Abstract
Pseudomonas sp. strain TXG6-1, a chitinolytic gram-negative bacterium, was isolated from a vegetable field in Taixing city, Jiangsu Province, China. In this study, a Pseudomonas chitinase C gene (PsChiC) was isolated from the chromosomal DNA of this bacterium using a pair of specific primers. The PsChiC gene consisted of an open reading frame of 1443 nucleotides and encoded 480 amino acid residues with a calculated molecular mass of 51.66 kDa. The deduced PsChiC amino acid sequence lacked a signal sequence and consisted of a glycoside hydrolase family 18 catalytic domain responsible for chitinase activity, a fibronectin type III-like domain (FLD) and a C-terminal chitin-binding domain (ChBD). The amino acid sequence of PsChiCshowed high sequence homology (> 95%) with chitinase C from Serratia marcescens. SDS-PAGE showed that the molecular mass of chitinase PsChiC was 52 kDa. Chitinase assays revealed that the chitobiosidase and endochitinase activities of PsChiCwere 51.6- and 84.1-fold higher than those of pET30a, respectively. Although PsChiC showed little insecticidal activity towards Spodoptera litura larvae, an insecticidal assay indicated that PsChiC increased the insecticidal toxicity of SpltNPV by 1.78-fold at 192 h and hastened death. These results suggest that PsChiC from Pseudomonas sp. could be useful in improving the pathogenicity of baculoviruses.
Keywords: catalytic domain, chitin-binding domain, fibronectin type-like domain, gene cloning, insecticidal activity
Introduction
Chitin, an insoluble polymer of N-acetyl-D-glucosamine (GlcNAc), is an important component of insect exoskeletons, crustacean shells and fungal cell walls. Chitin is the second most abundant component of biomass in nature after cellulose and can be degraded to GlcNAc monomers by chitinase. Chitinases are found in a wide range of organisms, including bacteria, fungi, plants, insects, crustaceans and some vertebrates. Chitinases, including endochitinase, exochitinase, β-N-acetylglucosaminidase and chitobiase, play important physiological and ecological roles in chitin metabolism (Flach et al., 1992; Felse and Panda, 1999). Many microbial species produce chitinase and digest chitin as a carbon and energy source. Bacteria in particular can produce high levels of chitinolytic enzymes. Based on their amino acid sequence similarities, chitinases are grouped into families 18 and 19 of glycoside hydrolases (Henrissat and Bairoch, 1993). Family 19 chitinases include classes I, II and IV of plant chitinases (Collinge et al., 1993; Levorson and Chlan, 1997; Gomez et al., 2002) and family 18 chitinases include most of the chitinases from bacteria, fungi, insects, animals and plants (class III and V chitinases).
Spodoptera litura multicapsid nucleopolyhedrovirus (SpltNPV) is highly specific and infects only a single host, the cotton leaf worm (Spodoptera litura). This insect is an economically important polyphagous pest in China, India and Japan, where it causes considerable economic loss to many vegetable and field crops. SpltNPV has been successfully applied in large scale as a commercial biological insecticide against the cotton leaf worm in China (Pang et al., 2001). However, like other biopesticides, SpltNPV has several potential limitations for broad commercial use, such as slow speed of kill (ranging from five days to more than two weeks), short field stability, very narrow host specificity, susceptibility to UV light, short shelf-life and high production costs (Inceoglu et al., 2006; Mills and Kean, 2010). Although there is considerable information on bacterial chitinase genes, there have been no reports on the chitinase genes from Pseudomonas species. In this study, we isolated a bacterium with chitinase activity from a vegetable field and identified this strain as a Pseudomonas species. To further characterize the chitinase genes from this bacterium, we initially cloned the chitinase C gene and analyzed the resulting sequence. Subsequently, the Pseudomonaschitinase C gene (PsChiC) was expressed in Escherichia coli BL21(DE3) using a 6xHis-fusion protein expression system and the synergistic effects of PsChiC in combination with SpltNPV on larvicidal activity against S. liturawere evaluated.
Material and Methods
Isolation of chitinase-producing microorganisms
To isolate chitinase-producing microorganisms, soil was collected from a vegetable field in Taixing city, Jiangsu Province, China, crushed and suspended in sterile water with shaking for 2 h. After centrifugation at 8000 × g for 10 min, the supernatant was diluted and spread on synthetic medium containing 1.4% agar. Analysis of the 16S rDNA sequence, as well as biochemical and physiological characterization of the isolated strain were done by the TAKARA Company, China.
