Abstract
Telomerase is often upregulated during initiation and/or progression of human tumors, suggesting that repression of telomerase might inhibit cancer growth or progression. Here, we report that BRG1, the ATPase subunit of the SWI/SNF chromatin remodeling complex, is a general suppressor of hTERT transcription in human cancer cells. While overexpression of BRG1 inhibits hTERT transcription, depletion of BRG1 stimulates transcription of hTERT, leading to higher telomerase activity and longer telomeres. Chromatin-immunoprecipitation assays revealed that BRG1 binds to the transcription start site (TSS) of the hTERT promoter and forms a ternary complex with histone deacetylase 2 (HDAC2). BRG1 remodels chromatin structure to facilitate the action of HDAC2, leading to deacetylation of H3K9ac and H4ac at the TSS and suppression of hTERT transcription. On the other hand, β-catenin binds to the TSS and stimulates hTERT transcription. Thus, BRG1/HDAC2 and β-catenin constitute a manipulative apparatus at the TSS to play opposite but complementary roles in regulating hTERT expression. These results uncover a yin-yang mechanism in modulating hTERT transcription and provide explanation for limited transcription of hTERT in human cancer cells. BRG1/HDAC2 may have a potential as an anti-cancer therapeutic and/or for reactivating cellular proliferative capacity in the context of in vitro tissue engineering.
Keywords: BRG1, chromatin remodeling, HDACs, telomerase, telomeres, β-catenin
Abbreviations
- HDACs
histone deacetylase
- TSS
transcription start site
- TSA
Trichostatin A
- SWI/SNF
SWItch/Sucrose NonFermentable
Introduction
Telomeres are specialized DNA-protein complexes at the ends of linear chromosomes that prevent fusion of chromosome ends, and inhibit enzyme-mediated degradation and illegitimate recombination of chromosomal DNA.1 In human cells, the DNA component of telomere is composed of ∼5–15 kb of double-stranded -TTAGGG/AATCCC- repeats. Six essential proteins (TRF1, TRF2, RAP1, TIN2, TPP1 and POT1) form a complex called shelterin that binds telomric DNA, and all play important roles in end protection,1 telomeric DNA damage response,2 sister chromatin cohesion and telomere length maintenance by telomerase.3–4 Also, it has been reported that methylated TRF2 can serve as a potential biomarker for cellular senescence.5 In proliferating human somatic cells that lack telomerase activity, telomeres shorten progressively over time, because ∼50–200 terminal bp fail to be replicated per replicative cycle.6,7 Once a human somatic cell telomere regresses to a critical length, the short telomere triggers a state known as ‘telomere crisis,’ followed by replicative senescence or apoptosis.8 In many cases, tumor cells (∼85%) and germline cells avoid telomere crisis by activating telomerase, a ribonucleoprotein in which the reverse-transcriptase activity of the protein component (hTERT) catalyzes de novo synthesis of telomeric repeats at chromosome ends using a telomere-specific RNA template (hTR).9,10 While hTR is highly expressed in all human tissues, expression of the catalytic component of human telomerase, hTERT, is tightly regulated,11 such that its expression is repressed in most normal human cells.12,13 There is great interest in understanding how hTERT expression is regulated in cancer cells, because of the possibility of exploiting such knowledge in order to prevent or limit cancer cell growth. Thus, hTERT is considered as an important target for novel cancer therapeutic agents or drugs.
The regulation of hTERT expression in human cancer cells has been the subject of much research, summarized briefly as follows. The hTERT gene is more than 40 kb long and has 16 exons and 15 introns.14 The hTERT promoter is rich in CpG islands, Sp1 sites 15,16 and binding sites for several positive and negative regulatory proteins.17 Positive regulators of hTERT transcription include the oncogene c-myc, Sp1, steroid hormone receptors, human papillomavirus 16 (HPV16) protein E6 and E6-associated protein (E6-AP)18; negative regulators of hTERT transcription include p53, pRb, E2F, Mad1, WT1 and MZF-2, and antiproliferative/ differentiation factors such as interferon-γ and TGF-β. Many of these factors act in a cell type-dependent manner and their function in regulating hTERT transcription is not yet fully understood. Clearly, there is a great deal of complexity and heterogeneity in how hTERT expression is regulated in different tissues and through development.
BRG1 is an ATPase and a core component of the SWI/SNF complex, which utilizes energy from ATP hydrolysis to disrupt or reposition nucleosomes and modify chromatin structure,19 thereby facilitating or inhibiting transcription of target genes or gene regions. It has been reported that the SWI/SNF complex can activate or repress transcription, depending on the presence or absence of specific accessory factors.20 Growing evidence indicates that the SWI/SNF complex plays a role in tumor suppression; for example inactivating mutations in BRG1 and several other SWI/SNF subunits are found in a variety of cancer cells.21,22 In addition, BRG1-haploinsufficient mice are tumor prone.23 These data suggest that BRG1 is a bona fide tumor suppressor.
Earlier reports indicated that BRG1 binds to β-catenin and promotes expression of Wnt-related genes24 and that BRG1 is able to positively modulates expression of β-catenin.25 Recently, it was also reported that β-catenin regulates expression of hTERT in stem cells and cancer cells26,27 through its ability to recruit Setd1a to and modulate the chromatin structure of the TERT promoter (Setd1a is a histone methyltransferase that trimethylates histone H3 at lysine 4 generating H3K4me3). Therefore, it is speculated that BRG1 may be involved in promoting hTERT expression. This, however, is inconsistent with the observation in a mouse model that depletion of BRG1 correlates with a significant increase in TERT expression 28 in optic nerve cells. Furthermore, TERT protein is thought to regulate Wnt signaling through binding to BRG1.29 Additional experiments are needed to clarify whether and how BRG1 and β-catenin influence TERT expression.
The present study demonstrates that BRG1 negatively regulates transcription of hTERT in human cervical cancer cells and that BRG1 and the SWI/SNF complex repress transcription of hTERT in several human tumor cell lines, suggesting that BRG1 may be a general suppressor of hTERT. BRG1-SWI/SNF complex binds to the transcription start site (TSS) of the hTERT promoter and interacts with HDAC2, leading to the deacetylation of H3K9ac and H4ac. In contrast, β-catenin stimulates hTERT transcription by recruiting Setd1a to the TSS; therefore, the BRG1-SWI/SNF complex and β-catenin play opposite but complementary roles in regulating hTERT. Negative regulation of hTERT by BRG1 is consistent with its function as a tumor suppressor.
