Summary
Cell fate acquisition is heavily influenced by direct interactions between master regulators and tissue-specific enhancers. However, it remains unclear how lineage-specifying transcription factors, which are often expressed in both progenitor and mature cell populations, influence cell differentiation. Using in vivo mouse liver development as a model, we identified thousands of enhancers that are bound by the master regulators HNF4A and FOXA2 in a differentiation-dependent manner, subject to chromatin remodeling, and associated with differentially expressed target genes. Enhancers exclusively occupied in the embryo were found to be responsive to developmentally regulated TEAD2 and coactivator YAP1. Our data suggest that Hippo signaling may affect hepatocyte differentiation by influencing HNF4A and FOXA2 interactions with temporal enhancers. In summary, transcription factor-enhancer interactions are not only tissue specific but also differentiation dependent, which is an important consideration for researchers studying cancer biology or mammalian development and/or using transformed cell lines.
Graphical abstract
Introduction
The recent generation of induced hepatocytes from both mouse and human fibroblasts via ectopic expression of transcription factors (TFs), Hepatocyte nuclear factor 1 or 4 alpha (HNF1/4A), and Forkhead box A2 or 3 (FOXA2/3) in fibroblasts highlights how critical our understanding of TF biology and in vivo hepatocyte differentiation is for the advancement of alternative liver disease therapies (Du et al., 2014; Huang et al., 2011; Huang et al., 2014; Sekiya and Suzuki, 2011; Simeonov and Uppal, 2014). It is well established that lineage-specifying TFs determine cell fate through interactions with tissue-specific enhancers. However, it is not clear how master regulators, expressed in both embryonic and adult liver, coordinate the gene-expression changes that are necessary for organ formation and mature hepatocyte cell function.
The adult liver is primarily composed of hepatocytes, the principal functional cell of the liver. This relative homogeneity has made it an ideal tissue for numerous historic gene-regulation studies. In contrast, the embryonic liver is home to a large population of hematopoietic precursors, which has previously hampered investigation of transcriptional regulation in hepatocyte progenitors, known as hepatoblasts. Hepatoblasts are bipotential cells that during development progressively differentiate into hepatocytes and cholangiocytes (bile duct cells). Maturing hepatocytes begin to express genes that control metabolism and detoxification, and produce serum proteins, bile, and hormones. It is notoriously difficult to maintain mature hepatocyte gene-expression profiles and cell functions in vitro (Raju et al., 2013), and consequently the role of TFs in hepatocyte differentiation and function is best demonstrated by in vivo studies.
Embryos devoid of HNF4A fail to reach gastrulation due to defects in the visceral endoderm. Early lethality can be rescued by provision of wild-type extraembryonic tissue via a tetraploid methodology (Duncan et al., 1997). In this context, liver specification occurs in Hnf4a null embryos; however, hepatocyte precursors fail to differentiate (Parviz et al., 2003). A later depletion of HNF4A by 6 weeks postpartum results in disrupted liver architecture; changes in serum, lipid, and urea levels; and ultimately a 70% mortality rate by 8 weeks of age (Hayhurst et al., 2001). Members of the Forkhead box, subfamily A, of TFs (FOXA1/2/3) have been shown to play major roles in animal development. Foxa2 null mice fail to form regions of definitive endoderm that give rise to liver (Lee et al., 2005), and conditional Foxa2 deletion in late fetal hepatocytes causes sensitivity to a cholic-acid-containing diet, triggering a toxic accumulation of bile salts, endoplasmic reticulum stress, and liver injury (Bochkis et al., 2009). FOXAs have a structural similarity to linker histories and are able to bind condensed chromatin, where they can either passively act as placeholders for recruitment of factors later in development or actively recruit other factors for transcriptional initiation (Sekiya et al., 2009; Zaret and Carroll, 2011). Despite their pioneer function, FOXA TFs have previously been shown to bind different target sites in diverse cell lines and tissues that affect distinct gene-expression repertoires (Lupien et al., 2008). Furthermore, cooperation with nuclear hormone receptor family members, namely, the estrogen and androgen receptors, has been shown to influence binding-site choice and account for gender-dependent susceptibility to hepatocellular carcinoma (Li et al., 2012).
Although it has been clearly demonstrated that HNF4A and FOXA2 are necessary for both acquisition of hepatoblast cell fate and maintenance of mature hepatocyte function, it is not yet understood how stably expressed TFs are able to fulfill such distinct roles during organ maturation. We hypothesized that identifying and comparing target genes in hepatoblasts and hepatocytes would enable us to better understand how these master regulators contribute to hepatocyte differentiation. Here, we show that HNF4A and FOXA2 interact with thousands of enhancer regions in a differentiation-dependent manner to orchestrate gene-expression changes required for liver development. Our data uncover how developmentally regulated enhancer switching is affected by Hippo signaling. We propose that dynamic TF-DNA binding may represent a common mechanism by which master regulators control the process of organ maturation during mammalian development.
Results
Widespread Combinatorial Role for HNF4A and FOXA2 in the Embryonic Liver
To gain a detailed understanding of how HNF4A and FOXA2 cooperate and interact with cis-regulatory elements during hepatoblast maturation in vivo, we performed a chromatin immunoprecipitation followed by next-generation sequencing (ChIP-seq) analysis on hepatoblasts from fetal liver at embryonic day 14.5 (E14.5). We purified hepatoblasts from resident hematopoietic cells by magnetic-activated cell sorting using a previously characterized hepatoblast marker, Delta-like 1 (DLK1) Tanimizu et al., 2003; Wei et al., 2013), which is found on the surface of HNF4A/FOXA2-positive cells in the liver at E14.5 (Figures 1A and S1). We sequenced DNA immunoprecipitated by HNF4A or FOXA2 antibodies (as described in Experimental Procedures), and identified a total of 11,703 HNF4A-bound and 9,398 FOXA2-bound regions (based on a false-discovery rate [FDR] of 0.01 to determine peak height thresholds) in the fetal liver (Table S1).
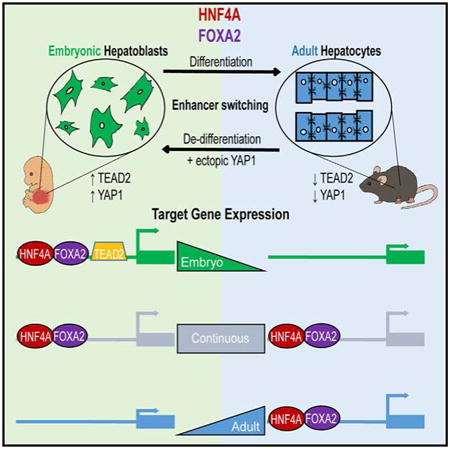
Figure 1. HNF4A and FOXA2 ChIP-Seq in Purified Fetal Hepatoblasts.

(A) Immunofluorescence analysis of cryosectioned mouse embryos at E14.5, showing colabeling of hepatoblasts with the cell-surface marker delta-like-1 (DLK1, green), hepatic nuclear factor 4a (HNF4A, red) and nuclei (DAPI, blue). Scale bar, 100 mm.
(B) Venn diagram representing the number of called peaks (FindPeaks, FDR < 0.01) for FOXA2 (white) and HNF4A (black) In hepatoblast ChIP-seq data sets. Peaks with >50 bp shared FOXA2 and HNF4A were considered to be common sites of enrichment (gray).
