells that are not involved in gametogenesis. NANOS, VASA, PL10 and other germline genes are also expressed in somatic stem cells (see below). Thus, what appears to differ between stem cells and germline cells are the upstream signals regulating the expression of genes such as VASA and NANOS, and not the expression of these genes themselves (Extavour et al., 2005). Extavour and Akam (2003) suggest that the complex suite of molecular characters, including many gene expression profiles, subcellular architecture of germ cells and possibly molecular mechanisms of regulating gene activity, is likely to have evolved only once as a homologous cell identity ‘program’. Nonetheless, this suite may also be expressed in other cells from different germ layers at different times and locations during development. There is a wide overlap of this molecular machinery between germ and somatic stem cell lineages, meaning that neither the mechanisms that trigger germ cell formation, nor the cells in which the program is elicited, are homologous (Extavour and Akam, 2003).
Are Non-Bilaterians the Answer for Sourcing Stem Cells?
A rapid expansion of research on stem cells has occurred in the last decade related to their use in the search for a cure for cancer and degenerative diseases (Funayama, 2013). Stem cells have been retrieved from the organs of most animals; however, we still know little about the evolution of stem cells. Are stem cells conserved throughout the evolution of metazoans? Do all animals rely on the same set of genes to specify their stem cells? An interesting characteristic of non-bilaterians is their capacity for regeneration and asexual reproduction. This characteristic may be related to the stem cells present throughout the life cycle. Can we learn something about the use of stem cells for our own regeneration?
Stem cells are usually defined by their capacity to self-renew and to generate other cell types. In metazoan adults, stem cells have a slow cycle and are undifferentiated and multipotent cells located in special microenvironments (niches) (Bosch et al., 2010). In vertebrate models, a large set of intrinsic and extrinsic cellular mechanisms balance self-renewal and differentiation in all stem cells (Hemmrich and Bosch, 2008). The transcription factor trio - NANOG, OCT4 and SOX2 - has been established as the core of the genetic machinery that governs multipotency. In addition, KLF-4 and MYC are also important to induce pluripotency in mammal stem cells. Are these genes found in the genome of non-bilaterians? If so, are they expressed and functional in non-bilaterian stem cells? Among the non-bilaterians, stem cells are most frequently studied in hydrozoans.
Hydra has three different stem cell lineages (the two epithelial cell lineages and the interstitial cells, also known as I-cells) that may be uni- or multipotent (Bosch et al., 2010; Plickert et al., 2012). I-cells are alleged to be the main stem cell source in hydrozoans, but their existence in other cnidarian lineages is improbable, as is the existence of a distinct stem cell population (Gold and Jacobs, 2013). Three of the five genes associated with pluripotency in differentiated somatic cells of mammals (NANOG, KLF4 and OCT4) were clearly not present in the genome of either Hydra or Nematostella (Chapman et al., 2010). However, MYC is expressed in I-cells of Hydra and is necessary for homeostasis of populations of these cells (Ambrosone et al., 2012). These findings indicate that some transcription factors enrolled in the development of stem cells in vertebrates were already in use in cnidarians (Gold and Jacobs, 2013). However, the analysis of other candidate genes and of the same genes in other cnidarians (including other hydrozoans and species of other classes) do not support the idea that cnidarian I-cells (and similar) are homologous to vertebrate stem cells (reviewed in Gold and Jacobs, 2013).
Sponges are famous for their capacity to de-differentiate their cells to form new individuals (Wilson, 1907; Funayama, 2013). Choanocytes and archaeocytes express two different paralogs of PIWI, and a paralog of Musashi is expressed in archaeocytes only (Funayama, 2010, 2013). PIWI proteins are known to maintain totipotency/ pluripotency/ multipotency of cells in various organisms, mostly in a small RNA (piRNA)-mediated manner, via several different molecular mechanisms (Grimson et al., 2008; Funayama, 2013). Yet Musashi has a role in maintaining stem cell state through RNA binding (Okamoto et al., 2012; Funayama, 2013). Not surprisingly, choanocytes and archaeocytes are capable of self-renewal and of generating different cell types in sponges (Funayama, 2013). Funayama (2013) argues that choanocytes are probably the stem cell ‘ancestor’ of sponges and all other animals. In fact, in calcareous sponges, fully differentiated functional sponges can regrow from fractions containing mainly choanocytes (E. Lanna & M. Klautau, unpublished results) reinforcing the hypothesis that these cells could be the former stem cell population of sponges. As choanocytes are cells dealing with many sources of DNA damage, it is possible that sponges had to rely on a secondary source of stem cells, which may have led to archaeocytes working as stem cells in sponges as well, mainly in demosponges (Funayama, 2013). These secondary stem cells could now be occupied solely in protecting genetic information while they moved in the mesohyl (a gelatinous layer between the choanocyte epithelium and the pinacocyte epithelium) of the sponges. Finally, ctenophores also have PIWI, VASA, PL10 and Bruno expressed not only in germ cells, but also in various localized populations of adult stem cells confined to somatic tissues and having nothing to do with the germline (Alié et al., 2011). Interestingly, while in cnidarians and sponges the stem cells are usually mobile and scattered across the organism body (suggesting the absence of a stem cell niche), in ctenophores stem cells are segregated in different and stable groups in various regions of the adult body, suggesting that in these animals a stem cell niche is present (Alié et al., 2011).