Results
BRG1 protein abundance is negatively-correlated with hTERT mRNA abundance in human cervical cancer cell lines
To investigate whether BRG1 and the SWI/SNF complex regulate transcription of TERT in human cells, BRG1 protein and hTERT mRNA were quantified in extracts from 4 human cervical tumor cell lines: C33A, SiHa, Caski, and HeLa. BRG1 western blot and q-PCR quantification of hTERT mRNA in these cell lines are shown in Figs. 1A and B, respectively. The results show that the abundance of BRG1 protein is inversely correlated with the abundance of hTERT mRNA in these 4 cell lines. hTERT mRNA is most abundant and BRG1 is least abundant in C33A cells, while BRG1 protein is most abundant and hTERT mRNA is least abundant in HeLa cells. Although it was previously reported that BRG1 interacts with β-catenin, which in turn positively regulates hTERT transcription,24,26 the data presented in Fig. 1 raised the possibility that BRG1 regulates hTERT expression independent of β-catenin.
Figure 1.
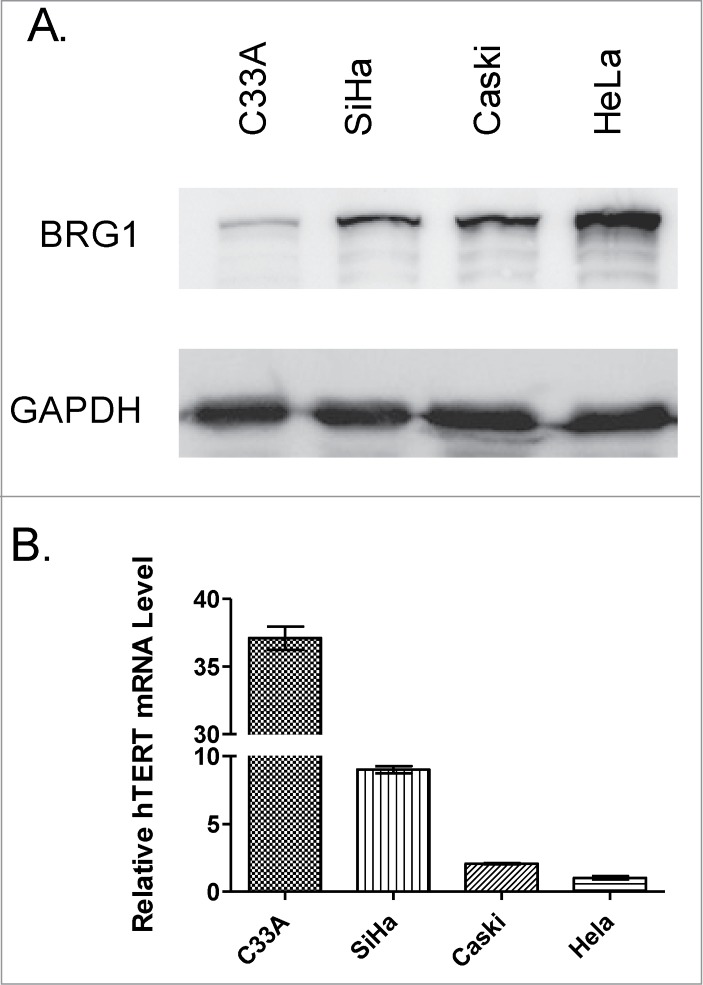
BRG1 protein and hTERT mRNA expression in human cervical cancer cell lines. (A) Western blot of BRG1 in cervical cancer cell lines with GAPDH as an internal control. (B) qRT-PCR data for hTERT mRNA in human cervical cancer cell lines. hTERT mRNA abundance was normalized to reference level in HeLa cells. Values are ±SD of three independent experiments.
Knockdown of BRG1 stimulates hTERT transcription in human cancer cells
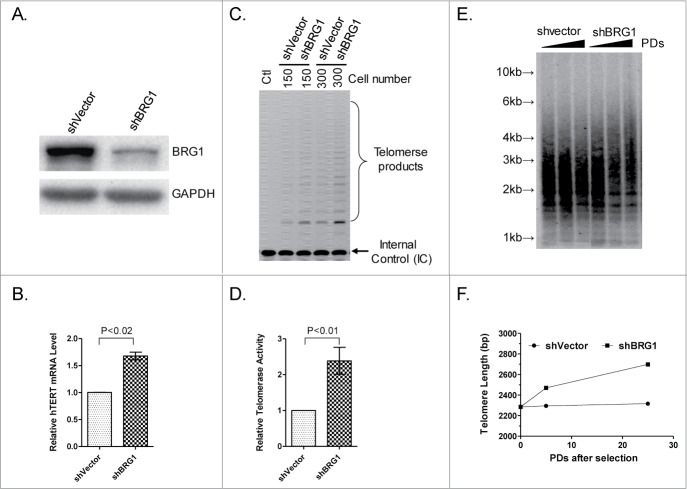
If BRG1 directly regulates hTERT expression, then it is predicted that stable knockdown of BRG1 will lead to an increase in abundance of hTERT mRNA. To test this idea, BRG1-deficient HeLa cells were constructed using shRNA, which reduced the level of BRG1 mRNA by ∼90% (Fig. 2A), with minimal effect on cell proliferation (Fig. S1). In these cells, qRT-PCR analysis showed that the hTERT mRNA was 1.7-fold more abundant than in control HeLa cells expressing shVector (Fig. 2B). This result is consistent with a previous study in BRG1-knockdown mice that the depletion of BRG1 resulted in a 1.5-fold increase in TERT expression.28
Figure 2.
Effect of knockdown of BRG1 on hTERT transcription in HeLa cells. (A) Western blot showing BRG1 protein after BRG1 knockdown by shBRG1. (B) qRT-PCR quantification of hTERT mRNA in BRG1-depleted cells; shVector was used as a control. (C) TRAP assay in BRG1-depleted (shBRG1) HeLa cells; shVector heat- inactivated samples (Ctl) were used as controls. (D) Quantitation of data in (C), Graph shows mean ± SD; Three independent experiments were performed. (E) TRF assay was performed in HeLa cells with or without BRG1-depletion; cells were cultured for different PDs (Population Doubling) after selection; ShVector was used as a control. (F) Quantification of data in (E). P values were calculated using the Student's t-test.
TRAP assay was used to determine whether hTERT mRNA abundance is an accurate measure of hTERT expression in BRG1 knockdown cells.30 The results show that telomerase activity is higher in BRG1-deficient cells than in control cells (Fig. 2C, D) such that a higher rate of telomere repeat synthesis as evidenced by increasing telomere length with population doublings (PDs) is observed (Fig. 2E, F). These results support the idea that BRG1 negatively regulates hTERT transcription and telomerase activity in human cancer cells.