(C) GO enrichment analysis of shared target genes Selected statistically significant enriched pathways are shown and graphed using the binomial log10 p value on the y axis.
(D) Read density histograms represent the enrichment of HNF4A and FOXA2 detected by ChIP-seq in embryonic hepatoblasts proximal to the hepatoblast biomarkers Gpc3 and Dlk1.
See also Figure S1 and Table S1.
Since regions bound by multiple TFs are more likely to function as tissue-specific enhancers (Hoffman et al., 2010), we determined the extent and location of HNF4A and FOXA2 colocalization prior to hepatocyte maturation. Approximately 30% of FOXA2 binding was found to occur at HNF4A-occupied sites (Figure 1B). Sites of coenrichment, like the majority of all HNF4A and FOXA2 sites, were frequently found within distal intergenic regions (Figure S1; Shin et al., 2009). Due to the remote location of many TF-binding sites, we associated cis-regulatory regions with target genes using previously established enhancer and promoter units and conducted a Gene Ontology (GO) analysis (Ashburner et al., 2000; Shen et al., 2012). This identified 2,957 genes as putative direct targets. Many genes were found to associate with processes integral to hepatocyte function, such as the lipid metabolic process (binominal p value = 4.5 × 10−48). Other target loci included TFs such as Prox1, Cited2, and Onecut1/2, which are linked to liver development (p = 1.01 × 10−102), cell number homeostasis (p = 1.10 × 10−39), integrin-mediated signaling pathway regulation (p = 1.5 × 10−42), and actin cytoskeleton organization (p = 3.47 × 10−169). Examples of shared HNF4A and FOXA2 binding events in embryonic liver are shown in Figure 1D for Gpc3 (top) and Dlk1 (bottom), both of which are associated with a hepatoblast cellular phenotype (Grozdanov et al., 2006; Tanimizu et al., 2003).
HNF4A and FOXA2 Bind the Genome in a Differentiation-Dependent Manner
To date, gene regulation in the embryonic liver has largely been examined on a locus-by-locus basis. For example, the regulatory domains for Alpha-fetoprotein (Afp), an established fetal liver biomarker, have been extensively studied (Godbout et al., 1988). A comparison of data from our genomic analysis of HNF4A and FOXA2 (Figure 2A) with the positions of enhancer regions previously shown to control transcription at the Afp locus in embryonic liver (E1 −2.25 to −2.5 kb, E2 −4.9 to −5.1 kb, E3 −6.25 to −6.65 kb; black lines below the histogram) showed a remarkable correlation (Figure 2A). Furthermore, when we examined this locus using adult liver HNF4A and FOXA2 chIP-seq data sets previously published by our group (Hoffman et al., 2010; Figure 2A), we confirmed that HNF4A and FOXA2 binding was lost at many of the Afp-associated regulatory regions upon hepatocyte differentiation. One site (indicated in gray) was occupied in both the embryonic and adult liver. In contrast, Prlr, which is associated with insulin regulation in the adult liver, was found to be targeted by HNF4A and FOXA2 exclusively in our adult data sets (Figure 2A).
Figure 2. HNF4A and FOXA2 Co-occupy Target Sites in a Differentiation-Dependent Manner.
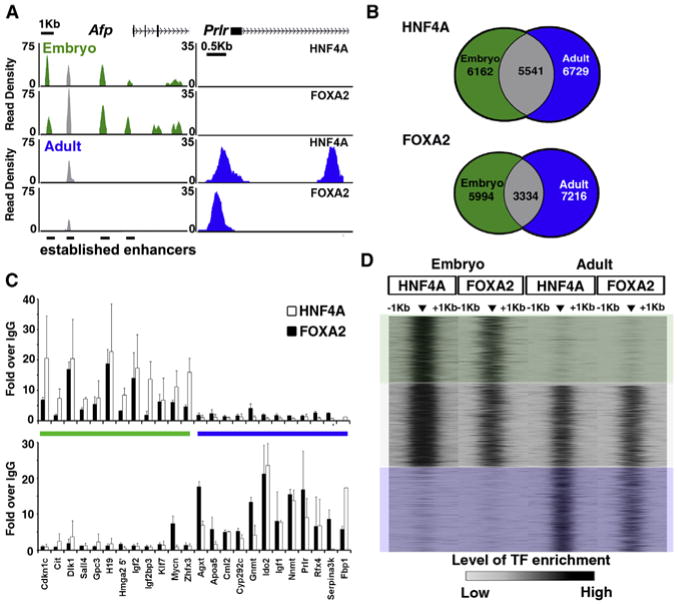
(A) Read density histograms proximal to the Afp (left) and Prlr (right) locus, displaying HNF4A and FOXA2 ChIP-seq data sets generated from embryonic hepatoblasts (green) and adult hepatocytes (blue).
(B) Venn diagram representing regions of enrichment in embryonic (green) and adult (blue) ChIP-seq data sets for HNF4A and FOXA2. Sites with >50 bp overlap between them were designated as common between data sets (gray).
(C) ChIP-qPCR verification of sites defined as embryonic (green bar) or adult (blue bar); data are represented as mean ± SEM from duplicate experiments.
(D) Heatmap representation of TF enrichment (black, high; gray, low) at all regions bound by both HNF4A and FOXA2 during at least one stage of liver development. Enrichment levels were profiled ±1 kb at a resolution of 50 bp from the peak center using SitePro in Galaxy/Cistrome toolbox. Boxes overlaying the heatmap highlight regions that were selected for further analysis and classified as embryonic only (green), continuous (gray), or adult only (blue).
See also Figure S2.
To establish the extent to which liver maturation is accompanied by changes in HNF4A and FOXA2 binding on a genome-wide scale, we compared chIP-seq profiles for HNF4A and FOXA2 embryonic and adult livers. A comparison of four data sets gave rise to 15 different types of enrichment profiles (Figure S2). A remarkable 55% of HNF4A-binding sites were uniquely occupied in either the embryonic or adult state, suggesting that these sites are regulated in a differentiation-dependent manner. FOXA2 also altered 60% of its DNA interaction sites during liver development (Figure 2B). This global switch of DNA-binding sites was unexpected and distinct from situations in which a TF binds only high-affinity sites in one cell type but both high- and low-affinity sites in another cell type.
To eliminate the possibility that these binding-site differences were simply artifacts that arose from the comparison of historic data sets with new ones, differential data processing, or imposed height cutoff thresholds, we used unthresholded data to profile the read enrichment of the HNF4A and FOXA2 data sets within 1 kb of differentially bound sites using SitePro within the Galaxy/Cistrome toolbox (Shin et al., 2009). We found no evidence to suggest that TF enrichment was artificially masked by imposed height cutoffs or that the TFs were binding to alternative sites in close proximity, confirming that we had identified differentiation-dependent regions of TF-DNA interactions (Figure 2D). In addition, to experimentally validate our finding, we carried out four biologically replicated ChIP quantitative PCR (chIP-qPCR) experiments on 24 sites that showed distinct patterns of enrichment in our chIP-seq data sets (Figure 2C). Cdknlc, Cit, Dlk1, Sall4, Gpc3, H19, Hmga2, Igf2, Igf2bp3, Klf7, Mycn, and Zfhx3 showed enriched binding of both HNF4A and FOXA2 in our embryonic, but not adult, chIP-seq data sets. In contrast, enhancers associated with the genes Agxt, ApoaS, Cml2, Cyp2c29, Gnmt, Ido2, Igf1, Nnmt, Prlr, Rfx4, Serpina3k, and Fbp1 showed enrichment in our adult chIP-seq HNF4A and FOXA2 data. Thus, differentiation-dependent TF binding occurs during liver development.