Strains, plasmids and media
Pseudomonas sp. TXG6-1 was grown in synthetic medium containing colloidal chitin (5 g/L), MgSO4•7H2O (0.3 g/L), KH2PO4 (0.3 g/L) and K2HPO4 (0.3 g/L) at 30 °C. Colloidal chitin was prepared from crude chitin (Sigma, USA) by the method of Roberts and Selitrennikoff (1988), with a few modifications. Escherichia coliDH5α and E. coli BL21(DE3) for gene cloning, were grown in LB medium at 37 °C. The vectors pGEM T-easy (Promega, USA) and pET30a+ were used for cloning and expression of the chitinase gene, respectively.
PCR cloning and DNA sequencing
Chromosomal DNA of Pseudomonas sp. TXG6-1 was isolated using an EZNA bacterial DNA kit (Omega Bio-Tek, USA). The 16S rRNA gene sequence of TXG6-1 was amplified with two conserved bacterial 16S rDNA PCR primers (27F and 1495R) and sequenced using an ABI PRISM 3100 Genetic Analyzer system (Applied Bio-systems, Inc., USA). The 16S rRNA gene sequence was determined and compared to those of related taxa retrieved from the PseudoMLSA Database (Bennasar et al., 2010), GenBanK and EMBL databases.
Based on the sequence of the bacterial chitinase ChiC gene, the following pair of primers was designed for PCR cloning: P1: 5′-AGC TGA TAT TGC CGG CGA G-3′ and P2: 5′-AAA TAG TTG TTT GCT TAG GTG CGG-3′. The PCR amplification conditions for each cycle were: 95 °C (1 min), 55 °C (1.5 min) and 72 °C (1 min) for a total of 30 cycles. The corresponding gene from TXG6-1 was then inserted into the pGEM T-easy vector and sequenced by the dideoxy chain-termination method in an ABI 3730 automatic sequencer. The nucleotide sequences have been deposited in the GenBank nucleotide sequence database under accession number GU724605 for PsChiC.
Using the identified gene fragment encoding the chitinase as template, PCR was done with primers P3: 5′-GAC CAT ATG GCA CCA ATA ACA TTA TT-3′ and P4: 5′-GAC CCA TGG GGC GAT GAG CTG CCA CAG-3′. The PCR product was cleaved with NdeI and NcoI, and then ligated into NdeI-NcoI digested pET-30a+ to construct the recombinant plasmid pET-PsChiC.
Bioinformatics analysis of PsChiC
Analysis of the DNA sequences was done using the DNAman software package (version 4.0, Lynnon Corporation, Pointe-Claire, QC, Canada) (Huang and Zhang 2004). Homology searches were done using the BLAST algorithms against various databases in GenBank. Protein domain predictions, including the determination of the glycosyl hydrolase family 18 domains, were obtained from the SMART 7 - protein domain annotation resource (Letunic et al., 2012) and from the SBASE Domain Prediction System. The theoretical pI/MW were computed using the ExPASy Bioinformatics Resources Portal (Atrimo et al., 2012).
Expression and purification of PsChiC
The plasmid pET-PsChiC was transformed in E. coli BL21 (DE3) competent cells using the CaCl2 heat shock method (Sambrook and Russell, 2001). The clones were streaked onto Luria-Bertani (LB) agar plates containing kanamycin (50 μg/mL) and the plates then incubated at 37 °C for 16 h. A single clone was selected and used to inoculate 10 mL of LB broth containing kanamycin (50 μg/mL) followed by incubation at 37 °C with vigorous agitation in a shaking incubator. Approximately 5 mL of overnight culture was used to inoculate 100 mL of LB medium containing kanamycin (50 μg/mL) in a 500 mL fermenter and the culture was grown at 30 °C with vigorous agitation. When cells reached an optical density of 0.6 at 600 nm, isopropyl-β-D-thiogalactoside (IPTG; 0.8 mM) was added to induce chitinase gene expression. After an appropriate period of induction (Fan et al., 2007), the cells were harvested by centrifugation (3,000 × g, 15 min, 4 °C), ruptured on ice in an Ultrasonic Cell Disruptor and either used immediately or stored at −20 °C. After centrifugation (10,000 × g, 20 min) to remove cells, the total cell extract (crude chitinase) containing the intracellular chitinase was collected for measurement of chitinase activity or subjected to His-Tag affinity purification.
Chitinase assay
Pseudomonas sp. TXG6-1 was grown in 100 mL of basal medium containing 0.1% K2HPO4 and 0.05% MgSO4•7H2O (pH 7.0), and supplemented with 0.5–2% (w/v) of colloidal chitin as the sole carbon/nitrogen (C/N) source. The bacteria were cultured aerobically in a 250 mL Erlenmeyer flask at 30 °C for three days on a rotary shaker (150 rpm). After centrifugation (3,000 × g, 4 °C, 15 min), the supernatants were collected for measurement of chitinase activity.