BRG1 regulates hTERT transcription in a SWI/SNF complex-dependent manner
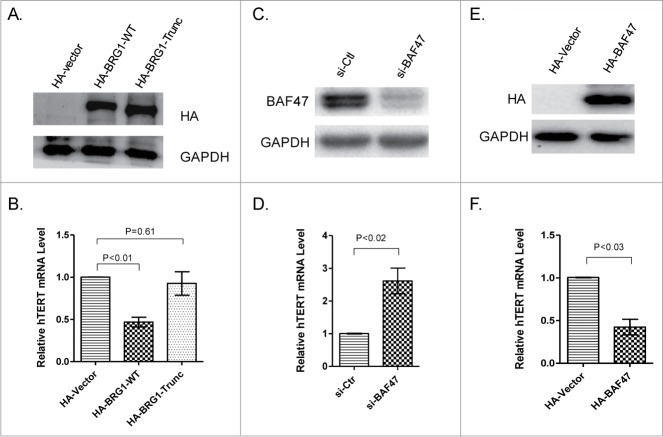
A BRG1 truncation mutant (ATPase-defective BRG1) was constructed and expressed in HeLa cells to determine whether BRG1 ATPase is required for its ability to inhibit hTERT transcription. The results show that overexpression of wild-type BRG1 but not of ATPase-defective BRG1 inhibits hTERT transcription in the tested cell line (Fig. 3A, B). These results suggest that ATP-dependent chromatin remodeling is required in order for BRG1 to regulate transcription of hTERT.
Figure 3.
Roles of BRG1 ATPase and SWI/SNF complex in hTERT transcriptional suppression. (A) HA-tagged BRG1 and HA-BRG1-Trunc (carrying deletion of ATPase domain) were overexpressed in HeLa cells. (B) Relative hTERT mRNA by qRT-PCR in cells overexpressing HA-tagged BRG1 and HA-BRG1-Trunc. (C) Western blot showing siRNA knockdown of BAF47 in HeLa cells. Scrambled sequence was used as a control (si-Ctl). (D) qRT-PCR of hTERT mRNA in BAF47-depleted and control cells. (E) Overexpression of HA-tagged wild-type BAF47 in HeLa cells. HA-Vector was used as a control. (F) qRT-PCR analysis of hTERT mRNA in cells overexpressing BAF47 and control cells. Mean ± SD are shown; Three independent experiments were performed. P values were calculated using the Student's t-test.
The human SWI/SNF chromatin remodeling complex contains either BRG1 or hBRM as the central catalytic ATPase and 10–12 BRG1-associated factors (BAFs).31-33 BAF47 is a SWI/SNF accessory factor that is required for the chromatin remodeling activity of the complex. To confirm that SWI/SNF contributes to BRG1-dependent inhibition of hTERT transcription, BAF47 was knocked down or overexpressed in human HeLa cells. Depletion of BAF47 correlated with higher abundance of hTERT mRNA, while overexpression of BAF47 correlated with decreased abundance of hTERT mRNA, relative to the relevant control cells (Figs. 3C-F). These data suggest that the role of BRG1 in regulating hTERT transcription is co-mediated by SWI/SNF complex including BAF47; in other words, BRG1 inhibits hTERT transcription in a SWI/SNF-dependent manner.
To extend the study to other human cell types, BRG1 was knocked down or overexpressed (wild type and ATPase-defective) in human breast cancer MDA-MB-231 cells (Fig. S2) and human embryonic kidney 293T cells (Fig. S3), and similar results were obtained, suggesting that BRG1-SWI/SNF may be a general suppressor in human cells.
BRG1-SWI/SNF complex locates to the TSS of hTERT promoter, and interacts with HDAC2
The role of the BRG1-SWI/SNF complex in chromatin remodeling is well established. However, the specific interaction between BRG-SWI/SNF and the hTERT promoter has not been characterized previously. Here, this interaction was examined over a 5 kb region surrounding the transcriptional start site (TSS) of the hTERT gene (i.e., 3 kb upstream to 2kb downstream of the TSS) using a standard ChIP assay, in which an antibody to pull down BRG1 and 5 PCR primer pairs (4 primer pairs targeting the TSS or upstream promoter region, and 1 primer pair targeting sequences downstream of the TSS; Fig. 4A) were used. The results show that BRG1-SWI/SNF interacts specifically with the TSS of the hTERT promoter (Fig. 4B). Here, the pair of TS primes covered 147 bp sequence at the vicinity of TSS (−107 to +36) (Fig. 4A). This suggests that the BRG1-SWI/SNF complex modulates the chromatin structure at or surrounding the TSS, and that this is the basis of its ability to inhibit transcription of the hTERT gene.
Figure 4.
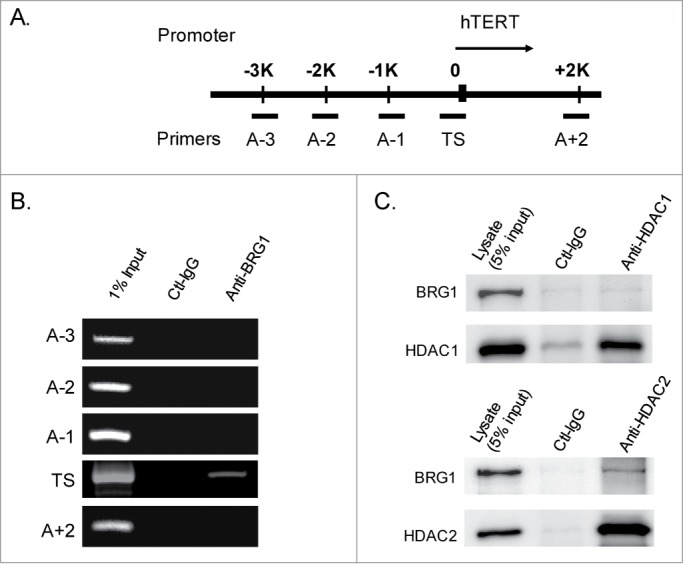
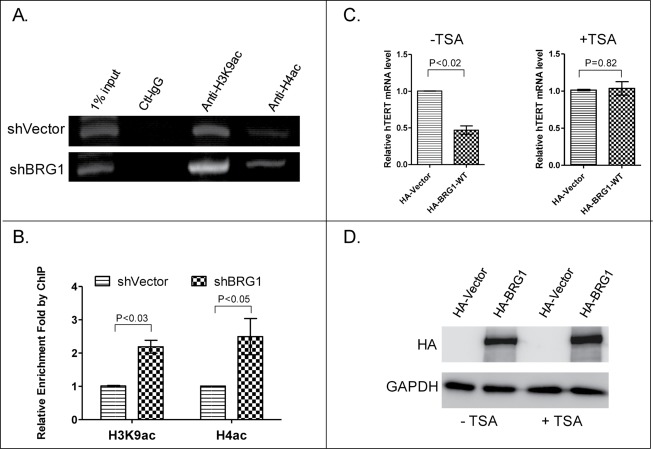
BRG1-SWI/SNF complex is localized to the TSS of hTERT and interacts with HDAC2. (A) Schematic diagram of hTERT promoter showing primers for ChIP analysis. (B) PCR of BRG1-immunopreciptated chromatin (ChIP) from HeLa cells. (C) Co-immunoprecipitation (Co-IP) of BRG1 with HDAC1 and HDAC2. Antibodies against HDAC1 and HDAC2 were used to pull down BRG1 from the lysate of HeLa cells.