Differentiation-Dependent Binding Sites Function as Temporal Enhancers
Genomic regions occupied by more than one TF have previously been shown to associate with tissue-specific gene expression (Hoffman et al., 2010). Consequently, to better understand the biological relevance and mechanisms underlying differentiation-dependent DNA binding, we interrogated regions in which both TFs were gained, lost, or maintained during development. We categorized sites as embryonic (HNF4A and FOXA2 enrichment in embryonic data sets only [891 sites]), continuous (HNF4A and FOXA2 enrichment in all data sets [1,050 sites]), or adult (HNF4A and FOXA2 enrichment in adult data sets only [1,094 sites] Figure 2D). Previous studies suggested that higher levels and a bimodal distribution pattern of monomethylated histone 3 lysine 4 (H3K4me1) correlate with enhancer function (Hoffman et al., 2010; Rada-Iglesias et al., 2011). To determine the activity of identified putative enhancer regions, we measured H3K4me1 at differentiation-dependent binding sites using our H3K4me1 chIP-seq data set generated from adult liver (Hoffman et al., 2010). Figure 3A shows a heatmap in which the levels of H3K4me1 surrounding each target site (±1 kb) are represented. We observed that regions occupied at both stages of differentiation (continuous) or exclusively in the adult showed the highest levels of H3K4me1 at each side of the central TF binding-site position with a central depletion in signal (a bimodal pattern). In contrast, embryonic sites that were no longer bound in the adult showed the highest levels of H3K4me1 in a central position (a monomodal pattern; Figure 3A). Moreover, the overall level of H3K4me1 was reduced in the embryonic sites (Figure 3B). These patterns were also observed in a chIP-seq analysis on adult perfused liver using a pan-acetylated H4 antibody (Figure S3).
Figure 3. Differentiation-Dependent Binding Sites Show Distinct Patterns of Enhancer-Associated H3K4me1.
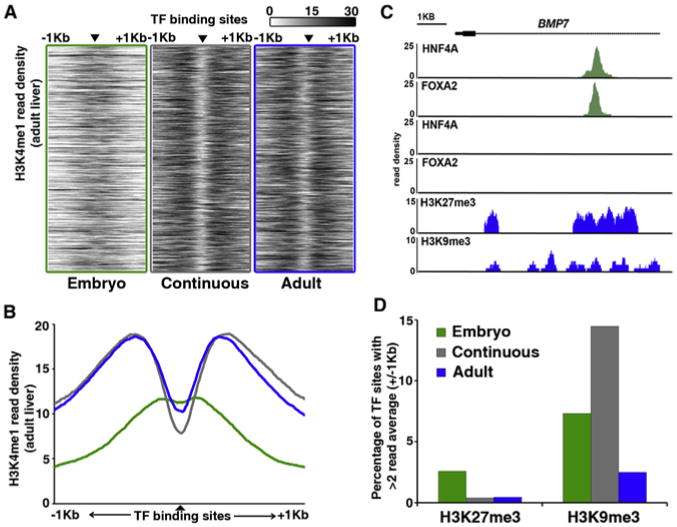
(A) Heatmap representation of H3K4me1 at regions bound by both HNF4A and FOXA2 (▲) in embryonic liver only (green, left), both embryonic and adult liver (gray, middle), or adult liver only (blue, right). Enrichment levels (white, low; black, high) were profiled ±1 kb at a resolution of 10 bp from the TF peak center, represented by the small black triangle. The vertical ordering of sites is random.
(B) Average enrichment profiles of H3K4me1 at differentiation-dependent HNF4A and FOXA2 binding sites (green line, embryo; gray line, continuous; blue line, adult).
(C) Read density histograms proximal to the BMP7 locus, displaying HNF4A and FOXA2 ChIP-seq data sets generated from embryonic hepatoblasts (green), and HNF4A, FOXA2, H3K27me3, and H3K9me3 ChIP-seq data sets generated from adult hepatocytes (blue).
(D) Percentage of differentiation-dependent binding sites showing >2 average read enrichment for repressive histone modifications H3K27me3 or H3K9me3 within 2 kb regions surrounding TF-binding sites.
See also Figure S3.
To determine whether embryonic-specific TF-binding sites are masked in adult liver via chromatin compaction, we measured the enrichment patterns of the repressive histone modifications H3K27me3 and H3K9me3 using chIP-seq. We found that only a few embryonic enhancers were enriched for H3K27me3 and H3K9me3 in the adult liver (3% and 7%, respectively), an example of which is shown in Figure 3C. Furthermore, an analysis of publically available DNase I hypersensitivity data (Shen et al., 2012) confirmed that >60% of embryonic sites remain accessible in the adult liver (data not shown). Together, these data suggest that enhancer switching is not simply a consequence of Polycomb-mediated chromatin compaction. This finding is consistent with the established role of FOXA2 as a pioneer factor that is able to bind compacted nucleosomes. In summary, our results show that H3K4me1 and H4ac signatures are distinct at differentiation-dependent binding sites, and further support the existence of widely dispersed, developmentally regulated, functional enhancers that are differentially bound by HNF4A and FOXA2 in the liver.
We next investigated the impact of temporal TF-DNA binding on associated target gene transcription by using RNA sequencing (RNA-seq) to profile the transcriptomes of both DLK1 + hepatoblasts and adult liver (Table S2). We used alternative expression analysis by sequencing (ALEXA-seq) (Griffith et al., 2010) to identify differentially expressed transcripts. More than 2,000 transcripts were enriched in adult hepatocytes compared with embryonic precursors (Embryo < Adult; Figure 4A), whereas more than 4,000 genes were more highly expressed in embryonic hepatoblasts (Embryo > Adult; Figure 4A). Histograms of the top-ranking differentially expressed transcripts, H19 (Embryo) and Serpina3K (Adult), are shown as examples in Figure 4B. To identify the most differentially expressed pathways, we carried out a GO analysis (Eden et al., 2009). As anticipated, “cell cycle” was significantly enriched in proliferating embryonic hepatoblasts, whereas pathways including “oxidation reduction” and “metabolic process” dominated the transcriptome of mature hepatocytes. Interestingly, embryonic samples were also significantly enriched for transcripts associated with cell component organization, adhesion, and differentiation (Figure 4C). In contrast to previous studies by Kyrmizi et al. (2006), we did not observe statistically significant differential expression of Hnf4a and Foxa2 transcript levels during liver maturation. This is likely because they used whole embryonic livers, whereas we used isolated hepatoblasts. Furthermore, we detected only minor contributions from the Hnf4a P2 promoter (Figure S4; Torres-Padilla et al., 2001).
Figure 4. Differential Gene Expression during Hepatocyte Maturation Is Associated with Differentiation-Dependent HNF4A and FOXA2 Binding.