The recent interest in the evolution of the molecular machinery regulating the development of stem cells has revealed that genes conventionally treated as germline markers are also involved in stem cell regulation in non-bilaterians and basal bilaterians (Alié et al., 2011; Funayama, 2013; Gold and Jacobs, 2013). As indicated by Alié et al. (2011), there are two hypotheses for the occurrence of these genes in stem cells: (i) VASA, NANOS and other genes are expressed in stem cell regions because the stem cells have the potential to generate germ derivatives or (ii) these genes are part of the animal toolkit for the maintenance of stem cells, independent of the fate of their progeny. An investigation using the ctenophore Pleurobrachia pileus demonstrated the expression of germline genes in stem cell regions that never form gametes, favoring the second hypothesis (Alié et al., 2011). Therefore, it seems that the germline toolkit was shared with stem cells early in the evolution of the animals, but was later restricted to regulate one of the functions. A possible explanation for this hypothesis is that some of these genes (e.g. PIWI) work to protect the genome through transposon silencing (Alié et al., 2011).
We are currently unable to answer whether the stem cell systems of animals are homologous or appeared several times during metazoan evolution. The current results indicate, however, that the genetic control of multipotency has a deep ancestry, but the ways in which stem cells are produced and maintained are probably not homologous across the Metazoa (Gold and Jacobs, 2013).
Final Remarks
Here, I have briefly reviewed some of the recent findings related to non-bilaterian development that are helping us to understand animal evolution. I have shown that research on organisms belonging to different evolutionary lineages is usually complementary, and that sometimes it may raise conflicting results. The divergence in some aspects of development highlights the necessity to increase the number of non-bilaterian models for evo-devo studies. Thus, paraphrasing Gold and Jacobs (2013): no single clade of cnidarians, sponges or ctenophores (much less any single species) can represent these phyla as a whole. Although the number of models is already significant (see Table 1), more models are still needed. We should begin investigating the next wave of organisms, paying particular attention to those that will be of the greatest interest for comparative studies (either within or between classes of these organisms) (Steele, 2005).
Another important technological development to test the hypotheses raised by comparative studies will be the establishment of manipulative techniques to alter gene expression (e.g. RNAi, morpholinos) in these lineages and the cultivation of mutant strains. With the exception of a few cnidarians (Table 1), little advance in this area has yet been achieved for sponges, placozoans and ctenophores. Some attempts have already been made to knockdown genes in sponges. Soaking sponges in solutions containing RNAi or feeding them with bacteria expressing RNAi proved to affect the phenotype of two different species (Rivera et al., 2011). Although it was a cost-effective technique to silence genes in sponges, it has not yet been applied in new studies. The most anticipated technique for gene editing, known as TALENs and CRISPR/Cas, will certainly provide a solid framework for functional genetic studies in every group of animals (Giles and Averof, 2014). This advance may overcome the necessity of functional studies on a wide variety of models, which is necessary if we are to understand more deeply how the body plans of the animals evolved. Finally, a robust phylogeny is mandatory if we are to understand the evolution of development in metazoans.
Almost ten years have passed since Martindale (2005) wrote: "Unraveling the intricate details of the embryonic regulation of axial patterning and gastrulation events in embryos of basal metazoan taxa will yield a rich source of information about the evolution of animal body plans." Many advances have been achieved, especially in the discovery of genes related to the development of these organisms. Nevertheless, functional and experimental investigations are still needed, and a thorough knowledge of the variation in embryogenesis and other aspects in different classes of these phyla is still needed to understand the evolution of animal body plans. The next years will be an exciting period for those interested in the early evolution of animals.
Acknowledgments
I thank the two anonymous reviewers who have significantly contributed to the improvement of the original manuscript. The Laboratório de Embriologia e Biologia Reprodutiva (LEBR) of the Instituto de Biologia of UFBA is financially supported by grants from CNPq, FAPESB and PROPCI-UFBA/PRODOC.
Footnotes
Associate Editor: Igor Schneider
References