Chitinase activity was measured using a chitinase assay kit (Sigma-Aldrich), according to the manufacturer’s instructions. The exochitinase (N-acetyl-β-glucosaminidase), chitobiosidase and endochitinase activities in the harvested culture medium were determined using p-NPGlcNAc, p-NP-(GlcNAc)2 and p-NP-(GlcNAc)3 as substrates, respectively. The release of p-nitrophenol was monitored at 405 nm. A molar extinction coefficient for p-nitrophenol of ε = 17,700 M−1cm−1 was used to calculate the product concentration. Protein concentrations were measured by the method of Lowry et al. (1951) using bovine serum albumin as the standard.
Insecticidal bioassay
The synergistic effects of PsChiC on the insecticidal activity of Spodoptera litura nucleopolyhedrovirus (SpltNPV) on second-instar larvae of S. litura were studied using a surface contamination bioassay in 24-well trays. SpltNPV (1 × 107 particles) mixed with an artificial diet was added to the 24-well trays. Crude chitinase (20 μL, 50 mU) was soaked into each cube of artificial diet. For the control treatment, water instead of SpltNPV was added to the artificial diet. Three replicates were done for each treatment and each replicate contained 30 larvae. The plates were incubated at 26 ± 1 °C, 85% humidity and a 12 h light/dark photoperiod. The confidence intervals and other regression parameters were determined using the SPSS program.
Results
Identification of Pseudomonas TXG6-1
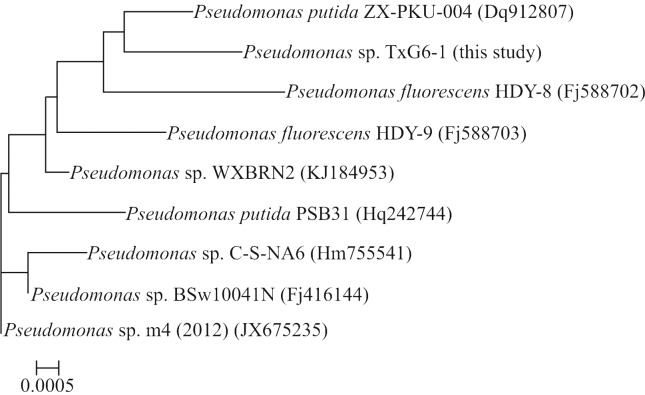
For morphological and biochemical classification, the microscopic characteristics of the Pseudomonas strain were analyzed with a Nikon H-600L system (Nikon, Japan). The sequences of TXG6-1 16S rRNA (1400 bp) were provisionally identified and classified based on BLASTN analysis against several 16S rRNA gene databases (PseudoMLSA, GenBank and EMBL). A homology search indicated that Pseudomonas strain TXG6-1 had > 99% sequence identity with Pseudomonas putidaZX-PKU-004 (GenBank accession number DQ912807). Figure 1 shows a Neighbor-Joining phylogenetic tree inferred from the sequences of the isolate and related organisms and clearly indicates that the isolate belongs to the genus Pseudomonas.
Figure 1. Rooted neighbor-joining distance matrix tree derived from the 16S rRNA gene sequences of TXG6-1 and related bacterial. Accession numbers for the 16S rRNA gene sequences used are given in parentheses after the species and strain names. The scale bar represents a 0.05% nucleotide substitution rate based on Kimura’s two-parameter model.
Bioinformatic comparison of PsChiC
The open reading frame of PsChiC had A+T = 40.4% and G+C = 59.6%. PsChiC consisted of 480 amino acids, with a predicted molecular mass of 51.66 kDa and pI of 5.36. Amino acid sequence analysis revealed that PsChiC shared high sequence homology with several Serratia and Pseudomonas chitinases, such as Serratia marcescens chitinase C1 (99.17%, accession no. BAA76623; 98.54%, CAF74787; 98.13%, ABI79318), Pseudomonas aeruginosa PA7 chitinase (66.25%; ABR83459) and Pseudomonas fluorescens Pf-5 chitinase (65.83%; AAY91366). Bacterial family 18 chitinases can be divided into three subfamilies (A, B and C) based on the amino acid sequence similarity of their catalytic domains (Suzuki et al., 1999). The major structural difference deduced from comparison of the amino acid sequences of subfamilies A and B is that chitinases in subfamily A have an insertion domain between the seventh and eighth β-strands of the (α+β)8 -barrel catalytic domain that is absent in subfamily B chitinases. PsChiC has the catalytic domain of subfamily B instead of subfamily A. Prediction of the functional domains of the protein revealed a glycoside hydrolase family 18 catalytic domain, a fibronectin type III-like domain (FLD) and a chitin-binding domain (ChBD).
