Abstract
Genome integrity is fundamental for cell survival and cell cycle progression. Important mechanisms for keeping the genome intact are proper sister chromatid segregation, correct gene regulation and efficient repair of damaged DNA. Cohesin and its DNA loader, the Scc2/4 complex have been implicated in all these cellular actions. The gene regulation role has been described in several organisms. In yeast it has been suggested that the proteins in the cohesin network would effect transcription based on its role as insulator. More recently, data are emerging indicating direct roles for gene regulation also in yeast. Here we extend these studies by investigating whether the cohesin loader Scc2 is involved in regulation of gene expression. We performed global gene expression profiling in the absence and presence of DNA damage, in wild type and Scc2 deficient G2/M arrested cells, when it is known that Scc2 is important for DNA double strand break repair and formation of damage induced cohesion. We found that not only the DNA damage specific transcriptional response is distorted after inactivation of Scc2 but also the overall transcription profile. Interestingly, these alterations did not correlate with changes in cohesin binding.
Keywords: cohesin network, DNA double strand break, microarray, Scc2, transcription profile
Abbreviations
- DI-cohesion
damage induced cohesion
- DSB DNA
double strand break
- FDR
false discovery rate
- HO
homothallic switching endonuclease
- S
supplementary
- SCC
sister chromatid cohesion
- SMC
structural maintenance of chromosomes
- WT
wild type
Introduction
Preserved genome integrity is fundamental for cell survival and cell cycle progression. One important mechanism for keeping the genome intact is proper segregation of sister chromatids, an event that requires the cohesin complex, which holds the sister chromatids together from the time of their formation during replication until separation at anaphase.1,2 Cohesin, together with condensin and the Smc5/6 complex, form a family of large multi-subunit complexes built from Structural Maintenance of Chromosome (SMC) proteins.1,2 The cohesin complex consists of Smc1 and Smc3, as well as the non-SMC proteins Mcd1 (also called Scc1) and Scc3 (also called Irr1), and forms a ring-like structure.1,2 In addition, the accessory proteins Pds5 and Wpl1 have in Saccharomyces cerevisiae been shown to interact with the complex via Scc3 and be important for cohesion maintenance.3 DNA loading of cohesin, that happens prior to DNA replication in all organisms analyzed, depends on the protein complex Scc2/4 (human NIPBL/MAU2).4-8 Cohesion is then established during S phase, in strong connection to replication and depending on acetylation of Smc3 by the acetyltransferase Eco1.9-11 During the G2/M phase cohesion is maintained, to finally be dissolved at anaphase through cleavage of Mcd1 by separase.1
Two fractions of cohesin exist in the cell, one that is stably bound, and one that is constantly moving on and off the chromosomes.12,13 After S phase has been completed the latter will not become cohesive unless DNA is damaged. This reestablishment of cohesion in response to DNA damage in G2 is called Damage Induced (DI)-cohesion and like S phase cohesion it depends on Scc2 and Eco1 for loading and establishment.14,15 DI-cohesion is formed both proximal to the DNA double strand break (DSB) and throughout the genome.14,15
Cohesin and Scc2/4 are in addition to their role in chromosome segregation, essential for correct DNA repair as well as DNA damage checkpoint activation and have also been shown to influence gene expression.16 This was first described in Drosophila melanogaster where Nipped B, the fly homolog of yeast Scc2, was found to be involved in long-range enhancer-promoter communications.17 In Caenorhabditis elegans and Xenopus laevis, MAU2 the homolog of yeast Scc4, has been implicated in neuronal development.18 In mammalian cells cohesin has been shown to frequently bind to the same sites as the chromatin insulator protein CCCTC-binding factor (CTCF), where it is important for its function as a chromatin insulator.19 This is potentially of medical importance since heterozygous loss-of-function mutations in NIPBL, the human Scc2 homolog, has been found to be the major cause of the development syndrome Cornelia de Lange syndrome (CdLS).20 Since CdLS is characterized by developmental rather than chromosome segregation defects, it has been suggested to be caused by transcriptional dysfunctions, possibly due to alterations in cohesin chromatin binding dynamics.21 Both cell lines from CdLS patients and cells from a mouse CdLS model (Nipbl+/−) display altered transcription profiles compared with control cells.21,22 In S. cerevisiae, cohesin was initially suggested to influence gene expression through control of their nuclear position.16 Thus, cohesin in yeast was believed to function as a boundary element at the silent loci of HMR, involved in mating type switching.23 Recent evidence is now expanding the gene regulation function of cohesin also in yeast. Inactivation of Mcd1 in the G1 phase of the cell cycle caused altered expression of a number of genes with related function in a coordinated fashion.24 Furthermore, proteins in the cohesin network were reported to regulate gene expression during yeast meiosis,25,26 and Smc1 and Eco1 were shown to also promote rRNA production.24,27
Here, we set out to further investigate whether inactivation of Scc2 will influence the transcription program also in yeast, using a temperature sensitive SCC2 allele (scc2–4) under conditions known to inactivate both sister chromatid cohesion and DNA repair.14,28 We chose to study the effect of transiently disabling Scc2 in G2/M arrested cells when cohesin and its loader are important for homologous recombination based DSB repair and DI-cohesion. In this fashion we also avoid the alteration in transcription caused by inactivation of cohesin loading during G1 and establishment of cohesion in S phase. We investigated the gene transcription profiles in wild type (WT) and Scc2 deficient cells in the absence and presence of DNA damage and conclude that Scc2 is indeed instrumental for correct gene expression also in budding yeast, both globally and in response to a single DSB. Interestingly, these alterations did not correlate with changes in cohesin binding.
Results
Gene expression profiling of wild type and Scc2 deficient cells in S. cerevisiae
To compare the gene expression profiles of WT and Scc2 deficient cells in the absence and presence of DNA damage we employed 2 sets of S. cerevisiae strains. The first set consisted of one WT strain and one harboring the temperature sensitive allele for SCC2 (scc2–4). In the second set both the WT and the scc2–4 strain also carried a single recognition site for the yeast HO (HOmothallic switching endonuclease) enzyme, allowing induction of a single specific DSB at chosen position in the genome. All strains are otherwise genetically identical, and contain the HO endonuclease under control of the inducible galactose promoter (pGAL-HO). The HO activity normally promotes mating type switching by cleavage of the MAT locus on Chr. III. Here we introduced the HO recognition site centrally on the right arm of Chr. VI. The endogenous HO cleavage site at the MAT locus was deleted to avoid the specific gene expression program it activates.29 The experimental procedure is depicted in Figure 1A. In short, log phase yeast cells were arrested in G2/M. The temperature was raised and 30 minutes later galactose was added to induce one DSB in cells containing the recognition site for the HO enzyme. Ninety minutes later, cells were harvested, RNA isolated and cDNA synthesized for analysis of global gene expression by GeneChip Yeast genome arrays (5841 transcripts/array). Induction of the DSB was confirmed by pulse field gel electrophoresis and the G2 arrest by FACS (Fig. 1B and C). This experimental setup facilitates 4 possible comparisons: scc2–4 versus WT cells in absence and in the presence of break, WT cells in the presence vs. absence of break, as well as scc2–4 cells in the presence compared to absence of break.
Figure 1.
