Abstract
Expression of fusion proteins such as MBP fusions can be used as a way to improve the solubility of the expressed protein in E. coli (Fox and Waugh, 2003; Nallamsetty et al., 2005; Nallamsetty and Waugh, 2006) and as a way to introduce an affinity purification tag. The protocol that follows was designed by the authors as a first step in the purification of a recombinant protein fused with MBP, using fast protein liquid chromatography (FPLC). Cells should have been thawed, resuspended in binding buffer, and lysed by sonication or microfluidization before mixing with the amylose resin or loading on the column. Slight modifications to this protocol may be made to accommodate both the protein of interest and the availability of equipment.
1. Theory
Affinity chromatography separates proteins on the basis of an interaction between a protein and a specific ligand. The binding of the protein to a ligand attached to a matrix is reversed by either competition or by decreasing the affinity with pH and/or ionic strength. Affinity chromatography is an ideal first purification step due to its selectivity and high capacity. It exploits biological protein functions such as antibody–antigen recognition (e.g., protein A), lectin–polysaccharide binding, nucleic acid–heparin interactions, and recombinant fusion tags of proteins, such as maltose-binding protein and gluthathione S-transferase (see Purification of GST-tagged proteins), or metal chelators such as 6× histidine tags (see Purification of His-tagged proteins). Affinity chromatography can also be used as a way to remove serine proteases such as thrombin and Factor X, utilizing their affinity for benzamidine sepharose (see other methods for affinity purification of proteins on Hydroxyapatite Chromatography: Purification Strategies for Recombinant Proteins, Protein Affinity Purification using Intein/Chitin Binding Protein Tags, Immunoaffinity purification of proteins or Strep-tagged protein purification).
Affinity chromatography of MBP fusion proteins can be performed on an FPLC system with an amylose column or batch-wise using amylose agarose resin, followed by a step elution (Fig. 1). The purification of an MBP-fusion protein exploits the natural affinity of MBP for α-(1–4) maltodextrin. However, the MBP fusion tag should be considered mainly as a way to improve the solubility of a protein rather than as an affinity tag for the column to ease purification (for other ways to improve solubility, see Explanatory Chapter: Troubleshooting protein expression: what to do when the protein is not soluble). Only two buffers are required: a binding buffer and an elution buffer. They only differ in the presence of 10 mM maltose in the latter.
Figure 1.
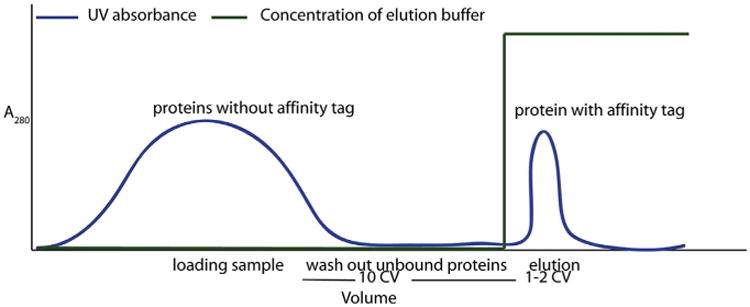
Chromatograph of a purification of an MBP-tagged fusion protein using amylose beads.
Many vectors are available to construct fusion proteins with MBP. Some of them target the fusion protein to the cytoplasm and others to the periplasmic space. Usually, higher yields of expression have been achieved in the cytoplasm than in the periplasm. Many of these vectors are available with a variety of sites for protease cleavage and may be obtained through academic laboratories (Fox and Waugh, 2003; Kapust and Waugh, 1999; Geisbrecht et al., 1998) and commercial vendors (e.g., New England Biolabs and Invitrogen).