Chromatin remodeling can also be mediated by the action of histone modifying enzymes.34 For example, BRG1 and Histone deacetylases (HDACs) are thought to coordinately remodel chromatin in mouse cells.35 To extend this to human cancer cells in the present study, HDAC1 or HDAC2 was immunoprecipitated from cell extracts, and the resulting protein aggregate was tested for the presence of BRG1. The results show that HDAC2, but not HDAC1, interacts with BRG1 in immunoprecipitates (Fig. 4C). Accordingly, chromatin-immunoprecipitation (ChIP) assays demonstrated that HDAC2 (Fig. 6C), but not HDAC1 (data not shown), locates to the TSS of hTERT promoter.
Figure 6.
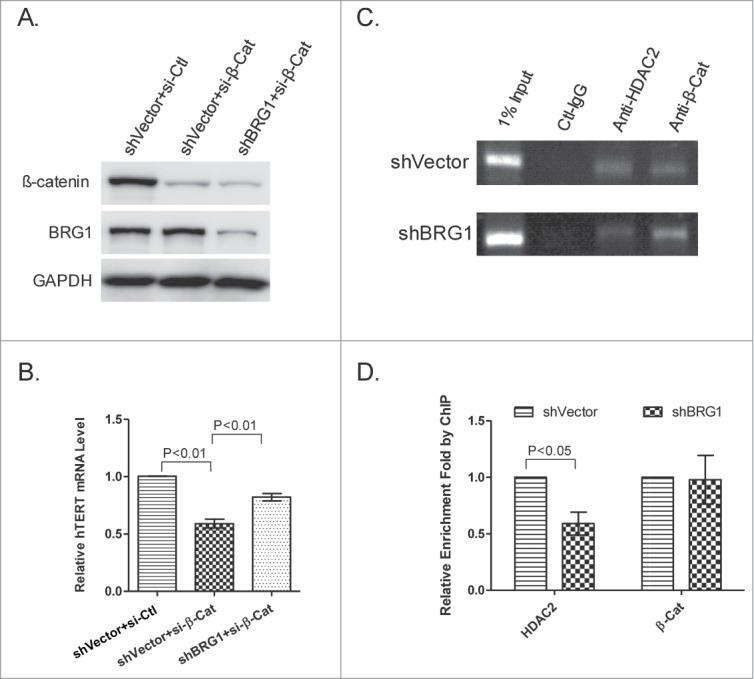
β-catenin and BRG1 regulate hTERT independently. (A) siRNA knockdown of β-catenin in the presence and absence of BRG1. BRG1 was depleted by shRNA as described above and shVector was used as a control. Western blot was used to determine the abundance of BRG1 and β-catenin. (B) qRT-PCR of hTERT mRNA in HeLa cells with β-catenin knocked down or with both β-catenin and BRG1 knocked down. (C) qPCR analysis of HDAC2- or β-catenin-immunopreciptated chromatin (ChIP) in BRG1-depleted HeLa (shBRG1) and in control cells (sh-Vector). (D) Quantitation of data in (C). Mean ± SD are shown; Three independent experiments were performed. P values were calculated using the Student's t-test.
Deacetylation of H3K9 and H4 at the TSS
The results of a ChIP experiment at hTERT promoter using antibody to H3K9ac and H4ac revealed that H3K9 and H4 are acetylated in the vicinity of the hTERT TSS, and the degree of acetylation significantly increased in BRG1-depleted cells (Fig. 5A, B), suggesting that BRG1-SWI/SNF can silence an actively transcribed promoter by promoting its deacetylation by HDAC2. If this is correct, it is expected that, Trichostatin A (TSA), an organic compound that selectively inhibits HDACs, will reverse BRG1-dependent inhibition of hTERT transcription. Data presented in Fig. 5C confirm this prediction, and demonstrate that TSA does not cause a concomitant decrease in expression of BRG1 (Fig. 5D). Therefore, we propose a model in which the BRG1-SWI/SNF complex remodels chromatin in the vicinity of the hTERT TSS and suppresses hTERT transcription in an HDAC2-dependent manner (see Fig. 7 and discussion for details).
Figure 5.
Quantification of H3K9ac and H4ac at the TSS of hTERT promoter with or without depletion of BRG1. (A) qPCR analysis of H3K9ac and H4ac-immunopreciptated chromatin in BRG1-depleted HeLa (shBRG1) and control cells (shVector). (B) Quantitation of data in (A). Mean ± SD are shown; Three independent experiments were performed. (C) qRT-PCR of hTERT mRNA in HeLa cells with or without exposure to TSA and with or without BRG1-overexpression (HA-BRG1 vs HA-Vector). (D) Western blot for BRG1 in HeLa cells treated with or without TSA. P values were calculated using the Student's t-test.
Figure 7.
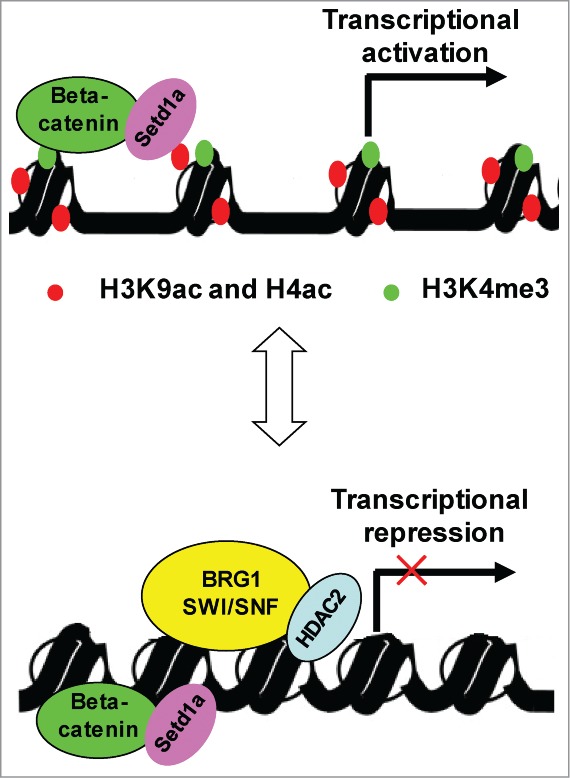
Proposed working model of BRG1-SWI/SNF complex (see text for details).