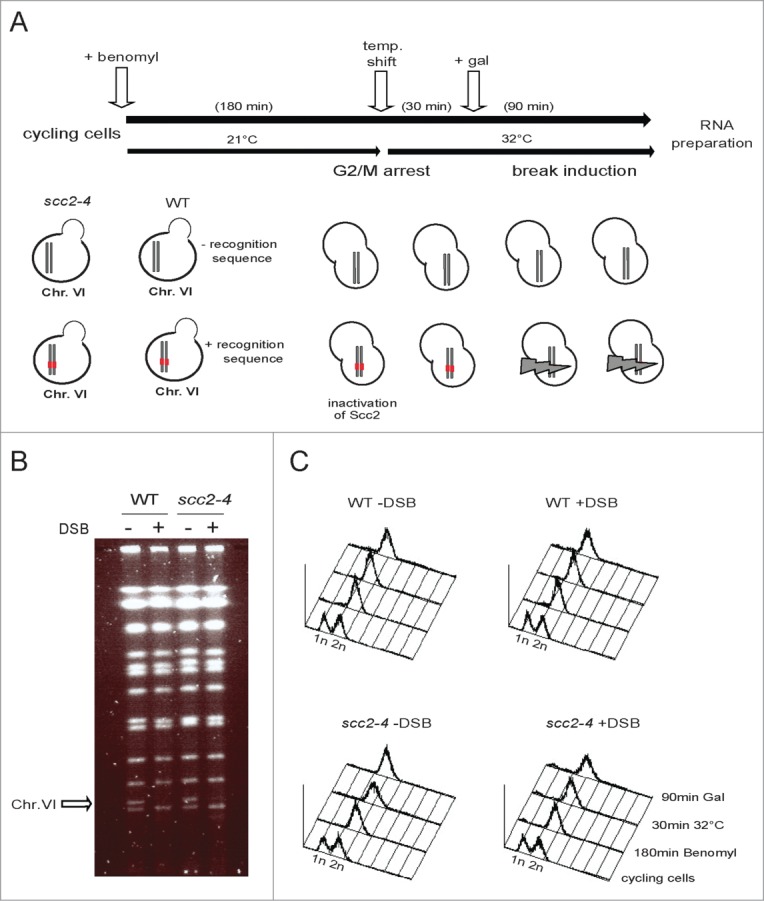
Experimental set up. (A) Schematic illustration of the experimental system used throughout this study. Pairs of S. cerevisiae strains, either WT or harboring the scc2–4 ts allele, genetically identical in all other aspects except for the presence or absence of the recognition site for the HO enzyme, were grown in YEP media supplemented with 2% raffinose at 21°C, and arrested in G2/M. A temperature raise to 32°C for 30 minutes, renders Scc2 dysfunctional before galactose addition, to induce one DSB or not. After 90 minutes break induction, cells were collected, total RNA prepared, cDNA synthesized and fragmented before hybridization to GeneChip Yeast genome 2.0 Array. (B) Pulse-field gel electrophoresis (PFGE) for verification of pGAL-HO break induction. The arrow points at Chr. VI. (C) FACS profiles of indicated yeast strains at indicated time points.
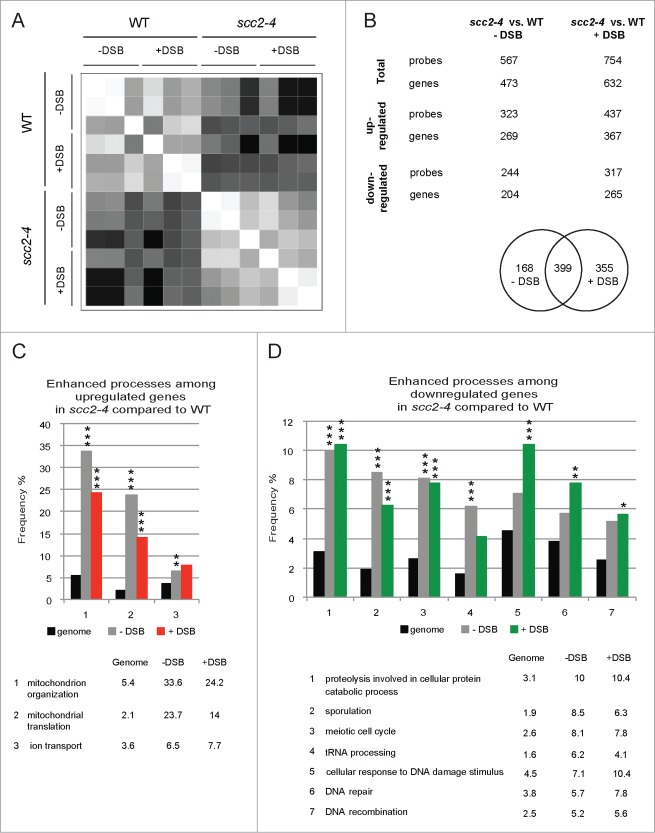
Samples were collected and prepared as described (Fig. 1). For comparisons between the different conditions gene expression data were pre-processed using limma and the rma (Robust Multichip Average) procedure.30-32 Samples were tested for differential expression (FDR ≤ 0.05) between different strains, and within strains but between absence and presence of DSB. A correlation graph is presented in Figure 2A where the strongest correlation is depicted white and the weakest black. The best correlations were found within the groups of either WT or scc2–4 cell, indicating that the effect of the induced DSB within the same cell type was diluted in the much larger lack of correlation between WT and scc2–4 cells (Fig. 2A).
Figure 2.
Inactivation of Scc2 causes global changes in gene expression. (A) Correlation plot illustrating correlations between WT and Scc2 deficient cells and induction of DSB. White color represents perfect correlation (ρ = 1) and black represents no correlation (ρ=0). Higher correlation is seen within each cell type, independent of DSB induction. (B) Summary of the number of probes/genes significantly affected when comparing WT and scc2–4 cells in the absence and presence of DSB. (C and D) Bioinformatic analysis using SGD GO slim mapping. Up- and downregulated genes were sorted according to biological process. Processes were regarded as significantly enhanced if FDR ≤ 0.05 (*, P ≤ 0.05, **, P ≤ 0.01, ***, P ≤ 0.005). (C) Biological processes of upregulated genes in Scc2 deficient cells compared to WT. (D) Biological processes of downregulated genes in Scc2 deficient cells compared to WT.
Since the most prominent effect on gene expression appeared to depend on lack of functional Scc2 we initially focused on comparing the transcription profiles of scc2–4 and WT cells. Among the 5841 open reading frames (transcripts) examined, 567 probe sets, corresponding to 473 genes, were significantly affected in the absence of break. 57% of these genes were upregulated and 43% downregulated (Fig. 2B; data set S1). In the presence of break 754 probe sets, corresponding to 632 genes, were differentially expressed when comparing scc2–4 cells versus WT cells and of these 58% were up- and 42% downregulated (Fig. 2B; data set S2). Common for both conditions were 399 probe sets that were differentially expressed in Scc2 deficient cells compared with WT, leaving 168 probe sets uniquely affected in the absence (data set S3) and 355 in the presence of break (data set S4).
The data sets obtained when comparing scc2–4 cells to WT cells, consisting of 473 genes differentially expressed in the absence of break, and 632 in the presence of break, were analyzed using Saccharomyces Genome Database Gene Ontology (SGD GO) slim mapping according to biological process.33 The genes where divided into up- and downregulated genes (Tables S2–S5) and processes that showed a significant enhancement compared with genome frequency (FDR ≤ 0.05) were considered further. As seen in Figure 2C and D, most of the genes with altered expression in scc2–4 cells compared to WT cells, are involved in processes that are affected independently of break induction, and presumably instead depend on the inactivation of Scc2, also apparent in the correlation graph (Fig. 2A). We found that a majority of the genes upregulated after inactivation of Scc2, in the presence or absence of break, represented mitochondrial processes. In contrast, among the downregulated genes, the processes DNA damage, DNA repair and DNA recombination were significantly enhanced in the presence of DSB compared to the genome frequency. This suggested that in the absence of functional Scc2 also the transcriptional response to DNA damage is abrogated (Fig. 2D). An additional indication for this was that several known DNA damage response genes, such as HUG1, PLM2, MAG1, DUN1 and RNR1 were upregulated in WT compared to scc2–4 cells, when analyzing the 355 probe sets uniquely affected in the presence of DSB (data set S4).