2. Equipment
FPLC (capable of reading UV absorbance at 280 nm)
Amylose column (or amylose resin)
Sidearm filtering flask
Filter holder
0.22 μm syringe filters
0.22 μm filters (for vacuum filtration assembly)
3. Materials
Tris base
Hydrochloric acid (HCl)
Sodium chloride (NaCl)
Dithiothreitol (DTT)
EDTA
Amylose resin (or amylose column)
3.1. Solutions & buffers
Step 1 Binding buffer.
| Component | Final concentration | Stock | Amount |
|---|---|---|---|
| Tris–HCl, pH 8.0 | 50 mM | 1 M | 50 ml |
| NaCl | 200 mM | 1 M | 200 ml |
| EDTA | 1 mM | 0.5 M | 2 ml |
Add water to 1 l and pass through a 0.22 μm filter
Optional: 0.1-mM Na azide, or reducing agents such as 1 mM DTT or β-mercaptoethanol can be added
Step 4 Elution buffer.
| Component | Final concentration | Stock | Amount |
|---|---|---|---|
| Tris–HCl, pH 8.0 | 50 mM | 1 M | 50 ml |
| NaCl | 200 mM | 1 M | 200 ml |
| EDTA | 1 mM | 0.5 M | 2 ml |
| Maltose | 10 mM | 0.5 M | 20 ml |
Add water to 1 l and pass through a 0.22 μm filter
Any reducing agents used in the binding buffer should also be used in theelution buffer
Tip Other buffers such as HEPES or MOPS are well tolerated. Buffers that contain concentrations of up to 10% glycerol, 5% ethanol, or acetonitrile are compatible with the beads
Tip EDTA should be removed by dialysis before cleaving with a protease that requires a divalent cation for activity (e.g., Ca+2)
4. Protocol
4.1. Duration
| Preparation | Varies |
| Protocol | About 2 h |
4.2. Preparation
Grow bacteria and induce for expression of the MBP fusion protein according to the expression system used (see Small-scale Expression of Proteins in E. coli). Harvest the cells by centrifugation and freeze the cell pellet at −80 °C (optional). Resuspend the cell pellet in about 10-ml binding buffer per gram of cells, lyse the cells (e.g., by sonication or French press), and centrifuge at 9000 × g to remove the cell debris. Keep in mind that typical binding capacities for amylose agarose columns are 3 mg of protein per milliliter of column resin.
Determine the maximum and optimal flow rates for the column and the FPLC.
Prepare the collection tubes to save fractions from Steps 2–4 for further analysis.
See Fig. 2 for the flowchart of the complete protocol.
Figure 2.
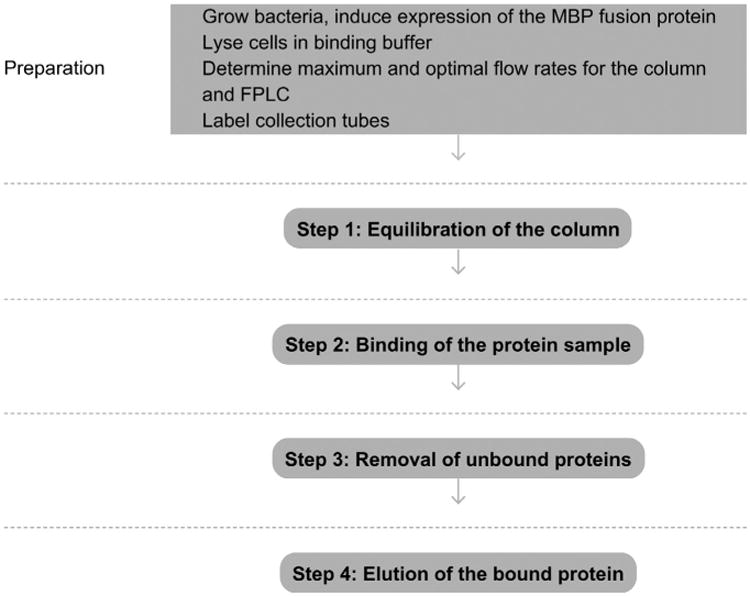
Flowchart of the complete protocol, including preparation.
5. Step 1 Equilibration of the Column
5.1. Overview
The column will be prepared for the binding of the protein.
5.2. Duration
15 min
1.1 Follow the manufacturer's instructions for removing the storage buffer of the column and placing the column in water.