- Adamska M, Degnan SM, Green KM, Adamski M, Craigie A, Larroux C, Degnan BM. Wnt and TGF-β expression in the sponge Amphimedon queenslandica and the origin of metazoan embryonic patterning. PLoS ONE. 2007;2:e1031. doi: 10.1371/journal.pone.0001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamska M, Degnan BM, Green K, Zwafink C. What sponges can tell us about the evolution of developmental processes. Zoology. 2011;114:1–10. doi: 10.1016/j.zool.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Adell T, Grebenjuk VA, Wiens M, Muller WEG. Isolation and characterization of two T-box genes from sponges, the phylogenetically oldest metazoan taxon. Dev Genes Evol. 2003;213:421–434. doi: 10.1007/s00427-003-0345-5. [DOI] [PubMed] [Google Scholar]
- Alié A, Manuel M. The backbone of the post-synaptic density originated in a unicellular ancestor of choanoflagellates and metazoans. BMC Evol Biol. 2010;10:e34. doi: 10.1186/1471-2148-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alie A, Leclére L, Jager M, Dayraud C, Chang P, Le Guyader H, Queinnec E, Manuel M. Somatic stem cells express Piwi and Vasa genes in an adult ctenophore: Ancient association of "germline genes" with stemness. Dev Biol. 2011;350:183–197. doi: 10.1016/j.ydbio.2010.10.019. [DOI] [PubMed] [Google Scholar]
- Ambrosone A, Marchesano V, Tino A, Hobmayer B, Tortiglione C. Hymyc1 downregulation promotes stem cell proliferation in Hydra vulgaris . PLoS One. 2012;7:e30660. doi: 10.1371/journal.pone.0030660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiel A, Houliston E. Three distinct RNA localization mechanisms contribute to oocyte polarity establishment in the cnidarian Clytia hemisphærica . Dev Biol. 2009;327:191–203. doi: 10.1016/j.ydbio.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Anderson PA. On the origins of that most transformative of biological systems-the nervous system. J Exp Biol. 2015;218:504–505. doi: 10.1242/jeb.119412. [DOI] [PubMed] [Google Scholar]
- Araújo H, Fontenele MR, da Fonseca RN. Position matters: Variability in the spatial pattern of BMP modulators generates functional diversity. Genesis. 2011;49:698–718. doi: 10.1002/dvg.20778. [DOI] [PubMed] [Google Scholar]
- Arendt D. The evolution of cell types in animals: Emerging principles from molecular studies. Nat Rev Genet. 2008;9:868–882. doi: 10.1038/nrg2416. [DOI] [PubMed] [Google Scholar]
- Borojevic R. Étude expérimentale de la différenciation des cellules de l’éponge au cours de son développement. Dev Biol. 1966;14:130–153. doi: 10.1016/0012-1606(66)90009-1. [DOI] [PubMed] [Google Scholar]
- Borojevic R, Levi C. Métamorphose artificielle de larves d’Éponges, aprés dissociation et réagrégation des cellules larvaires. C R Acad Sci Paris. 1964;259:4364–4366. [Google Scholar]
- Bosch TCG, Anton-Erxleben F, Hemmrich G, Khalturin K. The Hydra polyp: Nothing but an active stem cell community. Dev Growth Differ. 2010;52:15–25. doi: 10.1111/j.1440-169X.2009.01143.x. [DOI] [PubMed] [Google Scholar]
- Broun M, Gee L, Reinhardt B, Bode HR. Formation of the head organizer in hydra involves the canonical Wnt pathway. Development. 2005;132:2907–2916. doi: 10.1242/dev.01848. [DOI] [PubMed] [Google Scholar]
- Browne EN. The production of new hydranths in hydra by the insertion of small grafts. J Exp Zool. 1909;7:1–23. [Google Scholar]
- Brusca RC, Brusca GJ. Invertebrates. 2nd edition. Sinauer Ass. Inc; Sunderland: 2003. 888 [Google Scholar]
- Burton PM. Insights from diploblasts; the evolution of mesoderm and muscle. J Exp Zool. 2008;310B:5–14. doi: 10.1002/jez.b.21150. [DOI] [PubMed] [Google Scholar]
- Burton PM, Finnerty JR. Conserved and novel gene expression between regeneration and asexual fission in Nematostella vectensis . Dev Genes Evol. 2009;219:79–87. doi: 10.1007/s00427-009-0271-2. [DOI] [PubMed] [Google Scholar]
- Byrun CA, Martindale MQ. Gastrulation in the Cnidaria and Ctenophora. In: Stern C, editor. Gastrulation: From Cell to Embryo. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2004. pp. 33–49. [Google Scholar]
- Carroll SB, Grenier JK, Weatherbee SD. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. 2nd edition. Blackwell Publishing; Oxford: 2005. 258 [Google Scholar]
- Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D, et al. The dynamic genome of Hydra . Nature. 2010;464:592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AG, Chuchert PS, Marques AC, Jankowski T, Medina M, Schierwater BS. Medusozoan phylogeny and character evolution clarified by new large and small subunit rDNA data and an assessment of the utility of phylogenetic mixture models. Syst Biol. 2006;55:97–115. doi: 10.1080/10635150500433615. [DOI] [PubMed] [Google Scholar]
- Cummings SG, Bode HR. Head regeneration and polarity reversal in Hydra attenuata can occur in the absence of DNA synthesis. Rouxs Arch Dev Biol. 1984;194:79–86. doi: 10.1007/BF00848347. [DOI] [PubMed] [Google Scholar]