In response to DSB in WT or Scc2 deficient cells the statistical analysis showed that only 5 and 7 probe sets were affected respectively (data set S5 and S6). This most likely depends on the experiment being designed for examination of the general transcriptional difference between WT and Scc2 deficient cells. Thus, to understand the effect on the transcriptional response to DSB in the absence of functional Scc2 this had to be studied separately.
Transcriptional alterations induced by a single DSB
Several of the results from our initial experiment indicated that Scc2 would be important also for the transcriptional response induced by DSB formation. First, in the absence of DSB 473 genes were affected, while 632 genes were significantly affected in the presence of DSB, when comparing WT and Scc2 deficient cells. Second, in the group of downregulated genes comparing scc2–4 vs. WT cells in presence of DSB, multiple genes were sorted into processes such as cellular responses to DNA damage, DNA repair and DNA recombination (Fig. 2B and D). Lastly, in the WT compared with the scc2–4 cell population, multiple upregulated DNA damage response genes were found among the probe sets exclusively affected in the presence of DSB (data set S4).
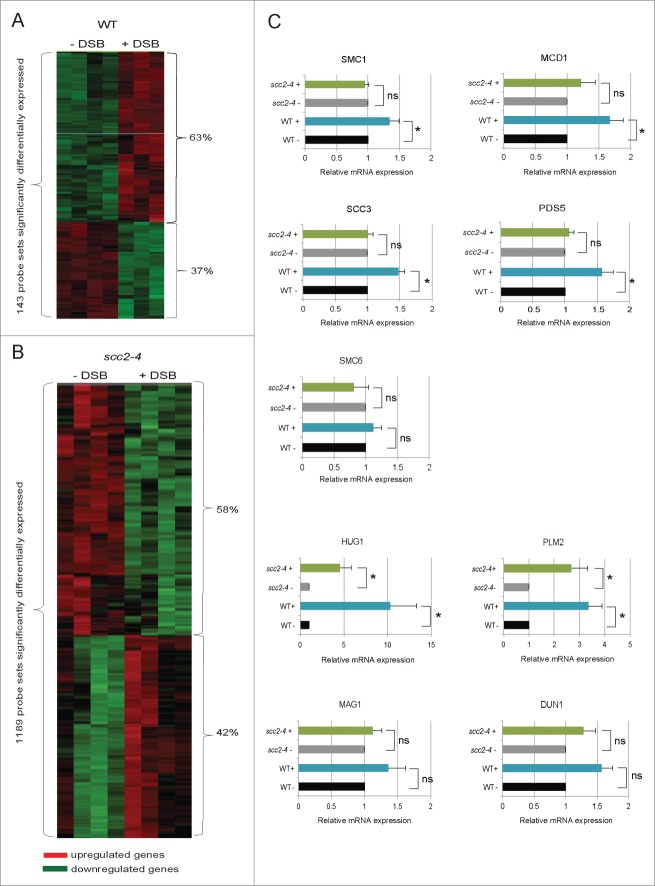
To be able to determine the transcriptional response caused by induction of a single DSB break in the absence of functional Scc2, we first tested if a single DSB would induce a typical transcriptional DNA damage response, analogous to what has been reported for IR, UV, MMS, HU, and 4-nitroquinone in WT cells.29,34,35 The same experimental procedure as described in Figure 1A was employed for WT cells alone. Among the 5841 open reading frames examined, 143 probe sets, corresponding to 113 genes, displayed significant differences in expression between absence and presence of one DSB with the false discovery rate, FDR ≤ 0.05 (data set S7). Of these were 63% up- and 37% downregulated (Fig. 3A; data set S7). A number of genes have previously been shown to be transcriptionally activated in response to different types of DNA damaging agents and function as part of the checkpoint response.29,34,35 We found that the majority of these genes were also induced in response to a single DSB on Chr. VI, namely HUG1, RNR1, RNR2, RNR3, RNR4, DIN7, DUN1, MAG1, RAD16, PLM2, RAD54 (data set S7). In addition, SMC1, MCD1 and SCC3, 3 out of the 4 genes encoding subunits of the core cohesin complex, as well as the cohesin accessory protein PDS5, were upregulated. Interestingly the same was true for SMC6, one of the subunits in the Smc5/6 complex that similar to cohesin has been shown to have DNA repair functions (data set S7).1,2 It has previously been reported that 2 clusters of genes, one induced and one repressed, are transcriptionally regulated in response to a range of different types of stressful conditions, including DNA damage. In this stereotyped response, called the environmental stress response (ESR), the repressed genes mainly encode ribosomal proteins, as well as genes involved in RNA metabolism, protein synthesis and different aspects of cell growth.36 This set of genes was found also here in the group of genes most evidently downregulated in response to DSB in WT cells (data set S5).
Figure 3.
For figure legend, see page 3651.Figure 3 (See previous page). Transcriptional alterations induced by a single DSB on Chr. VI. (A) Heatmap showing the 143 probe sets transcriptionally altered in response to a single DSB on Chr. VI (FDR ≤ 0.05) in WT cells. (B) Heatmap showing the 1189 probe sets transcriptionally altered in response to a single DSB on Chr. VI after inactivation of Scc2 (FDR ≤ 0.05). (A and B) Red represents genes that are upregulated and green those that are downregulated. (C) Relative gene expression of indicated genes was measured by qRT-PCR. Expression of respective gene for both WT and scc2–4 in the presence of break was related to its own absence of break sample that was set to 1. Data are mean values from 3 independent experiments with the respective deviation. Statistical significances are indicated by *, P ≤ 0.05, **, P ≤ 0.01, ***, P ≤ 0.005.
In conclusion, we found that one single DSB on Chr. VI indeed induced a number of genes previously reported to be transcriptionally activated upon DNA damage. Thus, the collected effect on the transcriptional response after DSB induction reflects a typical ESR pattern. Taken together, this convinced us that we have a suitable system for determination of the transcriptional alterations induced by a single DSB, and prompted us to investigate how transient absence of Scc2 specifically affects this response.
Influence of Scc2 on gene expression after DSB induction
In order to determine the importance of Scc2 for the transcriptional response to a DSB, we then performed an experiment following the same outline as in Figure 1A, but now analyzing only scc2–4 cells in the absence and presence of DSB. This microarray analysis showed that one single DSB on Chr. VI significantly affected the expression of 1189 probe sets, corresponding to 976 genes (Fig. 3B; data set S8). This is to be compared with 113 genes in WT cells (Fig. 3A; data set S7), using the same FDR cutoff ≤ 0.05. The analysis showed that 42% of the affected genes were upregulated and 58% downregulated (Fig. 3B; data set S8). Most of the genes that are known to be transcriptionally activated upon DNA damage and that we found in WT cells were upregulated also in Scc2 deficient cells (HUG1, RNR1, RNR2, RNR3, RNR4, DIN7, DUN1, MAG1, RAD16, PLM2, RAD54) (data set S8). A difference between WT and scc2–4 cells could be seen for genes encoding proteins of the SMC complexes. Both in WT and in Scc2 deficient cells, SCC3 was upregulated upon a HO break, but contrary to WT cells no significant change in expression of SMC1, MDC1, SMC6 or PDS5 was detected in scc2–4 cells (data sets S7 and S8). That all strains analyzed were evenly and consistently arrested in G2, and the DSB induction equally efficient in WT and scc2–4 strains, containing the recognition sequence for HO (data not shown), argues that the altered transcription profiles in scc2–4 cells compared with WT cells, in response to DSB induction, here depends on inactivation of Scc2.