The PsChiC N-terminal region (residues 25–305) showed similarity to the catalytic domains of chitinases belonging to glycosyl hydrolase family 18 (Henrissat and Bairoch, 1996; Henrissat, 1999). The region exhibited extensive similarity to the catalytic domains of several bacterial chitinases, such as Serratia marcescens chitinase C1 (97.51%) and Vibrio harveyi chitinase A (60.14%) (Figure 2). In this region, the deduced amino acid sequence from residues 134 to 141 (DGLDIDLE) was homologous to the active site motif of enzymes in glycosyl hydrolase family 18 ([DN]-G-[VFILM]-[DN]-[LFIMV]-[DN]-X-E) that form the fourth β-strand in the (β/α)8-barrel of their catalytic domain (Henrisssat and Bairoch, 1993; Aronson Jr et al., 2006). Furthermore, amino acid residues D-137, D-139 and E141 of PsChiC (corresponding to D-200, D-202 and E204 of Bacillus circulans chitinase ChiA) were well conserved and may play an essential role in chitinase activity (Watanabe et al., 1993).
Figure 2. Amino acid sequence alignment of the catalytic regions of PsChiC and several bacterial chitinases. PsChiC - Pseudomonas sp. TXG6-1 chitinase C (this study), VhChiA - Vibrio harveyichitinase A (AAC46383), SmChiC1 - Serratia marcescenschitinase C1 (CAF74787), BcChiD - Bacillus circulanschitinase D (P27050), HcChi - Hahella chejuensis KCTC 2396 chitinase (YP_432187) and MmChi - Microscilla marina ATCC 23134 chitinase (ZP_01688190). Black shading indicates amino acid residues that are identical. The active site motif is boxed.
As shown in Figure 3, the C-terminal region (residues 433 to 479) exhibited significant similarity not only to the ChBDs of various bacterial chitinases, but also to the C-terminal regions of Streptomyces griseus serine protease (Sidhu et al., 1994), and Vibrio alginolyticus deacetylase (Ohishi et al., 2000). In the ChBD region of many chitinases, aromatic amino acids (W and Y) are highly conserved and may play a crucial role during binding to the pyranosyl rings of N-acetylglucosamine residues in chitin (Morimoto et al., 1997).
Figure 3. Amino acid sequence alignment of the chitin-binding domains of PsChiC with those of other bacterial enzymes (protease, deacetylase and chitinases). PsChiC - Pseudomonas sp. TXG6-1 chitinase C (this study), BcChiA1 - Bacillus circulans chitinase A1 (AAA81528), CpChi18C - Clostridium paraputrificum chitinase 18C (BAD29950), SmChiC1 - Serratia marcescens GEI chitinase C1 (ACX42073), AcChiA - Acidobacterium capsulatum ATCC 51196 chitinase A (ACO33699), PaChi - PaeruginosaUCBPP-PA14 chitinase (ABJ11480), SgPrC - Streptomyces griseus serine protease (AAA26813), and VaDA1 - Vibrio alginolyticus deacetylase DA1 (CAC29092). Black shading indicates identical amino acid residues.

The middle region (residues 338 to 411) showed similarity to the fibronectin type III-like domain (Fn3) sequences found in the R-1 and R-2 regions of Bacillus circulans chitinase A1 (32.20% and 30.77% identity, respectively; Watanabe et al., 1990), and Chi74FLD1 and Chi74FLD2 of B. thuringiensis (16.22% and 24.32% identity, respectively; Barboza-Corona et al., 2003) (Figure 4). Fibronectin is a multifunctional extracellular matrix and plasma protein and plays a significant role in cell adhesion. Fibronectin type III-like domain (FLD) has been found in chitinase, cellulases, α-amylase and poly(3-hydroxybutyrate) (PHB) depolymerase (Little et al., 1994). Among bacterial enzymes capable of degrading insoluble substrates, the Fn3 domains are most frequently found in chitinases. The widespread occurrence of this domain in chitinases may have special relevance for the degradation of insoluble and crystalline polysaccharide chitin (Suzuki et al., 1999).
Figure 4. Alignment of the Fn3-like domain of PsChiC with those of other bacterial enzymes. The sequences of PsChiC Fn3-like are aligned with those of Serratia marcescens chitinase C1 (CAF74787), Bacillus circulans chitinase A1 (r1 and r2, AAA81528) and Bacillus thuringiensis serovar kenyae ChiA74 (Chi74FLD1 and Chi74FLD2, AAL17867). Black shading indicates identical amino acid residues.