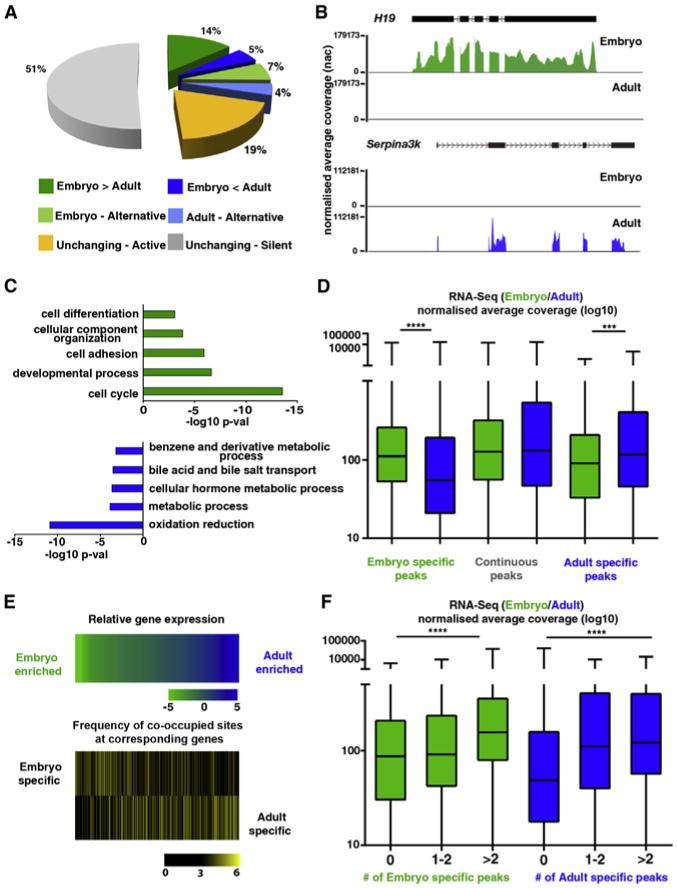
(A) Representation of transcriptome proportions subject to differential regulation (>2-fold difference with a Pearson correlation of <0.05), alternative transcription (inclusion of different exons and/or untranslated regions) in embryonic hepatoblasts (green) and adult hepatocytes (blue), or consistent (<2-fold change, yelow) in transcript evels between embryonic hepatoblasts and adult liver.
(B) RNA-seq profiles of the top-ranking differentially expressed genes (H19 and Serplna3k) in embryonic (green) and adult (blue) liver.
(C) GO analysis of differentially expressed transcripts in embryonic (green) and adult (blue) liver.
(D) Global expression levels in embryonic (green) and adult (blue) liver of putative target genes associated with HNF4A and FOXA2 in the embryo only (left), embryonic and adult liver (continuous, middle), or adult ony (right). The NAC is the cumulative base coverage of a feature normalized to feature length and library size, Significance was tested using Kruskal-Wallis one-way ANOVA with Dunn's post-test correction; ****p < 0.001, ***p < 0.005. Error bars in the boxplot represent minimum to maximum values.
(E) In the upper panel, relative expression levels (log10) of genes targeted by HNF4A and FOXA2 In a differentiation-dependent manner are ranked as either embryo-enriched (green) or adult livar-enriched (blue). Intensity in bottom two heatmaps (black, low; yellow, high) reflects the average number of differentiation-dependent DNA-binding sites within associated enhancer promoter unit divided by the average number of sites in an EPU included in the analysis (see aso Figure S4).
(F) Genes were categorized according to the associated number of differentiation-dependent enhancers and global gene expression levels were compared between those not bound specifically at the developmental stage indicated, targeted at one or two putative enhancers or bound at more than two sites. Significance was tested using Kruskal-Wallis one-way ANOVA with Dunn's post-test correction; ****p < 0.001. Error bars in the boxplot represent minimum to maximum values.
See also Table S2.
To explore how differentiation-dependent HNF4A and FOXA2 cobinding may affect temporal gene-expression patterns during hepatoblast maturation, we examined the expression level of putative target genes. Genes associated with “Embryonic” enhancers (cobound by HNF4A and FOXA2 in the embryo) exhibited a significant global decrease in expression during development, whereas expression of genes targeted by “Adult” enhancers (cobound by HNF4A and FOXA2 in the adult) showed a significant increase in the adult liver (p < 0.001 and p < 0.01, respectively). No significant change in continuously targeted genes was detected (Figure 4D).
Gene transcription is often influenced by TF occupancy at multiple enhancer regions (Ong and Corces, 2011). For example, Afp associates with a number of sites occupied by HNF4A and FOXA2 exclusively in the embryo in which it is highly expressed. To test for a relationship between the number of differentiation-dependent TF sites and differential gene-expression levels, we ranked expressed target genes by fold enrichment, from the highest in adult liver (blue) to the highest levels in embryonic liver (green) (Figure 4E; top heatmap). We calculated the number of differentiation-dependent binding sites (embryo, middle; adult, bottom) within corresponding gene regions and found that temporally expressed target genes were more highly associated with differentiation-dependent enhancers. Furthermore, when we applied a statistical analysis to this relationship, we determined that association with one or more differentiation-dependent binding sites is predictive of higher gene-expression levels at the appropriate developmental stage (p ≤ 0.001), suggesting that dynamic HNF4A and FOXA2 DNA binding influences the evolving hepatoblast-hepatocyte transcriptome during maturation. This association was also true for peaks occupied by HNF4A alone (Figure S4).
Motif Analysis of Temporal Enhancers Reveals an Embryonic-Specific Cofactor
Having demonstrated that HNF4A and FOXA2 bind to functional enhancer regions in a differentiation-dependent manner to regulate temporal gene-expression patterns, we investigated potential mechanisms underlying this enhancer-switching behavior. Previous studies suggested that local sequence conservation or motif affinity may influence the stability of TF binding in different tissues or conditions. Consequently, we compared sequences underlying sites co-occupied by HNF4A and FOXA2 at different stages of development. We found no strong differences in either sequence conservation or the underlying HNF4A or FOXA2 motifs between sites bound in a differentiation-dependent manner (Figure S5).
Although HNF4A and FOXA2 motifs were indistinguishable by our analysis, the enrichment or depletion of other TF binding may influence DNA accessibility and/or occupancy during development. Interestingly, we found that the motif related to the nuclear receptor FXR, which is more highly expressed in the adult, was enriched at adult specific sites (data not shown). However, we chose to focus on identifying TF motifs that underlie embryonic peaks, since many cofactors in the adult liver are already well established but comparatively little is known about the embryonic liver. To isolate differentially enriched motifs, we employed the motif-finding algorithm oPOSSUM (http://opossum.cisreg.ca/oPOSSUM3/). We examined embryonic-specific enhancer regions using adult-specific enhancers as a background data set (Subramanian et al., 2014). Reassuringly, our analysis detected TFs such as HNF1a, HNF1b, and CEBPa, which are known to work with HNF4A and FOXA2 during liver development (Figure 5A). FOXA2 was also among our candidates because embryonic enhancers showed a greater number of underlying FOXA2 motifs than adult-specific enhancers (Figure S5). Intriguingly, the TEAD1 motif was enriched. TEAD transcriptional activity is dependent on interactions with YAP1, a downstream component of the Hippo signaling cascade. This pathway controls liver size and is commonly amplified in human hepatocellular carcinoma (Camargo et al., 2007; Zender et al., 2006). We determined the quality of predicted TEAD1 motifs by generating a composite of all TEAD1 motifs at embryonic enhancer regions (Figure 5B). Since TEAD motifs are similar among family members, we verified the transcript levels of all four family members in embryonic and adult liver (Figure 5C). Tead2 was the most highly expressed family member in embryonic liver, being expressed at a level similar to that observed for Foxa2, and was the most dramatically downregulated in adult liver. Interestingly, the Tead2 locus itself is targeted by HNF4A in a developmentally regulated manner, highlighting a possible cofactor feedback mechanism (Figure 5D). TEAD2 protein was robustly detected in embryonic liver and its levels were decreased in adult liver (Figure 5E). Moreover, using an electrophoretic mobility shift assay (EMSA), we verified that TEAD2 can interact with predicted sites (such as the Sall4 locus). We detected binding of both HNF4A and TEAD2 upon incubation with nuclear extract isolated from 293T cells expressing MYC-tagged TFs and nuclear extracts from E14.5 livers.