- Dayraud C, Alie A, Jager M, Chang P, Le Guyader H, Manuel M, Queinnec E. Independent specialisation of myosin II paralogues in muscle vs. non-muscle functions during early animal evolution: A ctenophore perspective. BMC Evol Biol. 2012;12:e107. doi: 10.1186/1471-2148-12-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan BM, Vervoort M, Larroux C, Richards G. Early evolution of metazoan transcription factors. Curr Opin Genet Dev. 2009;19:591–599. doi: 10.1016/j.gde.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Derelle R, Manuel M. Ancient connection between NKL genes and the mesoderm? Insights from Tlx expression in a ctenophore. Dev Genes Evol. 2007;217:253–261. doi: 10.1007/s00427-007-0131-x. [DOI] [PubMed] [Google Scholar]
- De Robertis EM. Evo-Devo: Variations on ancestral themes. Cell. 2008;132:185–195. doi: 10.1016/j.cell.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann M, Wörheide G. Novel scenarios of early animal evolution - Is it time to rewrite textbooks? Integr Comp Biol. 2013;53:503–511. doi: 10.1093/icb/ict008. [DOI] [PubMed] [Google Scholar]
- Eckelbarger KJ, Larson R. Ultrastructure of the ovary and oogenesis in the jellyfish Linuche unguiculata and Stomolophus meleagris, with a review of ovarian structure in the Scyphozoa. Mar Biol. 1992;114:633–643. [Google Scholar]
- Edgecombe GD, Giribet G, Dunn CW, Hejnol A, Kristensen RM, Neves RC, Rouse GW, Worsaae K, Sorensen MV. Higher-level metazoan relationships: Recent progress and remaining questions. Org Divers Evol. 2011;11:151–172. [Google Scholar]
- Eitel M, Guidi L, Hadrys H, Balsamo M, Schierwater B. New insights into placozoan sexual reproduction and development. PLoS ONE. 2011;6:e19639. doi: 10.1371/journal.pone.0019639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ereskovsky AV. The Comparative Embryology of Sponges. Springer, Dordrecht, Heidelberg, London, New York; 2010. 329 [Google Scholar]
- Ereskovsky AV, Borchiellini C, Gazave E, Ivanisevic J, Lapebie P, Perez T, Renard E, Vacelet J. The Homo-scleromorph sponge Oscarella lobularis, a promising sponge model in evolutionary and developmental biology. Bioessays. 2009;31:89–97. doi: 10.1002/bies.080058. [DOI] [PubMed] [Google Scholar]
- Ereskovsky AV, Renard E, Borchiellini C. Cellular and molecular processes leading to embryo formation in sponges: Evidences for high conservation of processes throughout animal evolution. Dev Genes Evol. 2013;223:5–22. doi: 10.1007/s00427-012-0399-3. [DOI] [PubMed] [Google Scholar]
- Erwin DH, Laflamme M, Tweedt SM, Sperling EA, Pisani D, Peterson KJ. The Cambrian conundrum: Early divergence and later ecological success in the early history of animals. Science. 2011;334:1091–1097. doi: 10.1126/science.1206375. [DOI] [PubMed] [Google Scholar]
- Ewen-Campen B, Schwager EE, Extavour CGM. The molecular machinery of germ line specification. Mol Reprod Dev. 2010;77:3–18. doi: 10.1002/mrd.21091. [DOI] [PubMed] [Google Scholar]
- Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: Epigenesis and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- Extavour CG, Pang K, Matus DQ, Martindale MQ. vasa and nanos expression patterns in a sea anemone and the evolution of bilaterian germ cell specification mechanisms. Evol Dev. 2005;7:201–215. doi: 10.1111/j.1525-142X.2005.05023.x. [DOI] [PubMed] [Google Scholar]
- Finnerty JR, Pang K, Burton P, Paulson D, Martindale MQ. Origins of bilateral symmetry: Hox and dpp expression in a sea anemone. Science. 2004;304:1335–1337. doi: 10.1126/science.1091946. [DOI] [PubMed] [Google Scholar]
- Fortunato SA, Adamski M, Ramos OM, Leininger S, Liu J, Ferrier DE, Adamska M. Calcisponges have a ParaHox gene and dynamic expression of dispersed NK homeobox genes. Nature. 2014;514:620–623. doi: 10.1038/nature13881. [DOI] [PubMed] [Google Scholar]
- Freeman G. Experimental studies on embryogenesis in hydrozoans (Trachylina and Siphonophora) with direct development. Biol Bull. 1983;165:591–618. doi: 10.2307/1541468. [DOI] [PubMed] [Google Scholar]
- Fuchs B, Wang W, Graspeuntner S, Li Y, Insua S, Herbst EM, Dirksen P, Böhm A-M, Hemmrich G, Sommer F. Regulation of polyp-to-jellyfish transition in Aurelia aurita . Curr Biol. 2014;24:263–273. doi: 10.1016/j.cub.2013.12.003. [DOI] [PubMed] [Google Scholar]
- Funayama N. The stem cell system in demosponges: Insights into the origin of somatic stem cells. Dev Growth Differ. 2010;52:1–14. doi: 10.1111/j.1440-169X.2009.01162.x. [DOI] [PubMed] [Google Scholar]
- Funayama N. The stem cell system in demosponges: Suggested involvement of two types of cells: Archeocytes (active stem cells) and choanocytes (food-entrapping flagellated cells) Dev Genes Evol. 2013;223:23–38. doi: 10.1007/s00427-012-0417-5. [DOI] [PubMed] [Google Scholar]
- Galliot B, Quiquand M, Ghila L, de Rosa R, Miljkovic-Licina M, Chera S. Origins of neurogenesis, a cnidarian view. Dev Biol. 2009;322:2–24. doi: 10.1016/j.ydbio.2009.05.563. [DOI] [PubMed] [Google Scholar]
- Gilbert S. Developmental Biology. 9th edition. Sinauer Associates, Inc; Sunderland: 2010. 711 [Google Scholar]