Expression and purification of PsChiC
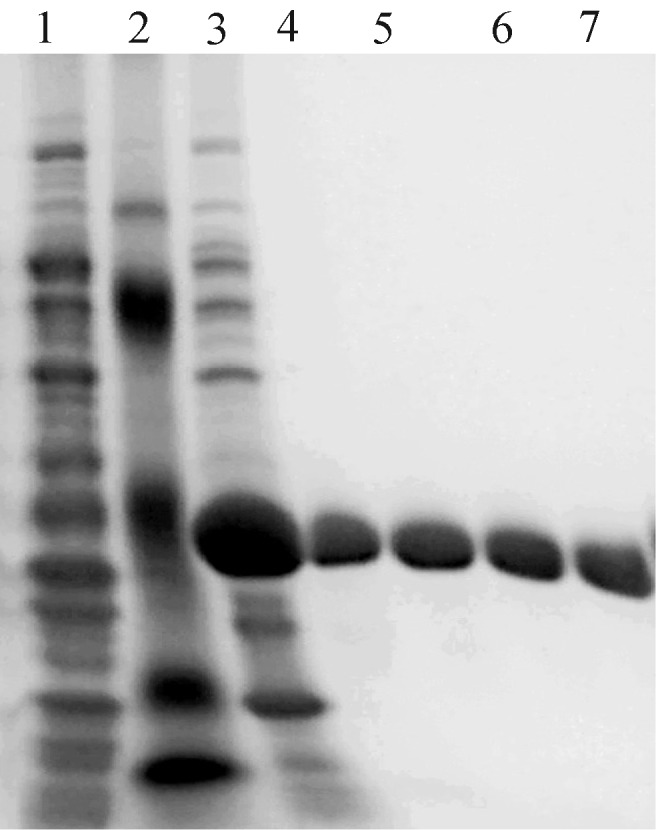
The PsChiC gene was subcloned into pET30a to construct the recombinant expression vector pETPsChiC. The identity of pETPsChiC was identified by restriction enzyme digestion and DNA sequencing. The protein in supernatant from E. coli BL21 (DE3) cultures grown in LB medium was purified through a His-Tag affinity resin designed for the recombinant chitinase. The enzyme activity and protein concentration of the fractions were determined. The molecular mass of the major band of purified PsChiC was estimated to be 52 kDa by SDS-PAGE (Figure 5). This molecular mass agreed well with the calculated molecular mass of 51.66 kDa based on the 480 amino acid sequence. Expression of the fusion protein was observed 1 h after induction, with maximal expression occurring at 6 h.
Figure 5. SDS-PAGE of recombinant PsChiC. The gel was stained with Coomassie brilliant blue R-250. Lane 1 - induced BL21(DE3)/pET30a, lane 2 - molecular mass markers (105, 71, 50, 35 and 25 kDa), lane 3 - BL21(DE3)/pETPsChiC induced for 4 h, lanes 4–7 - purified PsChiC.
Chitinase assay
Chitinolytic activities were quantified with a liquid assay using p-NP-(GlcNAc)n (n = 1–3) as substrates. The chitinase expressed in E. coli BL21(DE3)/ pETPsChiC showed hydrolytic activity towards the chitin tetrameric derivative (4-MU-(GlcNAc)3] and trimeric derivative [(4-MU-(GlcNAc)2], with practically no activity against the dimeric derivative (4-MU-GlcNAc) (Table 1). This finding indicated that PsChiC had both chitobiosidase and endochitinase activities, but no exochitinase activity.
Table 1. Chitinolytic activities of Pseudomonas chitinase C (PsChiC) expressed in E. coli.
| Sample | Protein | Chitinolytic activity (units/mg of protein) | ||
|---|---|---|---|---|
|
| ||||
| Endochitinase* | Chitobiosidase | Exochitinase | ||
| E. coli without vector | Secreted | 0.00 ± 0.00d | 0.05 ± 0.01c | 0.01 ± 0.01b |
| E. coli with pET-PsChiC | Secreted | 63.02 ± 0.11b | 21.33 ± 0.07b | 0.01 ± 0.01b |
| E. coli with pET-PsChiC | Intracellular | 34.42 ± 0.32c | 10.47 ± 0.04b | 0.02 ± 0.01b |
| Pseudomonas sp. TXG6-1 | Secreted | 15.26 ± 0.94a | 8.55 ± 0.66a | 1.73 ± 0.13a |
The data are expressed as the mean ± SD. Values with the same letter were not significantly different as determined by Tukey’s multiple range test (p < 0.05).
The substrates used for the enzymatic assays were: p-nitrophenyl N,N′-diacetyl-β-D-chitobioside (chitobiosidase activity), p-nitrophenyl N-acetyl-β-D-glucosaminide (exochitinase activity) and 4-nitrophenyl β-D-N,N′,N″-triacetylchitotriose (endochitinase activity). In all cases, formation of the reaction product (p-nitrophenol), which ionizes in basic pH, was monitored colorimetrically at 405 nm.