Figure 5. TEAD2 Cooperates with HNF4A and FOXA2 at Developmentally Regulated Embryonic Enhancers.
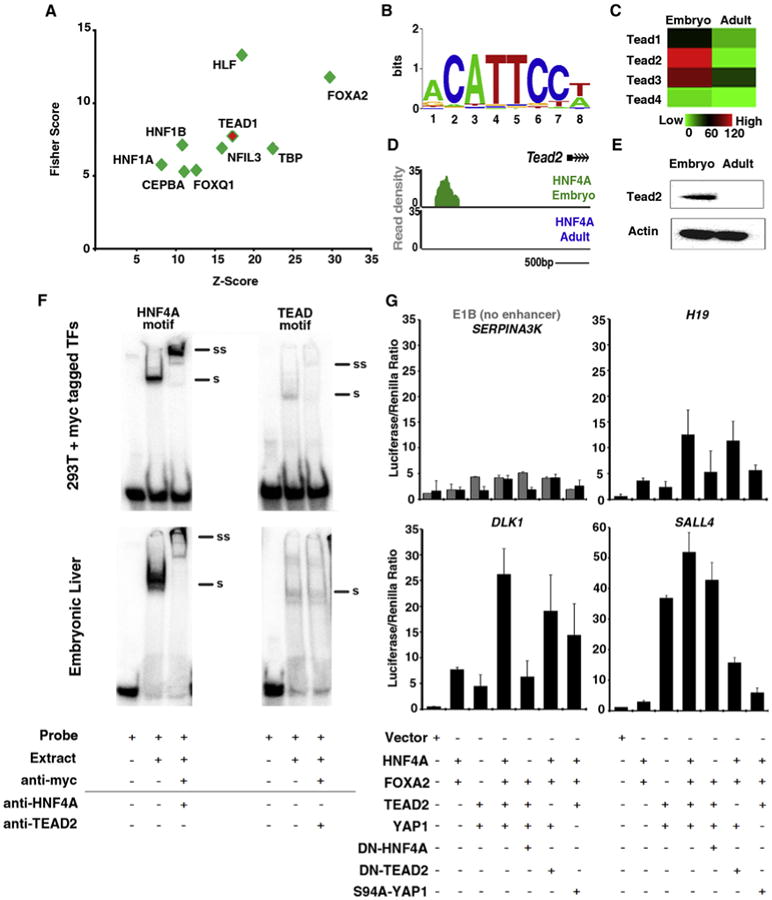
(A) Anchored combination site analysis using oPOSSUM. Motifs of expressed TFs enriched at embryonic enhancer regions were graphed according to Fisher and Z score values.
(B) Composite position-weighted matrix of all identified TEAD1 motifs at embryonic enhancer regions, generated using STAMP.
(C) Heatmap depicting NAC values for TEAD family members (red, high; green, low) in RNA-seq libraries made from DLK1 + hepatoblasts (Embryo) and adult liver (Adult).
(D) Read density histogram proximal to the Tead2 locus, displaying HNF4A binding in embryonic hepatoblasts (green), but not in adult hepatocytes (blue).
(E) Western blot analysis of protein isolated from embryonic (E14.5) and adult liver using anti-Tead2 and anti-actb.
(F) Nuclear extracts were isolated from 293T cells expressing myc-tagged TF or from E14.5 liver and incubated with P32-labeled probes containing TF-binding motifs found at the enhancer region associated with Sall4 and 3 mg of anti-myc (top panel), anti-HNF4A, or anti-TEAD2. Note that a shift (denoted by a black bar and “S”) was detected with both probes and in both 293T and E14.5 extracts. A supershift (denoted by a black bar and “SS”) was detected using anti-myc and anti-HNF4A (due to the poor quality of the antibody, no supershift to endogenous protein could be detected using anti-TEAD2).
(G) Embryonic enhancers containing TEAD motifs were cloned into luciferase vectors containing a minimal promoter (E1B) and transfected into 293T cells alongside a Renilla-containing construct. Expression vectors for HNF4A, FOXA2, TEAD2, YAP1, dominant-negative HNF4A (DN-HNF4A), DN-TEAD2, and mutant YAP (S94A YAP1) were cotransfected in different combinations. Luciferase assays were conducted 48 hr after transfection. Data shown are from duplicate experiments are represented as mean ± SEM.
See also Figure S5.
To determine whether TEAD binding can influence target gene expression, we cloned a selection of embryonic-specific enhancers into luciferase vectors. We observed limited activity when luciferase vectors containing no enhancer region (E1B promoter only) or the adult-specific enhancer for Serpina3K were cotransfected with TEAD2 and YAP1 expression plasmids (Figure 5G). However, enhancer regions predicted to contain TEAD motifs (associated with H19, Dlk1, and Sall4 genes) produced a >2-fold induction of activity in the presence of TEAD2 and YAP1 expression. Similar levels of induction were observed (∼2-fold) when HNF4A and FOXA2 were expressed. However, when all four factors were expressed, a >10-fold increase in enhancer activity was detected. The additive effect was diminished when a dominant-negative version of HNF4A or TEAD2 was substituted. A similar reduction was also seen upon addition of YAP S94A mutant (Hong et al., 2003). Interestingly, the enhancer associated with Sall4 showed a more robust response to TEAD2 and YAP1 than to HN4A and FOXA2, suggesting that it is highly dependent on Hippo signaling. Together, these data support a model in which TEAD2 and YAP1 can promote the expression of genes associated with a subset of HNF4A/FOXA2 embryonic enhancer regions.
To determine whether the presence of TEAD2 and YAP1 can influence HNF4A and FOXA2 recruitment to differentiation-dependent enhancers in vivo, we used an inducible transgenic mouse model in which YAP1 is expressed in the liver in response to doxycycline (Figure S6; Dong et al., 2007). After 2 weeks of treatment, we conducted a chIP-qPCR analysis of HNF4A and FOXA2 in livers of transgenic mice and age-matched controls (Figure 6A). We observed no changes in HNF4A and FOXA2 occupancy at sites occupied in both embryonic and adult liver (Afp enh2, Hnf4a, and Trf). At embryonic enhancer sites, we noted a small but significant increase (e.g., FOXA2 at Afp enh3 and HNF4A at Sall4; p ≤ 0.05). However, detection of HNF4A and FOXA2 occupancy notably decreased at the adult-specific enhancers Ido2 and Prlr (p ≤ 0.05) upon ectopic expression of YAP1. Consistent with a previously published microarray analysis of YAP-TG-induced livers (Dong et al., 2007), the expression levels of a number of genes associated with embryonic liver, including DIM, H19, and Afp, increased after YAP1 induction. However, we also noted a decrease in the expression of adult targets, including Ido2 and Nnmt. Figure 6B illustrates the fold change in enhancer occupancy detected by chIP-qPCR after induction of YAP1. Together, these results suggest that the presence of YAP1 in the nucleus influences HNF4A and FOXA2 binding-site choice and target gene expression.