BRG1 and β-catenin in regulating hTERT expression
It was reported that β-catenin binds to the TSS of TERT promoter and activates TERT expression through its interaction with Setda1.26 The role of β-catenin in regulating hTERT was examined in HeLa cells. β-catenin-targeted siRNAs used in this experiment depleted β-catenin by 85% and had a limited effect on BRG1 expression (Fig. 6A), and reduced hTERT expression to 60% of control (Fig. 6B). The attenuated expression of hTERT by knockdown of β-catenin could be partially rescued by decreased number of BRG1 (Fig. 6A, B). One model to explain these data is that BRG1 and β-catenin act independently, to negatively and positively regulate telomerase expression, respectively. To further explore the function of BRG1, HDAC2 and β-catenin at TSS of hTERT promoter, we carried out the ChIP assay using antibody to HDAC2 and β-catenin. As expected, both HDAC2 and β-catenin are located to the TSS (Fig. 6C). The knockdown of BRG1 significantly diminishes the presence of HDAC2 at the TSS, indicating that the location of HDAC2 to the TSS is dependent on BRG1 (Fig. 6C, D). In contrast, the abundance of BRG1 has no effect on β-catenin binding to the TSS, further arguing that BRG1 and β-catenin function at TSS independently. Consistent with this model, the knockdown of BRG1 marginally reduces the abundance of β-catenin, but significantly increases hTERT expression (Fig. 2A; Fig. S4A). Similarly, overexpression of BRG1 inhibits hTERT transcription without a concomitant change in abundance of β-catenin (Fig. 3A; Fig. S4B).
Discussion
BRG1-SWI/SNF and HDAC2 coordinately suppress hTERT transcription in human cancer cells
In eukaryotic cells, DNA is packaged into a chromatin. Heterochromatin is a tightly packed DNA, which is associated with many biological processes such as cellular senescence,36 DNA damage response37,38 and telomere position effect (TPE).39,40 While the regulation of gene expression is often mediated by transcription factors and/or others,41,42 heterochromatin plays a central role in it. Because of the “closed” conformation of heterochromatin, gene promoter regions remain in general inaccessible to transcription factors.43 Transcriptionally active eukaryotic chromatin adopts an “open” conformation which allows the gene transcription machinery and gene-specific transcription factors to access cognate gene regulatory sequences.44 Chromatin remodeling enzymes alter the transcriptional status of chromatin, by promoting ATP-dependent change in chromatin structure or/and by modulating the histone code and adding or removing epigenic marks, including post-translational acetyl, methyl, phosphoryl, and/or ubiquitin groups on histones.19,45
In present study, we provide evidence that the BRG1-SWI/SNF complex and HDAC2 coordinately inhibit expression of hTERT in human cancer cells. Our data show that BRG1 physically interacts with HDAC2 and that the association of HDAC2 with the hTERT TSS is dependent on BRG1 (Fig. 6C). Moreover, overexpression of BRG1 inhibits transcription of hTERT in the absence but not in the presence of HDAC2 inhibitor TSA (Fig. 5C), suggesting that HDAC2 is indispensable for BRG1 to suppress hTERT transcription. Furthermore, BRG1 ATPase is required for its ability to inhibit hTERT transcription (Fig. 3B). These data suggest a model in which BRG-SWI/SNF binds to the hTERT TSS and facilitates the action of HDAC2 for local deacetylation of H3K9ac and H4ac and may then be retained at the TSS to maintain transcriptional repression. A schematic representation of this model is presented in Fig. 7.
Positive and negative regulation of hTERT expression by β-catenin and BRG1
There are several features that make BRG1 distinct from other hTERT transcription factors.17 First, BRG1 represses hTERT expression in all tested cell lines including human cervical cancer cells, embryonic kidney 293T cells and human breast cancer MDA-MB-231 cells, indicating that BRG1 may represent a general suppresser of hTERT transcription in human cells. Second, both ATPase activity of BRG1 and HDAC2 are required for suppression of hTERT expression. Therefore, chromatin remodeling and histone alteration are imposed upon hTERT promoter by BRG1 in order for suppressing hTERT transcription. Third, BRG1 and β-catenin constitute the transcriptional switcher at the TSS of hTERT promoter that regulates hTERT expression in a bi-directional manner.
Our data suggest that BRG1 and β-catenin function independently in regulating hTERT transcription. In the absence of BRG1 and HDAC2, β-catenin stimulates hTERT transcription by recruiting setd1a, an H3 lysine methyltransferase that trimethylates H3K4 to generate H3K4me3 at the hTERT TSS.26 H3K4me3, H3K9ac and H4ac ensure an open conformation of chromatin that allows the gene transcription (Fig. 7). BRG1 complex remodels the chromatin conformation and facilitates the action of HDAC2 that deacetylates H3K9ac and H4ac, thus converting local chromatin into a compact (close) conformation that suppresses the transcription of hTERT (Fig. 7). Therefore, the transcriptional status of hTERT depends on not only the abundance of β-catenin, but also BRG1, HDAC2 and BRG1-associated factors such as BAF47 (Fig. 3C).
It has been reported that BRG1 interacts with β-catenin and participates in transcriptional activation of Wnt-related genes.24 However, this appears not to be true with regard to expression of hTERT (Fig. 2B; 3A). Thus, our current data support the model that BRG1 and β-catenin have independent and opposite effects on expression of hTERT.
BRG1 in hTERT expression and telomere length maintenance
In normal human cells, telomerase expression is silenced. In contrast, most cancer cells re-activate telomerase in order to prevent replicative senescence linked to telomere shortening.46 The mechanism leading to reactivation of the hTERT promoter in cancer cells is not fully understood. However, BRG1 may play a critical role in preventing hTERT reactivation, in that inactivating mutations in BRG1 occur frequently in cancer cells.21,22 The findings presented here reinforce the idea that BRG1 acts as a tumor suppressor in human cells.