A selection of interesting genes, as indicated by microarray to be significantly altered, were further analyzed by quantitative real time PCR (qRT-PCR) using cDNA isolated from WT and Scc2 deficient cells from the same experiment. The expression of this set of genes, mainly encoding cohesin components and DNA repair proteins interesting from a DSB repair perspective can be seen in Figure 3C. Among the genes encoding cohesin components, we could confirm that expression of 4 out of 5 genes tested (SCC3, SMC1, MCD1 and PDS5) showed a significant upregulation in WT cells, confirming the microarray result for these genes. For SMC6 the tendency was the same although not significant in qRT-PCR. Regarding the Scc2 deficient cells the microarray data indicated that there would be no effect on SMC1, MCD1, PDS5 or SMC6 and this was indeed confirmed using qRT-PCR (Fig. 3C). SCC3 on the other hand was upregulated in the microarray analysis, which was not the case using qRT-PCR where it showed the same pattern as the other genes (Fig. 3C).
In the microarray analysis several of the classical DNA damage response genes were significantly upregulated in both WT and Scc2 deficient cells. Among these HUG1 and PLM2 were significantly upregulated both in WT and in Scc2 deficient cells in response to DSB, also as determined by qRT-PCR. For the rest of the genes tested the tendency was clearly the same between the 2 types of analysis but not significant in qRT-PCR (Fig. 3C and data not shown).
Biological processes affected by break induction in WT and Scc2 deficient cells
Multiple DNA damage response genes were upregulated in response to DSB induction both in WT cells and in the absence of Scc2, however the overall transcriptional response was clearly different after inactivation of Scc2. Most apparently, 9 times as many genes were transcriptionally affected after break induction. In addition, from the initial experiment performed (Fig. 2D) it was apparent that in comparison with WT cells the collected transcriptional DNA damage response was downregulated in Scc2 deficient cells.
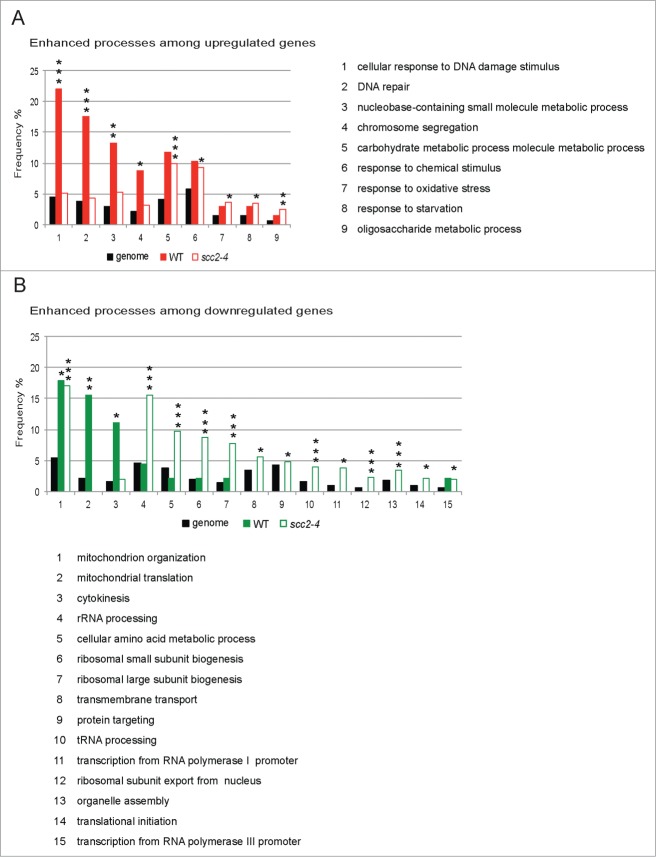
To get a better overall picture and understand how these different transcription profiles would affect the cellular functions we used the SGD GO slim mapping to analyze the separate WT and scc2–4 microarray data sets.33 The genes affected in response to a DSB, 113 in WT and 976 in Scc2 deficient cells, were divided into 2 groups, upregulated and downregulated, and sorted according to biological process (Tables S6–S9). Processes that showed a significant enhancement compared with genome frequency (FDR ≤ 0.05) were considered further. Rewardingly, 3 of the significantly enhanced processes in the group of upregulated genes in WT cells were cellular response to DNA damage stimulus, DNA repair and chromosome segregation. In Scc2 deficient cells completely different processes, such as responses to chemical stimuli, oxidative stress and starvation were enhanced (Fig. 4A, Table S6). When investigating the processes in the group of downregulated genes following break induction, “mitochondrion organization” is common for both WT and scc2–4 cells. However while additional mitochondrion processes are downregulated significantly in WT cells the repressed processes in Scc2 deficient cells instead mainly correspond to ribosomal small and large subunit biogenesis as well as tRNA processing, RNA polymerase I and III transcription, ribosome nuclear export and even translation, strongly indicated that in the absence of Scc2 the processes involved in ribosome production and function were impaired (Fig. 4B; Tables S7 and S9).
Figure 4.
A single DSB evokes different transcriptional programs in WT and scc2–4 cells. (A and B) SGD GO slim mapping was used to sort up- and downregulated genes according to biological process. Processes were regarded as significantly enhanced if FDR ≤ 0.05 (*, P ≤ 0.05, **, P ≤ 0.01, ***, P ≤ 0.005). (A) Frequencies for enhanced processes in the group of upregulated genes in response to DSB in WT and scc2–4 cells, in comparison with the frequency of the same processes in scc2–4 and WT cells, respectively. (B) Frequencies for enhanced processes in the group of downregulated genes in response to DSB in WT and scc2–4 cells, in comparison with the frequency of the same processes in scc2–4 and WT cells, respectively.
Taken together, this bioinformatic analysis indicated that inactivation of Scc2 clearly changed the gene response induced by a single DSB. This suggested to us that the expected transcriptional response after break induction is altered in scc2–4 cells. Furthermore the type of effect seen on all except very few genes, such as HUG1 and PLM2, are small but the effect is apparent on a large number of genes. This is a pattern similar to the transcriptional effects seen after knockdown of NIPBL, in NIPBL deficient CdLS patient cells and cells from Nipbl+/− mice.21, 22,37
Transcriptional alterations of DSB proximal genes
It has previously been reported that transcription of genes in close vicinity of the natural HO-cleavage site, localized at the MAT locus, are repressed in response to break induction. This was suggested to be due to resection of the DNA ends.29 In line with this, the microarray result for the genes surrounding the inserted HO recognition site on Chr. VI showed a similar pattern, where the majority of the genes in WT cells, LSB3, HIS2, CDC14, and PTR3 all showed reduced expression after break induction (data set S7; Fig. 5A). Somewhat surprisingly, we found that this was not true for the 2 genes located closest to the break on each side, where the expression of ULI1 was increased and no change in expression of ECO1 could be detected. Furthermore, when considering the microarray data for transcription over the same genes in Scc2 deficient cells, 3 out of the 6 closest genes (LSB3, HIS2, and PTR3) were not transcriptionally repressed after break induction (data set S8; Fig. 5A). In addition, ECO1 was shown to be upregulated after break induction in scc2–4 cells. However, CDC14 was repressed and ULI1 induced in response to the break in both WT and Scc2 deficient cells.
Figure 5.