Figure 6. Ectopic Expression of YAP1 in Adult Liver Induces Changes in HNF4A- and FOXA2-DNA Binding.
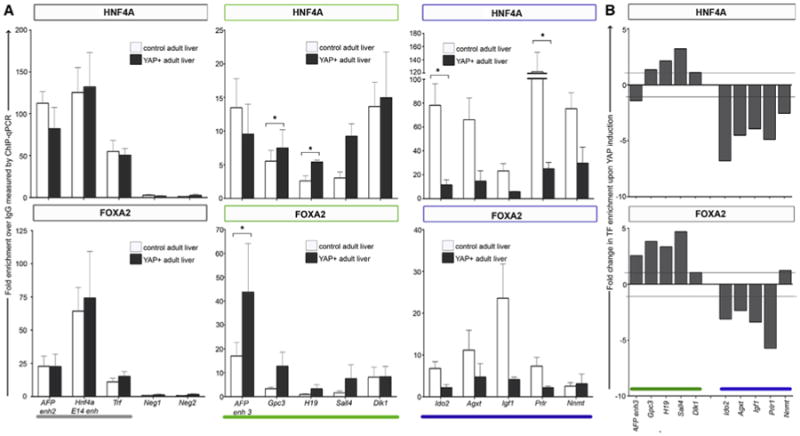
ChIP-qPCR analysis of HNF4A (top panel) and FOXA2 (lower panel) in control (white) and YAP-induced (gray) adult liver.
(A) TF enrichment is expressed as fold over control immunoprecipitation (normal rabbit IgG). The gray bar Indicates regions previously shown to be bound in both embryonic and adult liver (AFP enh2, HNF4a, and TRF). Neg1 and Neg2 function as negative control regions in which no enrichment was expected. Afp enh3, Gpc3, H19, Sall4, and Dlk1 (green bar) represent regions previously shown to be exclusively occupied in embryonic liver. Ido2, Agxt, Igf1, Prlr, and Nnmt (blue bar) represent regions previously shown to be exclusively occupied in adult liver. Data are represented as mean ± SEM from triplicate experiments. Multiple t tests were conducted to determine significance (*p < 0.05).
(B) ChIP-PCR analysis represented as fold change between control = 1 and YAP-induced liver.
Discussion
Here, we document how lineage-specifying TFs that are present in the nucleus throughout the differentiation process can contribute to the evolving transcriptomes of mammalian cells undergoing maturation in vivo. Using RNA-seq and chIP-seq libraries generated from embryonic hepatoblasts and adult liver, we have shown that the key hepatic TFs HNF4A and FOXA2 occupy enhancers and control target gene expression in a differentiation-dependent manner. Furthermore, we highlight how Hippo signaling influences HNF4A and FOXA2 enhancer switching, thereby affecting target gene expression and the status of hepatocyte differentiation.
It is notoriously difficult to propagate or even maintain hepatocytes in a functional differentiated state in vitro. Investigators have achieved partial success by developing 3D tissue culture models and using a multitude of extracellular matrix (ECM) components in the form of Matrigel (Raju et al., 2013). TEAD2 functions as a nuclear TF in the Hippo signaling pathway, which can regulate cell death, proliferation, and differentiation (reviewed in Yu and Guan, 2013). In the absence of Hippo pathway activity (e.g., at low cell density), YAP1 is shuttled to the nucleus, which enhances the ability of TEAD2 to induce target gene expression (Schlegelmilch et al., 2011). We propose a model in which reduced cell densities and disrupted cell-cell and cell-ECM contacts, all of which are required for in vitro propagation, likely cause nuclear localization of YAP1, which in turn influences HNF4A and FOXA2 binding-site choice. Consequently, genes associated with embryonic liver are activated and the expression levels of genes associated with fully differentiated, functional hepatocytes decline. Although the Hippo signaling cascade has previously been implicated in regulation of liver size and cancer development, only very recent studies using a mosaic YAP1 induction model have shown that nuclear YAP1 is able to induce hepatocyte dedifferentiation into clonogenic progenitors in vivo (Yimlamai et al., 2014). Our work further implicates Hippo signaling in hepatocyte differentiation.
Due to the difficulty of propagating hepatocytes in vitro, HEPG2, a hepatocellular carcinoma (HCC) cell line, is commonly used to represent hepatocytes. Interestingly, the ENCODE Project has conducted chIP-seq to examine HNF4A and TEAD4 in HEPG2 cells (https://www.encodeproject.org/experiments/ENCSROOOBRP/). Remarkably, 45% of TEAD4 binding sites overlap by at least 50 bp with HNF4A sites. Furthermore enhancers co-occupied by HNF4A and TEAD4 in HEPG2 cells were found at “embryonic” target genes, such as Sall4 and Dlk1, suggesting that the synergy we uncovered between TEAD2 and HNF4A could also be applicable to human liver development and HCC. The embryonic target gene Sall4 highlighted in our analysis is of particular interest because forced expression has been shown to inhibit hepatoblast differentiation along the hepatocyte lineage, whereas downregulation encourages bile duct formation (Oikawa et al., 2009). Furthermore, Sall4 was recently identified as a “targetable marker” for an aggressive subset of HCC (Yong et al., 2013).
HNF4A has previously been implicated in regulating tissue remodeling, which is required for normal liver formation. Ablation of HNF4A specifically in the embryonic liver causes loss of cohesive tissue architecture with disruption of cell-cell junction assembly (Battle et al., 2006; Parviz et al., 2003). In this work, we noted that members of the Focal Adhesion Kinase pathway featured prominently among the temporally expressed, embryonic-specific HNF4A target genes. Furthermore, a previous gene-expression analysis of postnatal heterozygous YAP mice indicated that ECM proteins become deregulated (Septer et al., 2012). Together, these studies suggest that significant crosstalk occurs between the hepatoblast microenvironment and HNF4A to regulate enhancer binding, target gene expression, and liver maturation.
Recent studies have examined the phenomenon of enhancer switching by master regulators using in vitro models of differentiation. For example, the pluripotency-associated pioneer factor OCT4 dramatically switches enhancers upon transition from an embryonic stem cell to an epiblast-like cell state (Buecker et al., 2014). Soleimani et al. (2012) showed that MYOD, a TF that is well known to induce muscle cell fate, can change binding sites between primary myoblast and induced myotube states. In contrast to our analysis, they demonstrated that subtle differences in motifs underlying differentiation-dependent sites enable competitive binding of a repressive TF. Temporal expression of a repressive factor may also explain why HNF4A and FOXA2 are unable to bind adult sites in the embryonic liver. Although we have found no evidence of this to date, it is possible that induction of YAP1 leads to activation of a repressive factor that can mask adult enhancer regions in the embryonic liver, thereby preventing premature differentiation.
In summary, our results highlight that lineage-specifying TFs undergo widespread enhancer switching during in vivo cell differentiation and organ development, which enables these key players to perform distinct yet important roles in both progenitor and mature cell populations. The data generated by this investigation into TF binding behaviors offer a rich resource that will continue to facilitate a greater understanding of both transcriptional regulation and hepatocyte differentiation.