Even in telomerase-positive human cancer cells, it is estimated that only 1 to 5 molecules of hTERT mRNA are present per cell 47; this suggests that transcriptional repression of hTERT may dominate, even after it has been partially relieved. In this context, the balance between activation and repression may be critical to telomere length in human cancer cells. Indeed, either overexpression 48 or knockdown of hTERT 49 can alternate telomere length in cancer cells, indicating precise regulation of hTERT transcription in proliferating cells. Although this balance might be expected to vary in a cell type specific manner, the present study shows that knockdown of BRG1 correlated with increased abundance of hTERT mRNA in human cervical cancer, breast cancer MDA-MB-231 cells and embryonic kidney 293T cells. This suggests that BRG1-SWI/SNF and HDAC2 may play a general role in repressing hTERT in rapidly proliferating cells. Future studies are warranted to explore whether BRG1 and/or the SWI/SNF complex can be exploited as a therapeutic target in cancer patients, or in vitro as a tool for tissue engineering and regenerative medicine.
Materials and Methods
Cell culture and plasmids
HeLa, C33A, Caski, SiHA, HEK293, 293T, MDA-MB-231 cells were obtained from Cell Resource Center of Peking Union Medical College and were cultured at 37°C under 5% CO2. HeLa, C33A, Caski, SiHA, HEK293 and 293T were grown in DMEM (Hyclone) with 10% fetal calf serum (PPA). MDA-MB-231 was grown in L15 (Gibco) with 10% fetal calf serum (PPA). Trichostatin A (TSA) from Sigma was dissolved in DMSO (Sigma) and added to cell culture medium at a final concentration of 0.5 μM. Cells were grown in the presence of TSA for 24 h and harvested. Control cells were incubated with DMSO.
pBabe-puro-BRG1 was obtained from Addgene (MA, USA). pCMV5-HA-BRG1 and pCMV5-HA-BRG1-Trunc was constructed by deleting DNA sequence from 668 to 75850 of pBabe-puro-BRG1. The BAF47 gene was amplified from HEK293 mRNA and cloned into the pCMV5-HA vector.
Gene silencing and overexpression
siRNA was transfected into target cells in a 6-well plate using Lipo2000 (Invitrogen), according to the manufacturer's instructions. siRNA against BAF47 (5′-GUCAGAGAAGGAGAACUCAdTdT-3′) was provided by Shanghai GenePharma Co., Ltd. The scrambled sequence was used as a control.
The double-stranded shRNA against BRG1 (forward sequence:
5′-GATCCACATGCACCAGATGCACAATTCAAGAGATTGTGCATCTGGTGCATGT TTTTTTGGAAA-3′, reverse sequence:
3′-GTGTACGTGGTCTACGTGTTAAGTTCTCTAACACGTAGACCACGTACAAAAAAACCTTTTCGA-5′) were first cloned into pSilence 2.1-U6 vector, and then subcloned into pFG12 vector to yield pFG12-shBRG1. Lentivirus was packaged in 293T cells using calcium phosphate transfection. Viral supernatants were collected and used to infect target cells. Empty pFG12 vector was used as a control. Infected cells were selected by FACS based on fluorescence. siRNA knockdown of β-catenin was carried out in HeLa cells using Lipo2000 (Invitrogen) transfection. Si-β-cat: CAGUUGUGGUUAAGCUCUUdAdC /AAGAGCUUAACCACAACUGd-AdC. For protein overexpression, target genes were cloned into pCMV5-HA vector and transfected into HeLa cells, MDA-MB-231 or 293T cells using Lipo2000 (Invitrogen). After 48 h, cells were harvested, and hTERT mRNA was quantified by qRT-PCR analysis.
Immunoprecipitation and western blot
Cells were lysed in IP lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM Na4P2O7, 1 mM C3H7O6P-Na2, 1 mM Na3VO4) containing protease inhibitors. After removing cell debris by centrifugation, the supernatants were incubated with anti-HDAC1 (Beyotime) or anti-HDAC2 (Proteintech) with agitation overnight at 4°C. Immunoprecipitation was performed at 4°C with protein-A/G agarose beads (Santa Cruz) and rabbit IgG was used as a control. Beads were washed 4 times with lysis buffer and then incubated with 1×SDS-PAGE loading buffer, boiled for 10 min. Protein samples were analyzed by SDS-PAGE and Western blot.
Chromatin immunoprecipitation (ChIP)
Cells were cross-linked with 1% formaldehyde for 10 min at room temperature, washed twice with cold PBS, resuspended in SDS lysis buffer (50 mM Tris-HCl, pH = 8.1, 10 mM EDTA, 1% SDS) and sonicated to generate DNA fragments of ∼500 bp in length. The supernatant was pre-cleared with Protein-A Agarose beads precoated with Salmon Sperm DNA (Millipore). ChIP was performed overnight at 4°C with anti-BRG1 or anti-β-catenin (Cell signaling technology), anti-HDAC1 (Beyotime), anti-HDAC2 (Proteintech), anti-H3K9ac (Sigma), anti-H4ac (Millipore) and IgG (Sangon, Shanghai, China). Protein-A agarose beads were washed 3 times, and eluted with 0.1M NaHCO3 and 1% SDS, followed by reverse cross-linking and phenol-chloroform extraction. DNA fragments were precipitated by ethanol in the presence of DNAmate (Takara). PCR was carried out to identify DNA fragments enriched in the complexes. The following primers were used to identify fragments of hTERT promoter: A-1: 5′-CGTTGTGGCTGGTGTGAG-3′, 5′-CAC-CCCAAATCTGTTAATCACC-3′; A-2: 5′-TCCACTGTTTCATTTGTTGGTT-3′, 5′-CCAGCCTGAGCAACAAGAGT-3′; A-3: 5′-CCAAACCTGTGGACAGAACC-3′, 5′-AGACTGACTGCCTCCATCGT-3′; TS: 5′-AGCCCCTCCCCTTC-CTTTCC-3′, 5′-AGCGCACGGCTCGGCAGC-3′; A+2: 5′-GTCGAGTGGACACGGTGAT-3′, 5′-AAGTTTATGCAAA-CTGGACAGGA-3′.
Quantitative real-time PCR
Total RNA was extracted from cells using RNAiso Plus Reagent (Takara) according to manufacturer's instructions. Briefly, 1.0 μg of total RNA was reverse-transcribed to cDNA using PrimeScript RT reagent Kit (Takara). An equal amount of cDNA was used for real-time PCR using Realtime PCR Master Mix (ABI). GAPDH was used as internal control for all experiments. The threshold cycle (CT) value was calculated using the Step One software V2.1 provided by ABI, based on the first maximum of the second derivative of the amplification curve of template. Relative expression level of hTERT mRNA was calibrated by GAPDH mRNA. The following primers were used for cDNA amplification: GAPDH-forward: 5′- TGTTGCCATCAATGACCCCTT-3′; GAPDH-reverse: 5′-CTCCACGACGTACTCAGCG-3′; hTERT-forward: 5′-GGAGCAAGTTGCAAAGCATTG-3′; hTERT-reverse: 5′-CCCACGACGTAGTCCATGTT-3′.