- Giles AF, Averof M. Functional genetics for all: Engineered nucleases, CRISPR and the gene editing revolution. EvoDevo. 2014;5:e43. doi: 10.1186/2041-9139-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold DA, Jacobs DK. Stem cell dynamics in Cnidaria: Are there unifying principles? Dev Genes Evol. 2013;223:53–66. doi: 10.1007/s00427-012-0429-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grens A, Mason E, Marsh JL, Bode HR. Evolutionary conservation of a cell fate specification gene: The Hydra achaete-scute homolog has proneural activity in Drosophila . Development. 1995;121:4027–4035. doi: 10.1242/dev.121.12.4027. [DOI] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadrys T, DeSalle R, Sagasser S, Fischer N, Schierwater B. The Trichoplax PaxB gene: A putative Proto-PaxA/B/C gene predating the origin of nerve and sensory cells. Mol Biol Evol. 2005;22:1569–1578. doi: 10.1093/molbev/msi150. [DOI] [PubMed] [Google Scholar]
- Harbison GR. On the classification and evolution of the Ctenophora. In: Conway Morris S, George JD, Gibson R, Platt HM, editors. The Origins and Relationships of Lower Invertebrates. Clarendon; Oxford: 1985. 400 [Google Scholar]
- Hemmrich G, Bosch TC. Compagen, a comparative genomics platform for early branching metazoan animals, reveals early origins of genes regulating stem-cell differentiation. Bioessays. 2008;30:1010–1018. doi: 10.1002/bies.20813. [DOI] [PubMed] [Google Scholar]
- Holland PWH. Evolution of homeobox genes. WIREs Dev Biol. 2013;2:31–45. doi: 10.1002/wdev.78. [DOI] [PubMed] [Google Scholar]
- Houliston E, Momose T, Manuel M. Clytia hemisphaerica: A jellyfish cousin joins the laboratory. Trends Genet. 2010;26:159–167. doi: 10.1016/j.tig.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Jager M, Chiori R, Alié A, Dayraud C, Quéinnec E, Manuel M. New insights on ctenophore neural anatomy: Immunofluorescence study in Pleurobrachia pileus (Müller, 1776) J Exp Zool B Mol Dev Evol. 2011;316:171–187. doi: 10.1002/jez.b.21386. [DOI] [PubMed] [Google Scholar]
- Jager M, Dayraud C, Mialot A, Quéinnec E, Le Guyader H, Manuel M. Evidence for involvement of Wnt signalling in body polarities, cell proliferation, and the neurosensory system in an adult ctenophore. PLoS ONE. 2013;8:e84363. doi: 10.1371/journal.pone.0084363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob W, Sagasser S, Dellaporta S, Holland P, Kuhn K, Schierwater B. The Trox-2 Hox/ParaHox gene of Trichoplax(Placozoa) marks an epithelial boundary. Dev Genes Evol. 2004;214:170–175. doi: 10.1007/s00427-004-0390-8. [DOI] [PubMed] [Google Scholar]
- Jenner RA, Wills MA. The choice of model organisms in evo-devo. Nat Rev Genet. 2007;8:311–319. doi: 10.1038/nrg2062. [DOI] [PubMed] [Google Scholar]
- Koizumi O. Nerve ring of the hypostome in Hydra: Is it an origin of the central nervous system of bilaterian animals? Brain Behav Evol. 2006;69:151–159. doi: 10.1159/000095204. [DOI] [PubMed] [Google Scholar]
- Kraus Y, Fritzenwanker JH, Genikhovich G, Technau U. The blastoporal organiser of a sea anemone. Curr Biol. 2007;17:R874–R876. doi: 10.1016/j.cub.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Kraus Y, Flici H, Hensel K, Plickert G, Leitz T, Frank U. The embryonic development of the cnidarian Hydractinia echinata . Evol Dev. 2014;16:323–338. doi: 10.1111/ede.12100. [DOI] [PubMed] [Google Scholar]
- Kunzel T, Heiermann R, Frank U, Muller W, Tilmann W, Bause M, Nonn A, Helling M, Schwarz RS, Plickert G. Migration and differentiation potential of stem cells in the cnidarian Hydractinia analysed in eGFP-transgenic animals and chimeras. Dev Biol. 2010;348:120–129. doi: 10.1016/j.ydbio.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Lanna E, Klautau M. Oogenesis and spermatogenesis in Paraleucilla magna (Porifera, Calcarea) Zoomorphology. 2010;129:249–261. [Google Scholar]
- Lapébie P, Gazave E, Ereskovsky A, Derelle R, Bézac C, Renard E, Houliston E, Borchiellini C. WNT/β-Catenin signalling and epithelial patterning in the homoscleromorph sponge Oscarella . PLoS ONE. 2009;4:e5823. doi: 10.1371/journal.pone.0005823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larroux C, Fahey B, Degnan SM, Adamski M, Rokhsar DS, Degnan BM. The NK homeobox gene cluster predates the origin of Hox genes. Curr Biol. 2007;17:706–710. doi: 10.1016/j.cub.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Leclère L, Jager M, Barreau C, Chang P, Le Guyader H, Manuel M, Houliston E. Maternally localized germ plasm mRNAs and germ cell/stem cell formation in the cnidarian Clytia . Dev Biol. 2012;364:236–248. doi: 10.1016/j.ydbio.2012.01.018. [DOI] [PubMed] [Google Scholar]
- Leclère L, Rentzsch F. RGM regulates BMP-mediated secondary axis formation in the sea anemone Nematostella vectensis . Cell Rep. 2014;9:1921–1930. doi: 10.1016/j.celrep.2014.11.009. [DOI] [PubMed] [Google Scholar]
- Lee PN, Pang K, Matus DQ, Martindale MQ. A WNT of things to come: Evolution of Wnt signaling and polarity in cnidarians. Semin Cell Dev Biol. 2006;17:157–167. doi: 10.1016/j.semcdb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Leininger S, Adamski M, Bergum B, Guder C, Liu J, Laplante M, Bråte J, Hoffmann F, Fortunato S, Jordal S, et al. Developmental gene expression provides clues to relationships between sponge and eumetazoan body plans. Nat Commun. 2014;5:e3905. doi: 10.1038/ncomms4905. [DOI] [PubMed] [Google Scholar]