Experimental Procedures
Animals
C57BL6/J and ApoE-rtTA; TRE-hYAP transgenic mice (Dong et al., 2007) were cared for under protocols approved by the Animal Care Committee of the University of British Columbia or the Johns Hopkins University IACUC. Three transgenic and three wild-type female mice (4 weeks old) were given a dose of 0.2 mg/ml doxycycline in drinking water for 2 weeks. Livers from these mice were flash-frozen before qRT-PCR, western blotting, and chIP-qPCR analysis were performed.
Purification of Fetal Hepatoblasts
Embryonic livers collected at E14.5 were incubated in 0.005% collagenase type I (STEMCELL Technologies), 0.05 U/ml dispase, 1 mM EDTA, and 10% fetal bovine serum for 40 min on a shaker at 215 rpm 37°C, and then incubated on ice for 10 min with 6 m FITC-conjugated, anti-DLK1 (D187-4; MBL International). Cells were magnetically separated using the EasySep FITC selection kit (STEMCELL Technologies). For RNA-seq, enriched populations were purified by fluorescence-activated cell sorting.
chIP-Seq and RNA-Seq
Perfused female adult liver or embryonic hepatoblasts were fixed prior to cell sorting in 1% formaldehyde for 10 min or 45 min (embryonic HNF4A ChIP only). ChIP was performed as described previously (Wederell et al., 2008) using 3 mg of anti-FOXA2 (Santa Cruz sc-6554), anti-HNF4A (Santa Cruz sc-8987), anti-H3K4me1 (Abeam ab8895), anti-H3K27me3 (Abcam Ab8580), anti-H3K9me3 (Abcam), or normal rabbit IgG (Santa Cruz). DNA was purified by 8% PAGE to obtain 100–300 bp fragments for library construction and sequenced on an lllumina Genome Analyzer as previously described (Robertson et al., 2007). Reads were aligned to the mouse reference genome (mm9) using ELAND and directionally extending to 200 bp in length. Clusters of overlapping extended reads were defined as enriched regions using FindPeaks2 (Fejes et al., 2008). The data sets were limited by applying thresholds as defined by an FDR cutoff of 0.01 and by trimming at 20% of maximal height to remove flanks and separate composite regions (Fejes et al., 2008). Input control samples (hepatoblasts and adult liver) were sequenced and analyzed using similar limits. Any regions of enrichment identified in input control samples were subtracted from the data sets. For RNA-seq analysis, RNA was prepared using TRIzol (Invitrogen) and sequenced on an Illumina Genome Analyzer as previously described (Griffith et al., 2010). The following NCBI data sets were used in this study: SRA008281, GSM751034, and GSM751035 (Hoffman et al., 2010; Tennant et al., 2013). Unthresholded TF chIP-seq data can be visualized in the UCSC Genome Browser using the following URLs.
Bioinformatic Analysis
Regions of enrichment (peaks) defined by chIP-seq analysis were designated as overlapping if >50 bp were shared. Data set comparisons were carried out using publically available Galaxy/Cistrome metaservers (Blankenberg et al., 2010; Goecks et al., 2010). CEAS was used to assess genomic distributions, SitePro (Shin et al., 2009) generated the composite histone modification profiles, and HNF4A and FOXA2 motif analysis was done using SeqPos (He et al., 2010). Heatmaps were generated using MeV (http://www.tm4.org/mev.html). Differential motif discovery was done using oPOSSUM (http://opossum.cisreg.ca/oPOSSUM3/) sequence-based anchored combination site analysis (Subramanian et al., 2014). Enriched motifs were identified in co-occupied embryonic sites using GC% matched co-occupied adult sites as background and an anchored HNF4A JASPAR CORE profile with a matrix match threshold of 85%. TF-binding sites were associated with target genes using previously established enhancer promoter units (Shen et al., 2012). RNA-seq analysis was carried out using the publically available ALEXA-seq pipeline (Griffith et al., 2010). Briefly, the cumulative base coverage of a feature was normalized to the feature length and library size, giving the “normalized average coverage” (NAC). Transcripts were considered differentially expressed if NAC values differed by a factor of 2 and their corrected p value was <0.05 (by Fisher's exact test). Data sets are accessible at http://www.alexaplatform.org/alexa_seq/Morgen/Summary.htm.
Cell Culture Assays
HNF4A and HNF4A DN expression vectors were provided by T. Leff (Hong et al., 2003). TEAD2 and YAP1 expression vectors were provided by J.F. Martin (Heallen et al., 2011). TEAD DN and YAP S94A expression vectors were previously described (Shen et al., 2012). Enhancer regions were PCR amplified and cloned into a PGL3 basic vector containing the E1B minimal promoter. TK-Renilla controlled for transfection efficiency. HEK293T were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Transfections were performed using PEI (Raju et al., 2013). Firefly and Renilla activity was measured with a Dual Luciferase Reporter Assay kit (Promega) and presented as the Firefly/Renilla ratio. Western blot analysis was carried out on nuclear extracts from pooled embryonic livers (E14.5) and female adult liver using anti-YAP1 (Cell Signaling), anti-TEF4 (Tead2; Novus Biologicals), and anti-beta actin (Santa Cruz). Nuclear extracts for EMSAs were prepared from 293T cells transfected with TEAD-myc/HNF4-myc or E14.5 as previously described (Labbé et al., 1998).
Supplementary Material
Highlights.
HNF4A and F0XA2 bind to the genome in a differentiation-dependent manner
Temporal enhancers regulate global gene expression during in vivo differentiation
Polycomb-mediated repression is not evident at temporal enhancers
TEAD2 and YAP1 enhance expression of embryonic target genes
Acknowledgments
The authors thank Amanda Kotzer for project management. Funding was provided by Genome Canada, Genome BC, and the Canadian Institutes for Health Research (FRN259575), with infrastructure support provided by the BC Cancer Foundation. Q.C. is a recipient of a Breast Cancer Research Postdoctoral Fellowship from the Department of Defense (BC093902). D.P. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Accession Numbers: The NCBI GEO accession number for the data reported in this paper is GSE54520.
Supplemental Information: Supplemental Information includes six figures and two tables and can be found with this article online at http://dx.doi.Org/10.1016/j.celrep.2014.08.046.