Insecticidal activity
Although crude PsChiC had little activity against S. litura larvae, co-culture with SpltNPV enhanced the insecticidal activity of the latter (Table 2). When crude PsChiC was mixed with 1×107 SpltNPV, some S. litura larvae began to die after 72 h while larvae fed an artificial diet of SpltNPV alone began to die at 96 h. The mortality rates were 91.1%, 51.1% and 17.8% at 192 h for S. litura larvae grown on artificial diet supplemented with 1×107 SpltNPV + 50 mU crude chitinase, 1×107 SpltNPV, and 50 mU crude chitinase, respectively. PsChiC alone was also insecticidal to S. litura larvae.
Table 2. Synergistic effects of Pseudomonas chitinase C gene (PsChiC) on the insecticidal activity of SpltNPV.
| Treatment (h)* | 72 | 96 | 120 | 144 | 168 | 192 |
|---|---|---|---|---|---|---|
| SpltNPV | 0Ij** | 2.22HIj | 10FGghi | 13.33DEFfgh | 20De | 51.11Bc |
| SpltNPV + PsChiC | 4.44GHIij | 12.22EFfgh | 15.56DEFfgh | 27.78Cd | 57.58Bb | 91.11Aa |
| PsChiC | 0Ij | 0Ij | 0Ij | 4.44GHIij | 8.89FGHhi | 17.78DEef |
| CK | 0Ij | 0Ij | 0Ij | 0Ij | 2.22HIj | 2.22HIj |
All data analyses were done using SPSS software.
Three replicates were done for each treatment and each replicate contained 30 larvae.
Numbers indicate mortality rate (%), uppercase letters indicate p < 0.01 and lowercase letters indicate p < 0.05. SpltNPV - Spodoptera litura multicapsid nucleopolyhedrovirus.
Discussion
The low yield of chitinase-producing strains is one of the major problems in the study and application of chitinases. The production of recombinant proteins in active form is therefore of great importance for studying the applications of these enzymes. Expression systems based on E. coli have been commonly used to express heterologous proteins because this bacterium is well characterized with regard to its molecular genetics, physiology and availability of different expression systems (Balbás, 2001). In this study, PCR primers specific for the chiC gene were used to amplify a chitinase gene fragment and to confirm the presence of the PsChiCgene in Pseudomonas strain TXG6-1. The chitinase PsChiC gene was transformed in an expression vector pET30a to construct the recombinant expression plasmid, pETPsChiC. The fusion protein PsChiC-His was successfully expressed in E. coli cells. The ability to produce this chitinase in E. coli will facilitate large-scale production and subsequent structural and functional studies of this protein.
Chitinases from similar or closely related species exhibit high sequence similarity (Henrissat, 1991). Based on the 16S rDNA sequence analysis, strain TXG6-1 belongs to the genus Pseudomonas, and comparison of the amino acid sequence of PsChiC from strain TXG6-1 with the chiC sequences of other bacterial chitinase indicated that PsChiC had a high level of homology (77.1–99.2%) with the corresponding enzymes from some gram-negative bacteria, including Serratia marcescens, S. proteamaculans, S. odorifera, P. aeruginosa and P. fluorescens. This finding suggests that the PsChiCgene of strain TXG6-1 and the chiC genes of these gram-negative bacteria were recently derived from the same ancestral chitinase gene.
The effective control of pest insect populations is an essential prerequisite for producing food and commodities for man and domestic animals. Chemical insecticides are highly effective in controlling pest insect populations. However, the use of chemical pesticides has led to several problems, including environmental contamination, the development of resistance to insecticides, the elimination of beneficial or non-target insects, and adverse effects on human health, including cancer and several immune system disorders (Andersen et al., 2012; Baldwin et al., 2013). The emergence of pesticide-resistant insect populations has also caused major outbreaks of secondary pests (Devine and Furlong, 2007). Thus, the search for safer agents and methods of plant protection has intensified in recent years.
The use of biological insecticides, alone or in combination with other agents and methods, in an integrated pest management system has gained momentum during the last 30 years (Ashour et al., 2007). Nucleopolyhedroviruses (NPVs) (family Baculoviridae) infect the larvae of many important lepidopteran pests and several of these viruses have been developed as commercial biopesticides (Moscardi, 1999). However, NPVs are of limited usefulness since their narrow spectrum of activity enables them to kill only certain insect species. NPVs are also not very resistant in the environment and require precise application practices since many of these pathogens are specific to young insect larval stages or are sensitive to radiation. The most important limitation is that the killing rate of NPVs is lower than that of chemical insecticides. Consequently, there is a need for agents to improve the speed of killing by NPVs so as to expand the use of NPVs. Several studies have demonstrated the potential use of chitinase for controlling insect pests because of the enzymes disrupting effect on the peritrophic membrane. In lepidopteran larvae, the chitinases (from AcMNPV and SpliMNPV-K1, respectively) act by disrupting the peritrophic membrane and increasing its permeability (Rao et al., 2004; Wang et al., 2013). As shown here, PsChiC alone had limited insecticidal activity against S. litura larvae but markedly enhanced the activity of SpltNPV against these larvae. This finding suggests that PsChiC could be useful in enhancing the pathogenicity of baculoviruses.