- Leys SP. Gastrulation in sponges. In: Stern C, editor. Gastrulation: From Cell to Embryo. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2004. pp. 23–31. [Google Scholar]
- Leys SP. Elements of a ‘nervous system’ in sponges. J Exp Biol. 2015;218:581–591. doi: 10.1242/jeb.110817. [DOI] [PubMed] [Google Scholar]
- Leys SP, Hill A. The physiology and molecular biology of sponge tissues. Adv Mar Biol. 2012;62:1–56. doi: 10.1016/B978-0-12-394283-8.00001-1. [DOI] [PubMed] [Google Scholar]
- Maldonado M, Bergquist PR. Porifera. In: Young CM, Sewell MA, Rice ME, editors. Atlas of Marine Invertebrate Larvae. Academic Press; San Diego: 2002. pp. 21–50. [Google Scholar]
- Maldonado M, Riesgo A. Reproduction in Porifera: A synoptic overview. Treb Soc Catalana Biol. 2008;59:29–49. [Google Scholar]
- Manuel M. Early evolution of symmetry and polarity in metazoan body plans. C R Biol. 2009;332:184–209. doi: 10.1016/j.crvi.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Marlow H, Matus DQ, Martindale MQ. Ectopic activation of the canonical wnt signaling pathway affects ectodermal patterning along the primary axis during larval development in the anthozoan Nematostella vectensis . Dev Biol. 2013;380:324–334. doi: 10.1016/j.ydbio.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale MQ. The evolution of metazoan axial properties. Nat Rev Genet. 2005;6:917–927. doi: 10.1038/nrg1725. [DOI] [PubMed] [Google Scholar]
- Martindale MQ, Henry JQ. Development and regeneration of comb plates in the ctenophore Mnemiopsis leidyi . Biol Bull. 1996;191:290–292. doi: 10.1086/BBLv191n2p290. [DOI] [PubMed] [Google Scholar]
- Martindale MQ, Henry JQ. Ctenophorans, the comb jellies. In: Gilbert S, Raunio AM, editors. Embryology: Constructing the Organism. Sinauer; Sunderland: 1997. pp. 87–111. [Google Scholar]
- Martindale MQ, Pang K, Finnerty JR. Investigating the origins of triploblasty: ‘Mesodermal’ gene expression in a diploblastic animal, the sea anemone, Nematostella vectensis (phylum, Cnidaria; Class Anthozoa) Development. 2004;131:2463–2474. doi: 10.1242/dev.01119. [DOI] [PubMed] [Google Scholar]
- Mendivil Ramos OM, Barker D, Ferrier DE. Ghost loci imply Hox and ParaHox existence in the last common ancestor of animals. Curr Biol. 2012;22:1951–1956. doi: 10.1016/j.cub.2012.08.023. [DOI] [PubMed] [Google Scholar]
- Momose T, Derelle R, Houliston E. A maternally localised Wnt ligand required for axial patterning in the cnidarian Clytia hemisphaerica . Development. 2008;135:2105–2113. doi: 10.1242/dev.021543. [DOI] [PubMed] [Google Scholar]
- Monteiro AS, Schierwater B, Dellaporta SL, Holland PWH. A low diversity of ANTP class homeobox genes in Placozoa. Evol Dev. 2006;8:174–182. doi: 10.1111/j.1525-142X.2006.00087.x. [DOI] [PubMed] [Google Scholar]
- Moroz LL. On the independent origins of complex brains and neurons. Brain Behav Evol. 2009;74:177. doi: 10.1159/000258665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz LL, Kocot KM, Citarella MR, Dosung S, Norekian TP, Povolotskaya IS, Grigorenko AP, Dailey C, Berezikov E, Buckley KM, et al. The ctenophore genome and the evolutionary origins of neural systems. Nature. 2014;510:109–116. doi: 10.1038/nature13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi N, Sogabe S, Degnan B. Evolutionary origin of gastrulation: Insights from sponge development. BMC Biol. 2014;12:e26. doi: 10.1186/1741-7007-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols SA, Dirks W, Pearse JS, King N. Early evolution of animal cell signaling and adhesion genes. Proc Natl Acad Sci USA. 2006;103:12451–12456. doi: 10.1073/pnas.0604065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen C. Six major steps in animal evolution: Are we derived sponge larvae? Evol Dev. 2008;10:241–257. doi: 10.1111/j.1525-142X.2008.00231.x. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Nakatsukasa M, Alie A, Masuda Y, Agata K, Funayama N. The active stem cell specific expression of sponge Musashi homolog EflMsiA suggests its involvement in maintaining the stem cell state. Mech Dev. 2012;129:24–37. doi: 10.1016/j.mod.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Pang K, Ryan JF, NISC Comparative Sequencing Program. Mullikin JC, Baxevanis AD, Martindale MQ. Genomic insights into Wnt signaling in an early diverging metazoan, the ctenophore Mnemiopsis leidyi . EvoDevo. 2010;1:e10. doi: 10.1186/2041-9139-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang K, Ryan JF, Baxevanis AD, Martindale MQ. Evolution of the TGF-b signaling pathway and its potential role in the ctenophore, Mnemiopsis leidyi . PLoS ONE. 2011;6:e24152. doi: 10.1371/journal.pone.0024152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perovic-Ottstadt S, Cetkovic H, Gamulin V, Schröder HC, Kropf K, Moss C, Korzhev M, Diehl-Seifert B, Müller IM, Müller WEG. Molecular markers for germ cell differentiation in the demosponge Suberites domuncula . Int J Dev Biol. 2004;48:293–305. doi: 10.1387/ijdb.041813sp. [DOI] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. Wnt signaling and the polarity of the primary body axis. Cell. 2009;139:1056–1068. doi: 10.1016/j.cell.2009.11.035. [DOI] [PubMed] [Google Scholar]