Telomere restriction fragment (TRF) assay
The size of terminal restriction fragments was evaluated by digesting genomic DNA with HifI and RsaI, separating the digested DNA on a 0.7% agarose gel, and then hybridizing the denatured, dried gel with a 32P-labeled telomeric probe.51
Telomerase assay
TRAP assay was performed as described previously.49,52 The telomerase products (6 bp ladder) and the 36 bp internal control (IC) bands were quantified using the ImageQuant software provided by the manufacturer. Relative telomerase activity was calculated as the intensity ratio of the TRAP ladder to that of the IC band.
Funding Statement
This work was supported by National Natural Science Foundation of China Grants (20921062, 30570960); and National Basic Research Program of China (2014CB964703); Guangdong Innovative Research Team Program (201001Y0104687244); and Shenzhen Science & Technology Program (ZDSY20120616222747467).
Disclosure of Potential Conflict of Interest
No potential conflicts of interest were disclosed.
Author Contributions
ZY, WS, and GY generated the hypotheses, designed experiments. WS, GY, Liu H performed experiments and generated and analyzed data. Huang L and Xue Y contributed to data interpretation. ZY and WS wrote the manuscript.
Acknowledgment
We thank Dr Miriam Sander for professional scientific editing.
Supplemental Materials
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 2005; 19:2100-10; PMID:16166375; http://dx.doi.org/ 10.1101/gad.1346005 [DOI] [PubMed] [Google Scholar]
- 2. Miller AS, Balakrishnan L, Buncher NA, Opresko PL, Bambara RA. Telomere proteins POT1, TRF1 and TRF2 augment long-patch base excision repair in vitro. Cell Cycle 2012; 11:998-1007; PMID:22336916; http://dx.doi.org/ 10.4161/cc.11.5.19483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature 2007; 445:559-62; PMID:17237767; http://dx.doi.org/ 10.1038/nature05469 [DOI] [PubMed] [Google Scholar]
- 4. Houghtaling BR, Canudas S, Smith S. A role for sister telomere cohesion in telomere elongation by telomerase. Cell Cycle 2012; 11:19-25; PMID:22157096; http://dx.doi.org/ 10.4161/cc.11.1.18633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mitchell TR, Zhu XD. Methylated TRF2 associates with the nuclear matrix and serves as a potential biomarker for cellular senescence. Aging (Albany NY) 2014; 6:248-63; PMID:24721747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature 1990; 345:458-60; PMID:2342578; http://dx.doi.org/ 10.1038/345458a0 [DOI] [PubMed] [Google Scholar]
- 7. Chow TT, Zhao Y, Mak SS, Shay JW, Wright WE. Early and late steps in telomere overhang processing in normal human cells: the position of the final RNA primer drives telomere shortening. Genes Dev 2012; 26:1167-78; PMID:22661228; http://dx.doi.org/ 10.1101/gad.187211.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shay JW, Wright WE. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis 2005; 26:867-74; PMID:15471900; http://dx.doi.org/ 10.1093/carcin/bgh296 [DOI] [PubMed] [Google Scholar]
- 9. Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science 1994; 266:2011-5; PMID:7605428; http://dx.doi.org/ 10.1126/science.7605428 [DOI] [PubMed] [Google Scholar]
- 10. Greider CW. Telomeres, telomerase and senescence. Bioessays 1990; 12:363-9; PMID:2241933; http://dx.doi.org/ 10.1002/bies.950120803 [DOI] [PubMed] [Google Scholar]
- 11. Cong YS, Wright WE, Shay JW. Human telomerase and its regulation. Microbiol Mol Biol Rev 2002; 66:407-25, table of contents; PMID:12208997; http://dx.doi.org/ 10.1128/MMBR.66.3.407-425.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ito H, Kyo S, Kanaya T, Takakura M, Inoue M, Namiki M. Expression of human telomerase subunits and correlation with telomerase activity in urothelial cancer. Clin Cancer Res 1998; 4:1603-8; PMID:9676833 [PubMed] [Google Scholar]
- 13. Takakura M, Kyo S, Kanaya T, Tanaka M, Inoue M. Expression of human telomerase subunits and correlation with telomerase activity in cervical cancer. Cancer Res 1998; 58:1558-61; PMID:9537264 [PubMed] [Google Scholar]
- 14. Cong YS, Wen J, Bacchetti S. The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum Mol Genet 1999; 8:137-42; PMID:9887342; http://dx.doi.org/ 10.1093/hmg/8.1.137 [DOI] [PubMed] [Google Scholar]
- 15. Horikawa I, Cable PL, Afshari C, Barrett JC. Cloning and characterization of the promoter region of human telomerase reverse transcriptase gene. Cancer Res 1999; 59:826-30; PMID:10029071 [PubMed] [Google Scholar]
- 16. Takakura M, Kyo S, Kanaya T, Hirano H, Takeda J, Yutsudo M, Inoue M. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res 1999; 59:551-7; PMID:9973199 [PubMed] [Google Scholar]
- 17. Cukusic A, Skrobot Vidacek N, Sopta M, Rubelj I. Telomerase regulation at the crossroads of cell fate. Cytogenet Genome Res 2008; 122:263-72; PMID:19188695; http://dx.doi.org/ 10.1159/000167812 [DOI] [PubMed] [Google Scholar]
- 18. Sanduja S, Kaza V, Dixon DA. The mRNA decay factor tristetraprolin (TTP) induces senescence in human papillomavirus-transformed cervical cancer cells by targeting E6-AP ubiquitin ligase. Aging (Albany NY) 2009; 1:803-17; PMID:20157568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res 2011; 21:396-420; PMID:21358755; http://dx.doi.org/ 10.1038/cr.2011.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tyler JK, Kadonaga JT. The "dark side" of chromatin remodeling: repressive effects on transcription. Cell 1999; 99:443-6; PMID:10589670; http://dx.doi.org/ 10.1016/S0092-8674(00)81530-5 [DOI] [PubMed] [Google Scholar]
- 21. Wong AK, Shanahan F, Chen Y, Lian L, Ha P, Hendricks K, Ghaffari S, Iliev D, Penn B, Woodland AM, et al. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res 2000; 60:6171-7; PMID:11085541 [PubMed] [Google Scholar]