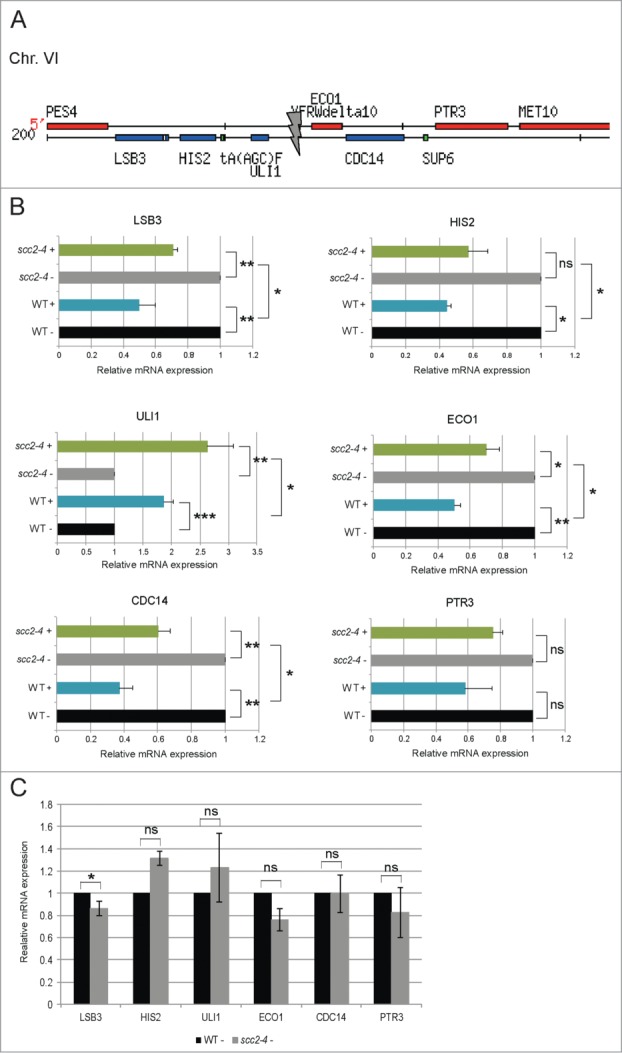
Scc2 is important for repression of DSB proximal genes. (A) Schematic illustration of the region immediately surrounding the DSB on Chr. VI (adapted from Sacchcaromyces Genome Database). Gray arrow points at the HO recognition site. (B) Relative gene expression of indicated genes was measured by qRT-PCR. Expression of respective gene for both WT and scc2–4 in the presence of break was related to its own absence of break sample that was set to 1. (C) Relative gene expression of indicated genes was measured by qRT-PCR. Expression of respective gene for scc2–4 cells in the absence of break was related to the WT absence of break sample that was set to 1. (B and C) Data are mean values from 3 independent experiments with the respective deviation (*, P ≤ 0.05, **, P ≤ 0.01, ***, P ≤ 0.005).
We then performed qRT-PCR on samples from WT and Scc2 deficient cells collected simultaneously, and analyzed expression of these DSB proximal genes. All genes except ULI1 then appeared to be repressed in response to DSB in WT cells, although PTR3 not significantly (Fig. 5B). In Scc2 deficient cells, the same pattern was observed. However the reduced expression in response to DSB was significantly less pronounced, compared with WT cells. This suggested that Scc2, or cohesin its target for loading, are important for proper downregulation of transcription in the direct vicinity of the DSB. The reason for the fairly strong upregulation of ULI1 in both WT and scc2–4 cells is enigmatic and we do currently not have any obvious explanation for that. Notably, the DSB proximal genes investigated here were not differently affected in the absence of DSB induction as analyzed by microarray or qRT-PCR (Fig. 5C; data set S1).
Alterations in gene expression due to Scc2 inactivation are not caused by changes in genome wide cohesin binding
Cohesin is dynamically interacting with chromatin throughout the cell cycle.12,13 The fraction of the cohesin pool that is stably bound is assumed to be responsible for the cohesion function.13 Scc2 has been shown to be responsible for chromatin loading of cohesin, not only in G1, but also in G2 phase, and in response to break induction. New cohesin is recruited to the region around the break and DI-cohesion is formed DSB proximally and genome wide.38 In human cells it was previously reported that cohesin binding correlated with changes in gene expression.21 We thus asked if the transcriptional dysregulation, detected here in Scc2 deficient cells, could be an effect of deficient cohesin loading and or dynamics in G2/M in the absence of Scc2, both in the DSB region and genome wide.
To investigate this, we performed chromatin immunoprecipitation followed by DNA sequencing (ChIP seq). The experimental setup used was identical to the one used for the microarray study (Fig. 1A), but in the end of the experiment ChIP was performed on Flag-tagged Scc1 (Scc13xFLAG). No apparent difference in cohesin binding could however be detected, rather the Scc13xFLAG was found at previously identified CARS (cohesin associated regions) genome wide in both WT and scc2–4 cells (Fig. 6A). The DSB proximal binding was as previously reported reduced after inactivation of Scc2 (Fig. 6B). Altogether the altered transcription profiles detected in Scc2 defective cells can not be based on removal of, or formation of new cohesin binding regions at new positions in the genome that depend on Scc2.
Figure 6.
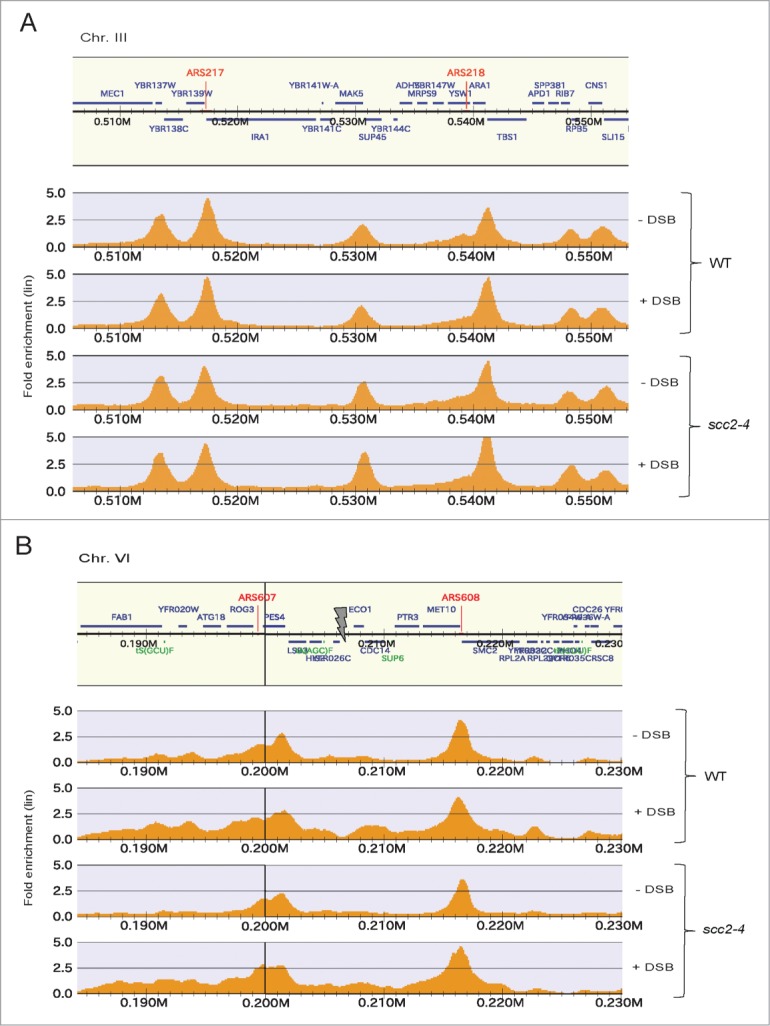
Cohesin binding genome wide and at the Chr. VI DSB. (A and B) Chromosomal association of Flag-tagged Scc1 analyzed by ChIP sequencing in WT and scc2–4 cells in the presence and absence of DSB induction at Chr. VI as indicated. Orange peaks display significant chromosomal binding sites where the x-axes show chromosomal positions and the y-axes show linear fold enrichment. (A) Shown is a representative undamaged region of Chr. III. (B) Shown is the region immediately surrounding the DSB on Chr. VI. Gray arrow points at the HO break site.