- Philippe H, Derelle R, Lopez P, Pick K, Borchiellini C, Boury-Esnault N, Vacelet J, Renard E, Houliston E, Queinnec E, et al. Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 2009;19:706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- Philippe H, Brinkmann H, Lavrov DV, Littlewood DTJ, Manuel M, Wörheide G, Baurain D. Resolving difficult phylogenetic questions: Why more sequences are not enough. PLoS Biol. 2011;9:e1000602. doi: 10.1371/journal.pbio.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatgorsky J, Kozmik Z. Cubozoan jellyfish: An Evo/Devo model for eyes and other sensory systems. Int J Dev Biol. 2004;48:719–729. doi: 10.1387/ijdb.041851jp. [DOI] [PubMed] [Google Scholar]
- Plickert G, Jacoby V, Frank U, Müller WA, Mokady O. Wnt signaling in hydroid development: Formation of the primary body axis in embryogenesis and its subsequent patterning. Dev Biol. 2006;298:368–378. doi: 10.1016/j.ydbio.2006.06.043. [DOI] [PubMed] [Google Scholar]
- Plickert G, Frank U, Müller WA. Hydractinia, a pioneering model for stem cell biology and reprogramming somatic cells to pluripotency. Int J Dev Biol. 2012;56:519–534. doi: 10.1387/ijdb.123502gp. [DOI] [PubMed] [Google Scholar]
- Podar M, Haddock SHD, Sogin ML, Harbison GR. A molecular phylogenetic framework for the phylum Ctenophora using 18S rRNA genes. Mol Phylogenet Evol. 2001;21:218–230. doi: 10.1006/mpev.2001.1036. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Renfer E, Amon-Hassenzahl A, Steinmetz PR, Technau U. A muscle-specific transgenic reporter line of the sea anemone, Nematostella vectensis . Proc Natl Acad Sci USA. 2010;107:104–108. doi: 10.1073/pnas.0909148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch F, Guder C, Vocke D, Hobmayer B, Holstein TW. An ancient chordin-like gene in organizer formation of Hydra . Proc Natl Acad Sci. 2007;104:3249–3254. doi: 10.1073/pnas.0604501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch F, Fritzenwanker JH, Scholz CB, Technau U. FGF signalling controls formation of the apical sensory organ in the cnidarian Nematostella vectensis . Development. 2008;135:1761–1769. doi: 10.1242/dev.020784. [DOI] [PubMed] [Google Scholar]
- Richards GS, Simionato E, Perron M, Adamska M, Vervoort M, Degnan BM. Sponge genes provide new insight into the evolutionary origin of the neurogenic circuit. Curr Biol. 2008;18:1156–1161. doi: 10.1016/j.cub.2008.06.074. [DOI] [PubMed] [Google Scholar]
- Riesgo A, Farrar N, Windsor PJ, Giribet G, Leys SP. The analysis of eight transcriptomes from all poriferan classes reveals surprising genetic complexity in sponges. Mol Biol Evol. 2014;31:1102–1120. doi: 10.1093/molbev/msu057. [DOI] [PubMed] [Google Scholar]
- Rivera AS, Hammel JU, Haen KM, Danka ES, Cieniewicz B, Winters IP, Posfai D, Wörheide G, Lavrov D, Knight SW, et al. RNA interference in marine and freshwater sponges: Actin knockdown in Tethya wilhelma and Ephydatia muelleri by ingested dsRNA expressing bacteria. BMC Bioltechnol. 2011;11:e67. doi: 10.1186/1472-6750-11-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JF. Did the ctenophore nervous system evolve independently? Zoology. 2014;117:225–226. doi: 10.1016/j.zool.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Ryan JF, Baxevanis AD. Hox, Wnt, and the evolution of the primary body axis: Insights from the early-divergent phyla. Biol Direct. 2007;13:e37. doi: 10.1186/1745-6150-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JF, Burton PM, Mazza ME, Kwong GK, Mullikin JC, Finnerty JR. The cnidarian-bilaterian ancestor possessed at least 56 homeoboxes: Evidence from the starlet sea anemone, Nematostella vectensis . Genome Biol. 2006:R64. doi: 10.1186/gb-2006-7-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JF, Pang K, NISC Comparative Sequencing Program. Mullikin JC, Martindale MQ, Baxevanis AD. The homeodomain complement of the ctenophore Mnemiopsis leidyi suggests that Ctenophora and Porifera diverged prior to the ParaHoxozoa. EvoDevo. 2010;1:e9. doi: 10.1186/2041-9139-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JF, Pang K, Schnitzler CE, Nguyen A-D, Moreland RT, Simmons DK, Koch BJ, Francis WR, Havlak P, Smith SA, et al. The genome of the ctenophore Mnemiopsis leidyiand its implications for cell type evolution. Science. 2013;342:12425921–1242592-8. doi: 10.1126/science.1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saina M, Genikhovich G, Renfer E, Technau U. BMPs and chordin regulate patterning of the directive axis in a sea anemone. Proc Natl Acad Sci. 2009;106:18592–18597. doi: 10.1073/pnas.0900151106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierwater B, de Jong D, DeSalle R. Placozoa and the evolution of Metazoa and intrasomatic cell differentiation. Int J Biochem Cell Biol. 2009;41:370–379. doi: 10.1016/j.biocel.2008.09.023. [DOI] [PubMed] [Google Scholar]
- Seipel K, Schmid V. Mesodermal anatomies in cnidarian polyps and medusae. Int J Dev Biol. 2006;50:589–599. doi: 10.1387/ijdb.062150ks. [DOI] [PubMed] [Google Scholar]