1.2 Equilibrate the column by running at least 8–10 column volumes of binding buffer over the beads. Check whether the UV absorbance reading (at 280 nm) is stable before proceeding.
1.3 Set the UV absorbance reading to zero.
5.3. Tip
If the protein of interest does not contain tryptophan, it will not absorb appreciably at 280 nm. Choose another wavelength, such as 260 nm for tyrosine, to monitor the protein.
5.4. Tip
If the UV absorbance reading has many spikes or is unusually high and DTT is present in the equilibration buffer, this may indicate that the DTT is oxidized. If DTT is used, it should be added immediately before purification. Do not use buffers containing DTT that are more than 1 day old.
See Fig. 3 for the flowchart of Step 1.
Figure 3.
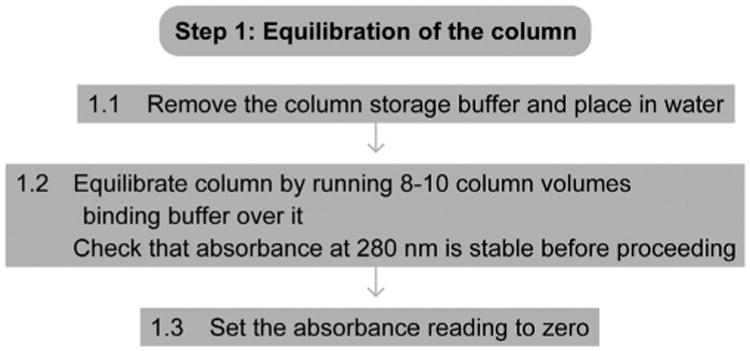
Flowchart of Step 1.
6. Step 2 Binding of the Protein Sample
6.1. Overview
The protein will be applied to the column.
6.2. Duration
30 min
2.1 Filter the protein through a 0.22-μm syringe filter.
2.2 Run the protein sample over the column.
6.3. Tip
If the protein sample contains many aggregates, the syringe filter may clog during filtering. If this occurs, simply use multiple filters to filter the sample.
6.4. Tip
If the binding between amylose and the MBP fusion protein is weak, consider alternative constructs with additional tags for affinity purification (see Purification of GST-tagged proteins, Purification of His-tagged proteins, Protein Affinity Purification using Intein/Chitin Binding Protein Tags, Strep-tagged protein purification).
See Fig. 4 for the flowchart of Steps 2 and 3.
Figure 4.
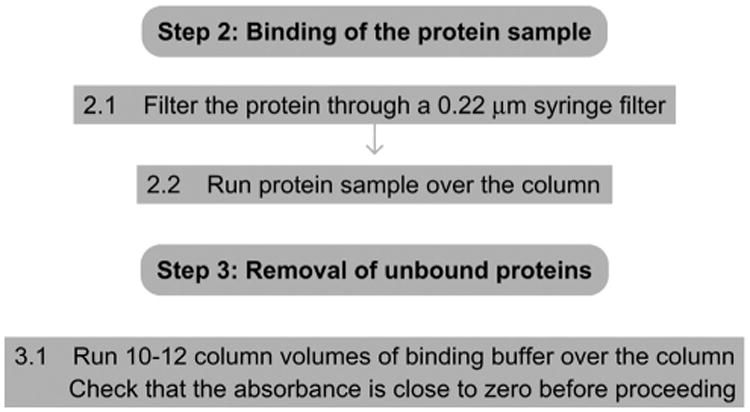
Flowchart of Steps 2 and 3.
7. Step 3 Removal of Unbound Proteins
7.1. Overview
Any remaining weakly interacting or nonbinding proteins will be removed.
7.2. Duration
10 min
3.1 Run at least 10–12 column volumes of binding buffer over the beads. Check whether the UV absorbance is zero or close to zero before proceeding.
8. Step 4 Elution of the Bound Protein
8.1. Overview
An isocratic elution using elution buffer (containing 10 mM maltose) will be performed to elute proteins with MBP tags.