Discussion
Cohesin has in budding yeast been shown to act as a boundary element at the silent loci of HMR, and this was suggested to be the sole gene regulatory function for cohesin in yeast.23 Later it was shown that inactivation of the cohesin subunit Mcd1 in the G1 phase of the cell cycle caused altered expression of a number of genes with related function in a coordinated fashion.24 In addition, the cohesin loader Scc2, regulates gene expression during yeast meiosis,25,26 and the cohesin protein Smc1 as well as the acetyltransferase Eco1 promote rRNA production.24,27 Altogether this points at a gene regulatory function for proteins in the cohesin network in yeast, similarly to higher eukaryotes.39
To further investigate this we have here studied whether inactivation of Scc2 has impact on the transcription program in yeast, using a temperature sensitive SCC2 allele (scc2–4) under conditions known to inactivate both sister chromatid cohesion and DNA repair.14,28 We chose to study the effect of transiently disabling Scc2 in G2/M arrested cells when cohesin and its loader are important for homologous recombination based DSB repair and DI-cohesion.
We found that most of the genes with altered expression in scc2–4 cells compared to WT cells, are involved in processes that are affected independently of break induction, and presumably instead depend on the inactivation of Scc2. The transcription profile that we identify in the absence of functional Scc2 is indeed very similar to what has been reported in studies of human and mouse, heterozygous for their Scc2 homologs, where a large number of genes are affected but with limited fold changes. The genes affected were sorted according to association with biological processes. This revealed, somewhat surprisingly, that in Scc2 deficient compared with WT cells, the main response in the group of upregulated genes involves mitochondrial organization and translation. The most significantly downregulated genes were spread on several processes. Interestingly we see that ribosomal small and large subunit biogenesis as well as tRNA processing, RNA polymerase I and III transcription, ribosome nuclear export and even translation are downregulated in the absence of functional Scc2. This is in line with the finding that Smc1 and Eco1, proteins in the cohesin network, have been implicated in promoting rRNA production and protein translation.27 In the same study a mutated version of Scc2 did not show the same phenotype as the mutated versions of Smc1 and Eco1 used, however since this Scc2 mutation is different from the one used here, the importance for the ribosomal processes could be common for all the proteins in the cohesin network.
Our attention was then drawn to the fact that processes such as DNA damage response, DNA repair and DNA recombination were significantly enhanced compared to their genome frequency, in the group of downregulated genes in Scc2 deficient cells in the presence of DSB.
We then concluded that a single DSB causes a typical DNA damage induced transcriptional response in WT cells. Indicative of this HUG1, which was the most differentially expressed gene, showing the lowest FDR in our analysis, has previously been shown to be one of the most transcriptionally induced genes in response DNA damage, irrespective of the type of DNA damage.40-42 Interestingly Hug1 is a negative regulator of the Mec1 pathway, instrumental for DSB signaling and formation of DI-cohesion.43 When inactivating the cohesin loader Scc2 on the other hand the described DNA damage response is altered. For example, several of the genes encoding cohesin components, such as SMC1, MCD1, SCC3 and PDS5 showed a significant upregulation in WT cells, which was not the case in Scc2 deficient cells. Importantly continuous MCD1 transcription has been shown to be important for formation of DI-cohesion in response to post replicative DSB.44 In addition it has been reported that the Mcd1 levels needed for cohesion formation in response to DSB are higher then for establishment of S phase cohesion.45
When Scc2 is inactivated in G2/M, the cohesin binding pattern is not in general altered in undamaged regions of the genome compared to S phase, indicating that the differences in gene response between WT and Scc2 deficient cells is not based on overt changes in cohesin binding. An exception for this is in the 50 kb region around the DSB where less cohesin is recruited in Scc2 deficient cells. The reduced repression of DSB proximal genes in response to the DSB induction that we see in scc2–4 cells could thus be due to the reduced recruitment of cohesin, provided cohesin is needed for the silencing of the DSB proximal genes. On the other hand, these genes have been suggested to be downregulated due to the resection of the break that facilitates single stranded DNA tails.46 This is puzzling, since we see less repression in the absence of Scc2 where there is also less cohesin, and intuitively more resection, unless the proposed active separase mediated removal of cohesin is instrumental for the initiation of the resection process. Cohesin that was never loaded cannot be removed.44 Alternatively, knowing that also Scc2 itself is recruited to the DSB proximal region38 it could analogous to the genome wide situation be a function for Scc2 in itself, independent of cohesin.
Potentially, Scc2/4 is responsible for correct transcriptional programming through shaping of the chromatin landscape. Interestingly the human homolog NIPBL, was shown to interact with HDAC and HP1,47,48 both important for modulating chromatin structure. Traditionally, cohesin and or the Scc2 homologues were implicated in long range promoter enhancer interactions. Thereby they may be important for correct gene expression.16 Such interactions were believed to in essence not take place in budding yeast with its very small sized genome, and the possible function for cohesin pathway proteins in gene regulation would have to be based on a different mechanism. However budding yeast is not devoid of 3-dimensional interactions between different regions of the genome. On the contrary, an intricate network of inter- and intra-chromosomal interactions is evident also in the yeast genome.49,50
We have here performed gene expression profiling of WT and Scc2 deficient cells, both in the absence and presence of DNA damage. We conclude that transient inactivation of Scc2, during a G2 specific cell cycle arrest, has large effects on the general transcription profile compared with WT cells. When studying the effect of DSB induction in Scc2 deficient cells in isolation, this reveals that Scc2 indeed influences also the transcriptional response caused by induction of a single DSB. This effect is clear both globally and in the DSB proximal region. Knowing that Scc2 is important for gene regulation also in yeast sets the basis for the possibility to perform mechanistic studies of the same.
Materials and Methods
Yeast strains
All strains used are haploid and of W303 origin (ade2–1, trp1–1, can1–100, leu2–3, leu112, his3–11, 15, ura3–1, RAD5). Genetic modifications and names of the individual strains are listed in Table S1. Deletions of genes were performed by conventional one-step replacement of the open reading frame in question, with kanamycin (kanMX6), or nourseothricin (natMX4) resistance.51 Insertion of the HO-cleavage site at Chr. VI and the Flag tagging of Scc1 (SCC1-his6-flag3:kanMX6) was performed as described.14
Experimental setup
The same experimental procedure was used for microarray, ChIP seq and the qRT-PCR analysis. Pairs of WT and scc2–4 S. cerevisiae strains, genetically identical in all other aspects except for the presence of the recognition site for the HO enzyme or not, were grown in YEP media supplemented with 2% raffinose at permissive temperature (21°C) and arrested in G2/M by addition of benomyl. The temperature was then raised to 32°C for 30 minutes, restrictive for the scc2–4 ts allele. The HO enzyme under control of the GAL1 promoter was then activated by addition of 2% Galactose whereby a DSB was induced in cells containing the recognition site for HO enzyme on Chr. VI. Cells were collected 90 minutes later.