Acknowledgments
This work was supported by the Special Fund for Agro-scientific Research in the Public Interest (201303028), Natural Science Fund project in Jiangsu Province (BK2011678) and the Doctorate Fellowship Foundation of Nanjing Forestry University.
Footnotes
Associate Editor: Célia Maria de Almeida Soares
References
- Andersen HR, Wohlfahrt-Veje C, Dalgård C, Christiansen L, Main KM, Nellemann C, Murata K, Jensen TK, Skakkebaek NE, Grandjean P. Paraoxonase 1 polymorphism and prenatal pesticide exposure associated with adverse cardiovascular risk profiles at school age. PloS One. 2012;7:e36830. doi: 10.1371/journal.pone.0036830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson NN, Jr, Halloran BA, Alexeyev MF, Zhou XE, Wang Y, Meehan EJ, Chen L. Mutation of a conserved tryptophan in the chitin-binding cleft of Serratia maracescens chitinase A enhances transglycosylation. Biosci Biotechnol Biochem. 2006;70:243–251. doi: 10.1271/bbb.70.243. [DOI] [PubMed] [Google Scholar]
- Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour MB, Ragheb DA, EI-Sheikh E-SA, Gomaa E-AA, Kamita SG, Hammock BD. Biosafety of recombinant and wild type nucleopolyhedrovirus as bioinsecticides. Int J Environ Res Public Health. 2007;4:111–125. doi: 10.3390/ijerph2007040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbás P. Understanding the art of producing protein and nonprotein molecules in E. coli . Mol Biotechnol. 2001;19:251–267. doi: 10.1385/MB:19:3:251. [DOI] [PubMed] [Google Scholar]
- Baldwin RA, Salmon TP, Schmidt RH, Timm RM. Wildlife pests of California agriculture: regional variability and subsequent impacts on management. Crop Protection. 2013;46:29–37. [Google Scholar]
- Barboza-Corona JE, Nieto-Mazzocco E, Velazquez-Robledo R, Salcedo-Hernandez R, Bautista M, Jimenez B, Ibarra JE. Cloning, sequencing, and expression of the chitinase gene chiA74 from Bacillus thuringiensis . Appl Environ Microbiol. 2003;69:1023–1029. doi: 10.1128/AEM.69.2.1023-1029.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasar A, Mulet M, Lalucat J, Garcia-Valdes E. PseudoMLSA: a database for multigenic sequence analysis of Pseudomonas species. BMC Microbiol. 2010;10:118. doi: 10.1186/1471-2180-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge DB, Kragh KM, Mikkelsen JD, Nielsen KK, Rasmussen U, Vad K., 1 Plant chitinases. Plant J. 1993;3:31–40. doi: 10.1046/j.1365-313x.1993.t01-1-00999.x. [DOI] [PubMed] [Google Scholar]
- Devine GJ, Furlong MJ. Insecticide use: contexts and ecological consequences. Agr Hum Values. 2007;24:281–306. [Google Scholar]
- Fan Y, Zhang Y, Yang X, Pei X, Guo S, Pei Y. Expression of a Beauveria bassiana chitinase (Bbchit1) in Escherichia coli and Pichia pastoris . Protein Expr Purif. 2007;56:93–99. doi: 10.1016/j.pep.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Felse PA, Panda T. Regulation and cloning of microbial chitinase genes. Appl Microbiol Biotechnol. 1999;51:141–151. doi: 10.1007/s002530051374. [DOI] [PubMed] [Google Scholar]
- Flach J, Pilet PE, Jolles P. What’s new in chitinase research? Experientia. 1992;48:701–716. doi: 10.1007/BF02124285. [DOI] [PubMed] [Google Scholar]
- Gomez L, Allona I, Casado R, Aragoncillo C. Seed chitinases. Seed Sci Res. 2002;12:217–230. [Google Scholar]
- Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. J Biochem. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B. Classification of chitinases modules. EXS. 1999;87:137–156. doi: 10.1007/978-3-0348-8757-1_10. [DOI] [PubMed] [Google Scholar]
- Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino-acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Bairoch A. Updating the sequence-based classification of glycosyl hydrolases. Biochem J. 1996;316:695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Zhang L. Rapid and sensitive dot-matrix methods for genome analysis. Bioinformatics. 2004;20:460. doi: 10.1093/bioinformatics/btg429. [DOI] [PubMed] [Google Scholar]