- Seipel K, Yanze N, Schmid V. Developmental and evolutionary aspects of the basic helix-loop-helix transcription factors Atonal-like 1 and Achaete-scute homolog 2 in the jellyfish. Dev Biol. 2004;269:331–345. doi: 10.1016/j.ydbio.2004.01.035. [DOI] [PubMed] [Google Scholar]
- Sinigaglia C, Busengdal H, Lerner A, Oliveri P, Rentzsch F. Molecular characterization of the apical organ of the anthozoan Nematostella vectensis . Dev Biol. 2015;398:120–133. doi: 10.1016/j.ydbio.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, Shoguchi E, Kawashima T, Hamada M, Hisata K, Tanaka M, Fujie M, Fujiwara M, Koyanagi R, Ikuta T, et al. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature. 2011;476:320–324. doi: 10.1038/nature10249. [DOI] [PubMed] [Google Scholar]
- Simion P, Bekkouche N, Jager M, Quéinnec E, Manuel M. Exploring the potential of small RNA subunit and ITS sequences for resolving phylogenetic relationships within the phylum Ctenophora. Zoology. 2015;118:102–114. doi: 10.1016/j.zool.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Simmons DK, Pang K, Martindale MQ. Lim homeobox genes in the ctenophore Mnemiopsis leidyi: The evolution of neural cell type specification. EvoDevo. 2012;3:e2. doi: 10.1186/2041-9139-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Varoqueaux F, Kittelmann M, Azzam RN, Cooper B, Winters CA, Eitel M, Fasshauer D, Reese TS. Novel cell types, neurosecretory cells, and body plan of the early-diverging metazoan Trichoplax adhaerens . Curr Biol. 2014;24:1565–1572. doi: 10.1016/j.cub.2014.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer RJ. The future of evo-devo: Model systems and evolutionary theory. Nat Rev Genet. 2009;10:416–422. doi: 10.1038/nrg2567. [DOI] [PubMed] [Google Scholar]
- Spemann H, Mangold H. Über Induktion von Embryo-nalanlagen durch Implantation artfremder Organisatoren. W Roux’s Arch Entwickl-Mech Org. Dev Genes Evol. 1924;100:599–638. [Google Scholar]
- Spring J, Yanze N, Josch C, Middel AM, Winninger B, Schmid V. Conservation of Brachyury, Mef2, and Snail in the myogenic lineage of jellyfish: A connection to the mesoderm of bilateria. Dev Biol. 2002;244:372–384. doi: 10.1006/dbio.2002.0616. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier MEA, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010a;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Larroux C, Lu DR, Mohanty K, Chapman J, Degnan BM, Rokhsar DS. Early evolution of the LIM homeobox gene family. BMC Biol. 2010b;8:e4. doi: 10.1186/1741-7007-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele RE. Genomics of basal metazoans. Integr Comp Biol. 2005;45:639–648. doi: 10.1093/icb/45.4.639. [DOI] [PubMed] [Google Scholar]
- Steinmetz PRH, Kraus JEM, Larroux C, Hammel JU, Amon-Hassenzahl A, Houliston E, Worheide G, Nickel M, Degnan BM, Technau U. Independent evolution of striated muscles in cnidarians and bilaterians. Nature. 2012;487:231–234. doi: 10.1038/nature11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau U, Bode HR. HyBra1, a Brachyury homologue, acts during head formation in Hydra. Development. 1999;126:999–1010. doi: 10.1242/dev.126.5.999. [DOI] [PubMed] [Google Scholar]
- Technau U, Scholz CB. Origin and evolution of endoderm and mesoderm. Int J Dev Biol. 2003;47:531–539. [PubMed] [Google Scholar]
- Technau U, Steele RE. Evolutionary crossroads in developmental biology: Cnidaria. Development. 2012;139:1447–1458. doi: 10.1242/dev.048959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt O, Collins AG, Pearse VB, Pearse JS, Ender A, Hadrys H, Schierwater B. Placozoa - no longer a phylum of one. Curr Biol. 2004;14:R944–R945. doi: 10.1016/j.cub.2004.10.036. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Fujisawa T, Holstein TW. Cnidarians and the evolutionary origin of the nervous system. Dev Growth Differ. 2009;51:167–183. doi: 10.1111/j.1440-169X.2009.01103.x. [DOI] [PubMed] [Google Scholar]
- Wiens M, Belikov SI, Kaluzhnaya OV, Adella T, Schroder HC, Perovic-Ottstadt S, Kaandorp JA, Muller WEG. Regional and modular expression of morphogenetic factors in the demosponge Lubomirskia baicalensis . Micron. 2008;39:447–460. doi: 10.1016/j.micron.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Wilson HV. On some phenomena of coalescence and regeneration in sponges. J Exp Zool. 1907;5:245–258. [Google Scholar]
- Windsor PJ, Leys SP. Wnt signaling and induction in the sponge aquiferous system: Evidence for an ancient origin of the organizer. Evol Dev. 2010;12:484–493. doi: 10.1111/j.1525-142X.2010.00434.x. [DOI] [PubMed] [Google Scholar]
- Wittlieb J, Khalturin K, Lohmann JU, Anton-Erxleben F, Bosch TC. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc Natl Acad Sci USA. 2006;103:6208–6211. doi: 10.1073/pnas.0510163103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wörheide G, Dohrmann M, Erpenbeck D, Larroux C, Maldonado M, Voigt O, Borchiellini C, Lavrov DV. Deep phylogeny and evolution of sponges (Phylum Porifera) Adv Mar Biol. 2012;61:1–78. doi: 10.1016/B978-0-12-387787-1.00007-6. [DOI] [PubMed] [Google Scholar]