Author Contributions: O.A. and P.A.H. designed the study. R.C. and S.L. prepared samples for ChIP and RNA-seq. A.C.K., W.W., and Y.Y. assisted with vector construction and luciferase assays. V.C.G. assisted with animal breeding and provided critical discussion. M.B., A.S.M., M.G., and G.A.R. mapped reads and called peaks for ChIP and transcript levels for RNA-seq libraries. Y.J. conducted library construction. D.P. and Q.C. provided YAP-TG mice. S.J.M.J., M.A.M., and P.A.H. provided essential resources. O.A. conducted all other experimental procedures, performed data analysis, and prepared the manuscript in partnership with P.A.H.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. The Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle MA, Konopka G, Parviz F, Gaggl AL, Yang C, Sladek FM, Duncan SA. Hepatocyte nuclear factor 4alpha orchestrates expression of cell adhesion proteins during the epithelial transformation of the developing liver. Proc Natl Acad Sci USA. 2006;103:8419–8424. doi: 10.1073/pnas.0600246103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. 2010;Chapter 19 doi: 10.1002/0471142727.mb1910s89. Unit 19.10.1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkis IM, Schug J, Rubins NE, Chopra AR, O'Malley BW, Kaestner KH. Foxa2-dependent hepatic gene regulatory networks depend on physiological state. Physiol Genomics. 2009;38:186–195. doi: 10.1152/physiolgenomics.90376.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buecker C, Srinivasan R, Wu Z, Calo E, Acampora D, Faial T, Simeone A, Tan M, Swigut T, Wysocka J. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Wang J, Jia J, Song N, Xiang C, Xu J, Hou Z, Su X, Liu B, Jiang T, et al. Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell Stem Cell. 2014;14:394–403. doi: 10.1016/j.stem.2014.01.008. [DOI] [PubMed] [Google Scholar]
- Duncan SA, Nagy A, Chan W. Murine gastrulation requires HNF-4 regulated gene expression in the visceral endoderm: tetraploid rescue of Hnf-4(-/-) embryos. Development. 1997;124:279–287. doi: 10.1242/dev.124.2.279. [DOI] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejes AP, Robertson G, Bilenky M, Varhol R, Bainbridge M, Jones SJ. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 2008;24:1729–1730. doi: 10.1093/bioinformatics/btn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godbout R, Ingram RS, Tilghman SM. Fine-structure mapping of the three mouse alpha-fetoprotein gene enhancers. Mol Cell Biol. 1988;8:1169–1178. doi: 10.1128/mcb.8.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J Galaxy Team. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith M, Griffith OL, Mwenifumbo J, Goya R, Morrissy AS, Morin RD, Corbett R, Tang MJ, Hou YC, Pugh TJ, et al. Alternative expression analysis by RNA sequencing. Nat Methods. 2010;7:843–847. doi: 10.1038/nmeth.1503. [DOI] [PubMed] [Google Scholar]
- Grozdanov PN, Yovchev MI, Dabeva MD. The oncofetal protein glypican-3 is a novel marker of hepatic progenitor/oval cells. Lab Invest. 2006;86:1272–1284. doi: 10.1038/labinvest.3700479. [DOI] [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, Martin JF. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman BG, Robertson G, Zavaglia B, Beach M, Cullum R, Lee S, Soukhatcheva G, Li L, Wederell ED, Thiessen N, et al. Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4A-, and PDX1-bound loci in islets and liver. Genome Res. 2010;20:1037–1051. doi: 10.1101/gr.104356.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YH, Varanasi US, Yang W, Left T. AMP-activated protein kinase regulates HNF4alpha transcriptional activity by inhibiting dimer formation and decreasing protein stability. J Biol Chem. 2003;278:27495–27501. doi: 10.1074/jbc.M304112200. [DOI] [PubMed] [Google Scholar]
- Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- Huang P, Zhang L, Gao Y, He Z, Yao D, Wu Z, Cen J, Chen X, Liu C, Hu Y, et al. Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell. 2014;14:370–384. doi: 10.1016/j.stem.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Kyrmizi I, Hatzis P, Katrakili N, Tranche F, Gonzalez FJ, Talianidis I. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev. 2006;20:2293–2305. doi: 10.1101/gad.390906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé E, Silvestri C, Hoodless PA, Wrana JL, Attisano L. Smad2 and Smad3 positively and negatively regulate TGF beta-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- Li Z, Tuteja G, Schug J, Kaestner KH. Foxal and Foxa2 are essential for sexual dimorphism in liver cancer. Cell. 2012;148:72–83. doi: 10.1016/j.cell.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T, Kamiya A, Kakinuma S, Zeniya M, Nishinakamura R, Tajiri H, Nakauchi H. Sall4 regulates cell fate decision in fetal hepatic stem/progenitor cells. Gastroenterology. 2009;136:1000–1011. doi: 10.1053/j.gastro.2008.11.018. [DOI] [PubMed] [Google Scholar]
- Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju R, Chau D, Verfaillie CM, Hu WS. The road to regenerative liver therapies: the triumphs, trials and tribulations. Biotechnol Adv. 2013;31:1085–1093. doi: 10.1016/j.biotechadv.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, Brummelkamp TR, Camargo FD. Yap1 acts downstream of a-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- Sekiya T, Muthurajan UM, Luger K, Tulin AV, Zaret KS. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 2009;23:804–809. doi: 10.1101/gad.1775509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Septer S, Edwards G, Gunewardena S, Wolfe A, Li H, Daniel J, Apte U. Yes-associated protein is involved in proliferation and differentiation during postnatal liver development. Am J Physiol Gastrointest Liver Physiol. 2012;302:G493–G503. doi: 10.1152/ajpgi.00056.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, Ren B. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Liu T, Manrai AK, Liu XS. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- Simeonov KP, Uppal H. Direct reprogramming of human fibroblasts to hepatocyte-like cells by synthetic modified mRNAs. PLoS ONE. 2014;9:e100134. doi: 10.1371/journal.pone.0100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimani VD, Yin H, Jahani-Asl A, Ming H, Kockx CE, van ljcken WF, Grosveld F, Rudnicki MA. Snail regulates MyoD binding-site occupancy to direct enhancer switching and differentiation-specific transcription in myogenesis. Mol Cell. 2012;47:457–468. doi: 10.1016/j.molcel.2012.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian K, Owens DJ, Raju R, Firpo M, O'Brien TD, Verfaillie CM, Hu WS. Spheroid culture for enhanced differentiation of human embryonic stem cells to hepatocyte-like cells. Stem Cells Dev. 2014;23:124–131. doi: 10.1089/scd.2013.0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimizu N, Nishikawa M, Saito H, Tsujimura T, Miyajima A. Isolation of hepatoblasts based on the expression of Dlk/Pref-1. J Cell Sci. 2003;116:1775–1786. doi: 10.1242/jcs.00388. [DOI] [PubMed] [Google Scholar]
- Tennant BR, Robertson AG, Kramer M, Li L, Zhang X, Beach M, Thiessen N, Chiu R, Mungall K, Whiting CJ, et al. Identification and analysis of murine pancreatic islet enhancers. Diabetologia. 2013;56:542–552. doi: 10.1007/s00125-012-2797-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Padilla ME, Fougere-Deschatrette C, Weiss MC. Expression of HNF4alpha isoforms in mouse liver development is regulated by sequential promoter usage and constitutive 3° end splicing. Mech Dev. 2001;109:183–193. doi: 10.1016/s0925-4773(01)00521-4. [DOI] [PubMed] [Google Scholar]
- Wederell ED, Bilenky M, Cullum R, Thiessen N, Dagpinar M, Delaney A, Varhol R, Zhao Y, Zeng T, Bernier B, et al. Global analysis of in vivo Foxa2-binding sites in mouse adult liver using massively parallel sequencing. Nucleic Acids Res. 2008;36:4549–4564. doi: 10.1093/nar/gkn382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Hou J, Alder O, Ye X, Lee S, Cullum R, Chu A, Zhao Y, Warner SM, Knight DA, et al. Genome-wide microRNA and messenger RNA profiling in rodent liver development implicates mir302b and mir20a in repressing transforming growth factor-beta signaling. Hepatology. 2013;57:2491–2501. doi: 10.1002/hep.26252. [DOI] [PubMed] [Google Scholar]
- Yimlamai D, Christodoulou C, Galli GG, Yanger K, Pepe-Mooney B, Gurung B, Shrestha K, Cahan P, Stanger BZ, Camargo FD. Hippo pathway activity influences liver cell fate. Cell. 2014;757:1324–1338. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong KJ, Gao C, Lim JS, Yan B, Yang H, Dimitrov T, Kawasaki A, Ong CW, Wong KF, Lee S, et al. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med. 2013;368:2266–2276. doi: 10.1056/NEJMoa1300297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;725:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