8.2. Duration
1h
4.1 Run 2-column volumes of elution buffer to elute the protein of interest. Collect fractions from the column.
4.2 Regenerate the beads according to the manufacturer's protocol.
4.3 Analyze fractions (loaded sample, flow through, washes, eluted protein) by SDS-PAGE (see One-dimensional SDS-Polyacrylamide Gel Electrophoresis (1D SDS-PAGE)). Pool peak fractions containing the purified protein.
4.4 Dialyze pooled fractions in the buffer required for protease cleavage.
4.5 If the fusion protein will be stored at −80 °C, either flash-freeze in liquid nitrogen and/or add 10% glycerol.
8.3. Tip
Most MBP fusion proteins elute in the first column volume; the suggested size of the fractions collected is 1/5 of the column volume.
8.4. Tip
To improve binding of the fusion protein to the amylase resin, consider removing all low-ionic detergents such as Triton X-100 and Tween-20. These have been shown to intefere with binding in some cases (see also Explanatory Chapter: Choosing the right detergent).
8.5. Tip
The affinity of amylose for MBP fusion constructs requires that the MBP portion of the protein be properly folded. The presence of denaturants such as urea and guanidinium chloride abolishes binding since they unfold MBP.
8.6. Tip
If the MBP tag will be cleaved (see Proteolytic affinity tag cleavage), do not subject the cleavage reaction to the column to separate the cleaved and the uncleaved proteins. Residual maltose will interfere with binding. Ion exchange chromatography (see Using ion exchange chromatography to purify a recombinantly expressed protein) is the preferred method to separate the cleaved and uncleaved species. E. coli MBP has a pI of 4.9 and binds efficiently to anion exchange resins at pH values higher than its pI. It has been observed that some proteins precipitate after being cleaved from MBP.
8.7. Tip
If the MBP fusion protein is insoluble, try expressing it at a lower temperature (e.g., 30 °C or 15 °C. See also Explanatory Chapter: Troubleshooting protein expression: what to do when the protein is not soluble).
8.8. Tip
Some, but not all, proteins require the addition of glycerol for freezing.
See Fig. 5 for the flowchart of Step 4.
Figure 5.
Flowchart of Step 4.
Footnotes
Referenced Protocols in Methods Navigator: Purification of GST-tagged proteins.
Purification of His-tagged proteins.
Hydroxyapatite Chromatography: Purification Strategies for Recombinant Proteins.
Protein Affinity Purification using Intein/Chitin Binding Protein Tags.
Immunoaffinity purification of proteins.
Strep-tagged protein purification.
Explanatory Chapter: Troubleshooting protein expression: what to do when the protein is not soluble.
Small-scale Expression of Proteins in E. coli.
One-dimensional SDS-Polyacrylamide Gel Electrophoresis (1D SDS-PAGE).
Explanatory Chapter: Choosing the right detergent.
Proteolytic affinity tag cleavage.
Using ion exchange chromatography to purify a recombinantly expressed protein.
Referenced Literature
- Fox JD, Waugh DS. Maltose-binding protein as a solubility enhancer. Methods in Molecular Biology. 2003;205:99–117. doi: 10.1385/1-59259-301-1:99. [DOI] [PubMed] [Google Scholar]
- Nallamsetty S, Austin BP, Penrose KJ, Waugh DS. Gateway vectors for the production of combinatorially-tagged His6-MBP fusion proteins in the cytoplasm and periplasm of Escherichia coli. Protein Science. 2005;14:2964–2971. doi: 10.1110/ps.051718605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallamsetty S, Waugh DS. Solubility-enhancing proteins MBP and NusA play a passive role in the folding of their fusion partners. Protein Expression and Purificatin. 2006;45:175–182. doi: 10.1016/j.pep.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Kapust RB, Waugh DS. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Science. 1999;8:1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbrecht BV, et al. Molecular characterization of Saccharomyces cerevisiae Delta3, Delta2-enoyl-CoA isomerase. Journal of Biological Chemistry. 1998;273:33184–33191. doi: 10.1074/jbc.273.50.33184. [DOI] [PubMed] [Google Scholar]