Pulse field gel electrophoresis
Pulse field gel electrophoresis was used for verification of induction of the Chr. VI DSB. Chromosomal DNA was prepared and separated on 1% agarose gel by Pulse Field Gel Electrophoresis as described28 (Biorad, Chef DRIII). For best separation in the size range of Chr. VI the gel was run at 14°C for 24 hr at 6 V/cm with a 35.4–83.55 s switch time and an included angle of 120°.
FACS analysis
FACS samples were prepared in essence as described5 and analyzed with a Becton Dickinson FACSCalibur, ensuring 10.000 events per sample.
RNA isolation, microarray and qRT-PCR
Total RNA was isolated after zymolase treatment for 1h at 30°C, using Invitrogen PureLink Micro-to-Midi Total RNA Purification System. Sample quantity and quality was assessed with a spectrophotometer (ND-1000, NanoDrop technologies) and Agilent Bioanalyzer 2100. For the microarray analysis, cDNA was synthesized and submitted to the Karolinska Institutet core facility for Bioinformatics and Expression Analysis (www.bea.ki.se), where hybridization to the GeneChip Yeast Genome 2.0 Array was performed. After probing and scanning, the quality of the images was checked. All arrays passed the Affymetrix quality control check. For the qRT-PCR analysis cDNA was prepared using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qRT-PCR was performed using SYBR green (Applied Biosystems) according to manufacturer's instructions (primer sequences available on request). Samples were then analyzed on ABI Prism 7000 sequence detection system (Applied Biosystems).
Analysis of gene expression data
Gene expression data was pre-processed using the limma package in R52 and the rma (Robust Multichip Average) procedure including background correction, quantile normalization and summarization. Then, each run was checked for technical variation and adjustments for batch effects were carried out.53 In the second experiment run (Fig. 3A), one sample was excluded because of non-biological variation. In each run, samples were tested for differential expression (FDR ≤ 0.05)54 between different strains, and within strains but between absence and presence of DSB. Selected probesets were annotated using DAVID software30,31 and NetAffxTM (Affymetrix), and heatmaps were generated using Cluster software and visualized in TreeView.32 Mathematica 7.0 or R 2.9.2 was used for all calculations. The unprocessed gene expression data can be found at the GEO database: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62494.
Categorization of genes according to biological process and enrichment analysis
To retrieve information of the ORF of interest, the Saccharomyces Genome Database (SGD, http://www.yeastgenome.org) was queried. Genes were categorized using the SGD GO Slim Mapper to define the biological processes in conjugation with GO annotations for yeast gene products curated by the SGD. P-values were calculated for enrichment categories using the Hypergeometric distribution with correction for multiple testing using FDR. Processes were regarded significantly enhanced if FDR ≤ 0.05.
ChIP sequencing
ChIP was performed on Flag-tagged Scc1 (Scc13xFLAG) with anti-Flag (Sigma) essentially as described,55 but after 30 min fixation with 1% formaldehyde at room temperature. The samples were then processed for ChIP sequencing (ChIP seq) as described.55 The unprocessed sequencing data can be found at the GEO database: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62494.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
The authors thank Professors Camilla Sjögren and Katsu Shirahige and Dr Yuki Katou for support and technical advice. The authors also thank Drs Elin Enervald and Christopher Bot for critically reading the manuscript.
Funding
This work was supported by a young investigator award from the Swedish Cancer Society to L Ström and grants from the Swedish Research Council, the Swedish Cancer Society, the KID program at the Karolinska Institutet, as well as the Jeansson's foundation, the Wibergs, Bergwalls, and the Karolinska Institute research foundations to L Ström
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Losada A, Hirano T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev 2005; 19:1269-87; PMID:15937217; http://dx.doi.org/ 10.1101/gad.1320505 [DOI] [PubMed] [Google Scholar]
- 2. Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol 2006; 7:311-22; PMID:16633335; http://dx.doi.org/ 10.1038/nrm1909 [DOI] [PubMed] [Google Scholar]
- 3. Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouet F, Underwood P, Metson J, Imre R, et al. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell 2009; 33:763-74; PMID:19328069; http://dx.doi.org/ 10.1016/j.molcel.2009.02.028 [DOI] [PubMed] [Google Scholar]
- 4. Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 1997; 91:47-57; PMID:9335334; http://dx.doi.org/ 10.1016/S0092-8674(01)80008-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 1997; 91:35-45; PMID:9335333; http://dx.doi.org/ 10.1016/S0092-8674(01)80007-6 [DOI] [PubMed] [Google Scholar]
- 6. Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 2004; 430:573-8; PMID:15229615; http://dx.doi.org/ 10.1038/nature02742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Nasmyth K. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell 2000; 5:243-54; PMID:10882066; http://dx.doi.org/ 10.1016/S1097-2765(00)80420-7 [DOI] [PubMed] [Google Scholar]
- 8. Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev 2008; 22:3089-114; PMID:19056890; http://dx.doi.org/ 10.1101/gad.1724308 [DOI] [PubMed] [Google Scholar]
- 9. Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev 1999; 13:320-33; PMID:9990856; http://dx.doi.org/ 10.1101/gad.13.3.320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev 1999; 13:307-19; PMID:9990855; http://dx.doi.org/ 10.1101/gad.13.3.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ivanov D, Schleiffer A, Eisenhaber F, Mechtler K, Haering CH, Nasmyth K. Eco1 Is a Novel Acetyltransferase that Can Acetylate Proteins Involved in Cohesion. Current biology : CB 2002; 12:323-8; PMID:11864574; http://dx.doi.org/ 10.1016/S0960-9822(02)00681-4 [DOI] [PubMed] [Google Scholar]
- 12. Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Current biology : CB 2006; 16:1571-8; PMID:16890534; http://dx.doi.org/ 10.1016/j.cub.2006.06.068 [DOI] [PubMed] [Google Scholar]
- 13. McNairn AJ, Gerton JL. Intersection of ChIP and FLIP, genomic methods to study the dynamics of the cohesin proteins. Chromosome Res: Int J Mol Supramol Evol Aspect Chromosome Biol 2009; 17:155-63; http://dx.doi.org/ 10.1016/j.cub.2006.06.068 [DOI] [PubMed] [Google Scholar]
- 14. Strom L, Lindroos HB, Shirahige K, Sjogren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell 2004; 16:1003-15; PMID:15610742; http://dx.doi.org/ 10.1016/j.molcel.2004.11.026 [DOI] [PubMed] [Google Scholar]
- 15. Unal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell 2004; 16:991-1002; PMID:15610741; http://dx.doi.org/ 10.1016/j.molcel.2004.11.027 [DOI] [PubMed] [Google Scholar]
- 16. Dorsett D, Strom L. The ancient and evolving roles of cohesin in gene expression and DNA repair. Curr Biol: CB 2012; 22:R240-50; http://dx.doi.org/ 10.1016/j.cub.2012.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rollins RA, Korom M, Aulner N, Martens A, Dorsett D. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol 2004; 24:3100-11; PMID:15060134; http://dx.doi.org/ 10.1128/MCB.24.8.3100-3111.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, Smith J, Bateman A, Krpic S, Hostert A, et al. Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance. PLoS Biol 2006; 4:e242; PMID:16802858; http://dx.doi.org/ 10.1371/journal.pbio.0040242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 2008; 132:422-33; PMID:18237772; http://dx.doi.org/ 10.1016/j.cell.2008.01.011 [DOI] [PubMed] [Google Scholar]