- Inceoglu AB, Kamita SG, Hammock BD. Genetically modified baculoviruses: A historical overview and future outlook. Adv Virus Res. 2006;68:323–360. doi: 10.1016/S0065-3527(06)68009-3. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levorson J, Chlan CA. Plant chitinase consensus sequences. Plant Mol Biol Rep. 1997;15:122–133. [Google Scholar]
- Little E, Bork P, Doolittle RF. Tracing the spread of fibronectin type III domains in bacterial glyco-hydrolases. J Mol Evol. 1994;39:631–643. doi: 10.1007/BF00160409. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Mills NJ, Kean JM. Behavioral studies, molecular approaches, and modelling: methodological contributions to biological control success. Biol Control. 2010;52:255–262. [Google Scholar]
- Morimoto K, Karita S, Kimura T, Sakka K, Ohmiya K. Cloning, sequencing, and expression of the gene encoding Clostridium paraputrificum chitinase ChiB and analysis of the functions of novel cadherin-like domains and a chitin-binding domain. J Bacteriol. 1997;179:7306–7314. doi: 10.1128/jb.179.23.7306-7314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscardi F. Assessment of the application of baculoviruses for control of Lepidoptera. Annu Rev Entomol. 1999;44:257–289. doi: 10.1146/annurev.ento.44.1.257. [DOI] [PubMed] [Google Scholar]
- Ohishi K, Murase K, Ohta T, Etoh H. Cloning and sequencing of the deacetylase gene from Vibrio alginolyticus H-8. J Biosci Bioeng. 2000;90:561–563. doi: 10.1016/s1389-1723(01)80041-4. [DOI] [PubMed] [Google Scholar]
- Pang Y, Yu J, Wang L, Hu X, Bao W, Li G, Chen C, Han H, Hu S, Yang H. Sequence analysis of the Spodoptera lituramulticapsid nucleopolyhedrovirus genome. Virology. 2001;287:391–404. doi: 10.1006/viro.2001.1056. [DOI] [PubMed] [Google Scholar]
- Rao R, Fiandra L, Giordana B, Eguileor Mde, Congiu T, Burlini N, Arciello S, Corrado G, Pennacchio F. AcMNPV ChiA protein disrupts the peritrophic membrane and alters midgut physiology of Bombyx mori larvae. Insect Biochem Mol Biol. 2004;34:1205–1213. doi: 10.1016/j.ibmb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Roberts WK, Selitrennikoff CP. Plant and bacterial chitinases differ in antifungal activity. J Gen Microbiol. 1988;134:169–176. [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2001. [Google Scholar]
- Sidhu SS, Kalmar GB, Willis LG, Borgford TJ. Streptomyces griseus protease C. A novel enzyme of the chymotrypsin superfamily. J Biol Chem. 1994;269:20167–20171. [PubMed] [Google Scholar]
- Suzuki K, Taiyoji M, Sugawara N, Nikaidou N, Henrissat B, Watanabe T. The third chitinase gene (chiC) of Serratia marcescens 2170 and the relationship of its product to other bacterial chitinases. Biochem J. 1999;343:587–596. [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Choi JY, Roh JY, Tao XY, Liu Q, Lee JH, Kim JS, Kim WJ, Je YH. Insecticidal activity of the chitinase from the Spodoptera litura nucleopolyhedrovirus. Entomol Res. 2013;43:63–69. [Google Scholar]
- Watanabe T, Kobori K, Miyashita K, Fujii T, Sakai H, Uchida M, Tanaka H. Identification of glutamic acid 204 and aspartic acid 200 in chitinase A1 of Bacillus circulans WL-12 as essential residues for chitinase activity. J Biol Chem. 1993;268:18567–18572. [PubMed] [Google Scholar]
- Watanabe T, Suzuki K, Oyanagi W, Ohnishi K, Tanaka H. Gene cloning of chitinase A1 from Bacillus circulans WL-12 revealed its evolutionary relationship to Serratia chitinase and to the type III homology units of fibronectin. J Biol Chem. 1990;265:15659–15665. [PubMed] [Google Scholar]
Internet Resources
- SMART 7 - protein domain annotation resource. [May 5;2014 ]. http://smart.embl-heidelberg.de/
- SBASE Domain Prediction System. [May 5;2014 ]. http://hydra.icgeb.trieste.it/sbase/
- ExPASy Bioinformatics Resources Portal. [May 5;2014 ]. http://www.expasy.org/cgi-bin/pi_tool.htmlserver