- 20. Liu J, Baynam G. Cornelia de Lange syndrome. Adv Exp Med Biol 2010; 685:111-23; PMID:20687500 [PubMed] [Google Scholar]
- 21. Liu J, Zhang Z, Bando M, Itoh T, Deardorff MA, Clark D, Kaur M, Tandy S, Kondoh T, Rappaport E, et al. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol 2009; 7:e1000119; PMID:19468298; http://dx.doi.org/ 10.1371/journal.pbio.1000119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kawauchi S, Calof AL, Santos R, Lopez-Burks ME, Young CM, Hoang MP, Chua A, Lao T, Lechner MS, Daniel JA, et al. Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/-) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet 2009; 5:e1000650; PMID:19763162; http://dx.doi.org/ 10.1371/journal.pgen.1000650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev 1999; 13:698-708; PMID:10090726; http://dx.doi.org/ 10.1101/gad.13.6.698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Skibbens RV, Marzillier J, Eastman L. Cohesins coordinate gene transcriptions of related function within Saccharomyces cerevisiae. Cell Cycle 2010; 9:1601-6; PMID:20404480; http://dx.doi.org/ 10.4161/cc.9.8.11307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin W, Jin H, Liu X, Hampton K, Yu HG. Scc2 regulates gene expression by recruiting cohesin to the chromosome as a transcriptional activator during yeast meiosis. Mol Biol Cell 2011; 22:1985-96; PMID:21508318; http://dx.doi.org/ 10.1091/mbc.E10-06-0545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin W, Wang M, Jin H, Yu HG. Cohesin plays a dual role in gene regulation and sister-chromatid cohesion during meiosis in Saccharomyces cerevisiae. Genetics 2011; 187:1041-51; PMID:21270391; http://dx.doi.org/ 10.1534/genetics.110.122358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bose T, Lee KK, Lu S, Xu B, Harris B, Slaughter B, Unruh J, Garrett A, McDowell W, Box A, et al. Cohesin proteins promote ribosomal RNA production and protein translation in yeast and human cells. PLoS Genet 2012; 8:e1002749; PMID:22719263; http://dx.doi.org/ 10.1371/journal.pgen.1002749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sjogren C, Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr Biol: CB 2001; 11:991-5; http://dx.doi.org/ 10.1016/S0960-9822(01)00271-8 [DOI] [PubMed] [Google Scholar]
- 29. Lee SE, Pellicioli A, Demeter J, Vaze MP, Gasch AP, Malkova A, Brown PO, Botstein D, Stearns T, Foiani M, et al. Arrest, adaptation, and recovery following a chromosome double-strand break in Saccharomyces cerevisiae. Cold Spring Harb Sym 2000; 65:303-14; http://dx.doi.org/ 10.1101/sqb.2000.65.303 [DOI] [PubMed] [Google Scholar]
- 30. Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003; 4:P3; PMID:12734009; http://dx.doi.org/ 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- 31. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44-57; PMID:19131956 [DOI] [PubMed] [Google Scholar]
- 32. Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 1998; 95:14863-8; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-9; PMID:10802651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, Brown PO. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell 2001; 12:2987-3003; PMID:11598186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Bassett DE, Jr., Hieter P, Vogelstein B, Kinzler KW. Characterization of the yeast transcriptome. Cell 1997; 88:243-51; PMID:9008165 [DOI] [PubMed] [Google Scholar]
- 36. Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 2000; 11:4241-57; PMID:11102521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zuin J, Franke V, van Ijcken WF, van der Sloot A, Krantz ID, van der Reijden MI, Nakato R, Lenhard B, Wendt KS. A cohesin-independent role for NIPBL at promoters provides insights in CdLS. PLoS Genet 2014; 10:e1004153; PMID:24550742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Strom L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjogren C. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 2007; 317:242-5; PMID:17626884 [DOI] [PubMed] [Google Scholar]
- 39. Dorsett D, Strom L. The ancient and evolving roles of cohesin in gene expression and DNA repair. Curr Biol: CB 2012; 22:R240-50; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Basrai MA, Velculescu VE, Kinzler KW, Hieter P. NORF5/HUG1 is a component of the MEC1-mediated checkpoint response to DNA damage and replication arrest in Saccharomyces cerevisiae. Mol Cell Biol 1999; 19:7041-9; PMID:10490641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Benton MG, Glasser NR, Palecek SP. The utilization of a Saccharomyces cerevisiae HUG1P-GFP promoter-reporter construct for the selective detection of DNA damage. Mutat Res 2007; 633:21-34; PMID:17618162 [DOI] [PubMed] [Google Scholar]
- 42. Mizukami-Murata S, Iwahashi H, Kimura S, Nojima K, Sakurai Y, Saitou T, Fujii N, Murata Y, Suga S, Kitagawa K, et al. Genome-wide expression changes in Saccharomyces cerevisiae in response to high-LET ionizing radiation. Appl Biochem Biotech 2010; 162:855-70. [DOI] [PubMed] [Google Scholar]
- 43. Ainsworth WB, Hughes BT, Au WC, Sakelaris S, Kerscher O, Benton MG, Basrai MA. Cytoplasmic localization of Hug1p, a negative regulator of the MEC1 pathway, coincides with the compartmentalization of Rnr2p-Rnr4p. Biochem Biophys Res Commun 2013; 439:443-8; PMID:24012676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McAleenan A, Clemente-Blanco A, Cordon-Preciado V, Sen N, Esteras M, Jarmuz A, Aragon L. Post-replicative repair involves separase-dependent removal of the kleisin subunit of cohesin. Nature 2013; 493:250-4; PMID:23178808 [DOI] [PubMed] [Google Scholar]
- 45. Heidinger-Pauli JM, Mert O, Davenport C, Guacci V, Koshland D. Systematic reduction of cohesin differentially affects chromosome segregation, condensation, and DNA repair. Curr Biol: CB 2010; 20:957-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev: MMBR 1999; 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lechner MS, Schultz DC, Negorev D, Maul GG, Rauscher FJ, 3rd. The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem Biophys Res Commun 2005; 331:929-37; PMID:15882967; http://dx.doi.org/ 10.1016/j.bbrc.2005.04.016 [DOI] [PubMed] [Google Scholar]
- 48. Jahnke P, Xu W, Wulling M, Albrecht M, Gabriel H, Gillessen-Kaesbach G, Kaiser FJ. The Cohesin loading factor NIPBL recruits histone deacetylases to mediate local chromatin modifications. Nucleic Acids Res 2008; 36:6450-8; PMID:18854353; http://dx.doi.org/ 10.1093/nar/gkn688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rodley CD, Bertels F, Jones B, O'Sullivan JM. Global identification of yeast chromosome interactions using Genome conformation capture. Fungal Genet Biol 2009; 46:879-86; PMID:19628047; http://dx.doi.org/ 10.1016/j.fgb.2009.07.006 [DOI] [PubMed] [Google Scholar]
- 50. Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. A three-dimensional model of the yeast genome. Nature 2010; 465:363-7; PMID:20436457; http://dx.doi.org/ 10.1038/nature08973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 1994; 10:1793-808; PMID:7747518; http://dx.doi.org/ 10.1002/yea.320101310 [DOI] [PubMed] [Google Scholar]
- 52. Smyth GK RG, Carey V, Dudoit S, Irizarry R, Huber W. Limma: linear models for microarray data. In: 'Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer, 2005; pages 397-420. [Google Scholar]
- 53. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007; 8:118-27; PMID:16632515; http://dx.doi.org/ 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- 54. Gentleman VJC R W, Huber R A, Irizarry S Dudoit. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. ISBN: 978-0-387-25146-2 (Print) 978-0-387-29362-2 (Online). [Google Scholar]
- 55. Kegel A, Betts-Lindroos H, Kanno T, Jeppsson K, Strom L, Katou Y, Itoh T, Shirahige K, Sjogren C. Chromosome length influences replication-induced topological stress. Nature 2011; 471:392-6; PMID:21368764; http://dx.doi.org/ 10.1038/nature09791 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.