- 22. Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 2011; 11:481-92; PMID:21654818; http://dx.doi.org/ 10.1038/nrc3068 [DOI] [PubMed] [Google Scholar]
- 23. Bultman SJ, Herschkowitz JI, Godfrey V, Gebuhr TC, Yaniv M, Perou CM, Magnuson T. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene 2008; 27:460-8; PMID:17637742; http://dx.doi.org/ 10.1038/sj.onc.1210664 [DOI] [PubMed] [Google Scholar]
- 24. Barker N, Hurlstone A, Musisi H, Miles A, Bienz M, Clevers H. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. Embo J 2001; 20:4935-43; PMID:11532957; http://dx.doi.org/ 10.1093/emboj/20.17.4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Griffin CT, Curtis CD, Davis RB, Muthukumar V, Magnuson T. The chromatin-remodeling enzyme BRG1 modulates vascular Wnt signaling at two levels. Proc Natl Acad Sci U S A 2011; 108:2282-7; PMID:21262838; http://dx.doi.org/ 10.1073/pnas.1013751108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoffmeyer K, Raggioli A, Rudloff S, Anton R, Hierholzer A, Del Valle I, Hein K, Vogt R, Kemler R. Wnt/beta-catenin signaling regulates telomerase in stem cells and cancer cells. Science 2012; 336:1549-54; PMID:22723415; http://dx.doi.org/ 10.1126/science.1218370 [DOI] [PubMed] [Google Scholar]
- 27. Zhang Y, Toh L, Lau P, Wang X. Human telomerase reverse transcriptase (hTERT) is a novel target of the Wnt/beta-catenin pathway in human cancer. J Biol Chem 2012; 287:32494-511; PMID:22854964; http://dx.doi.org/ 10.1074/jbc.M112.368282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, Liu L, Liu W, Wu LM, Mao M, et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell 2013; 152:248-61; PMID:23332759; http://dx.doi.org/ 10.1016/j.cell.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Park JI, Venteicher AS, Hong JY, Choi J, Jun S, Shkreli M, Chang W, Meng Z, Cheung P, Ji H, et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 2009; 460:66-72; PMID:19571879; http://dx.doi.org/ 10.1038/nature08137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wright WE, Shay JW, Piatyszek MA. Modifications of a telomeric repeat amplification protocol (TRAP) result in increased reliability, linearity and sensitivity. Nucleic Acids Res 1995; 23:3794-5; PMID:7479015; http://dx.doi.org/ 10.1093/nar/23.18.3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Trotter KW, Archer TK. Nuclear receptors and chromatin remodeling machinery. Mol Cell Endocrinol 2007; 265–266:162-7; PMID:17240047; http://dx.doi.org/ 10.1016/j.mce.2006.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. Embo J 1996; 15:5370-82; PMID:8895581 [PMC free article] [PubMed] [Google Scholar]
- 33. Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell 1999; 3:247-53; PMID:10078207; http://dx.doi.org/ 10.1016/S1097-2765(00)80315-9 [DOI] [PubMed] [Google Scholar]
- 34. Hassan AH, Neely KE, Vignali M, Reese JC, Workman JL. Promoter targeting of chromatin-modifying complexes. Front Biosci 2001; 6:D1054-64; PMID:11532604; http://dx.doi.org/ 10.2741/Hassan [DOI] [PubMed] [Google Scholar]
- 35. Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, Zhou B, Chang CP. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 2010; 466:62-7; PMID:20596014; http://dx.doi.org/ 10.1038/nature09130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kosar M, Bartkova J, Hubackova S, Hodny Z, Lukas J, Bartek J. Senescence-associated heterochromatin foci are dispensable for cellular senescence, occur in a cell type- and insult-dependent manner and follow expression of p16(ink4a). Cell Cycle 2011; 10:457-68; PMID:21248468; http://dx.doi.org/ 10.4161/cc.10.3.14707 [DOI] [PubMed] [Google Scholar]
- 37. Soria G, Almouzni G. Differential contribution of HP1 proteins to DNA end resection and homology-directed repair. Cell Cycle 2013; 12:422-9; PMID:23287531; http://dx.doi.org/ 10.4161/cc.23215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ball AR, Jr., Yokomori K. Damage site chromatin: open or closed? Curr Opin Cell Biol 2011; 23:277-83; PMID:21489773; http://dx.doi.org/ 10.1016/j.ceb.2011.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baur JA, Zou Y, Shay JW, Wright WE. Telomere position effect in human cells. Science 2001; 292:2075-7; PMID:11408657; http://dx.doi.org/ 10.1126/science.1062329 [DOI] [PubMed] [Google Scholar]
- 40. Lou Z, Wei J, Riethman H, Baur JA, Voglauer R, Shay JW, Wright WE. Telomere length regulates ISG15 expression in human cells. Aging (Albany NY) 2009; 1:608-21; PMID:20157543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell 2004; 119:157-67; PMID:15479634; http://dx.doi.org/ 10.1016/j.cell.2004.09.037 [DOI] [PubMed] [Google Scholar]
- 42. Cuadrado A, Remeseiro S, Gomez-Lopez G, Pisano DG, Losada A. The specific contributions of cohesin-SA1 to cohesion and gene expression: implications for cancer and development. Cell Cycle 2012; 11:2233-8; PMID:22617390; http://dx.doi.org/ 10.4161/cc.20318 [DOI] [PubMed] [Google Scholar]
- 43. Felsenfeld G, Groudine M. Controlling the double helix. Nature 2003; 421:448-53; PMID:12540921; http://dx.doi.org/ 10.1038/nature01411 [DOI] [PubMed] [Google Scholar]
- 44. Berger SL. The complex language of chromatin regulation during transcription. Nature 2007; 447:407-12; PMID:17522673; http://dx.doi.org/ 10.1038/nature05915 [DOI] [PubMed] [Google Scholar]
- 45. Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev 2002; 12:142-8; PMID:11893486; http://dx.doi.org/ 10.1016/S0959-437X(02)00279-4 [DOI] [PubMed] [Google Scholar]
- 46. Shay JW, Roninson IB. Hallmarks of senescence in carcinogenesis and cancer therapy. Oncogene 2004; 23:2919-33; PMID:15077154; http://dx.doi.org/ 10.1038/sj.onc.1207518 [DOI] [PubMed] [Google Scholar]
- 47. Yi X, Shay JW, Wright WE. Quantitation of telomerase components and hTERT mRNA splicing patterns in immortal human cells. Nucleic Acids Res 2001; 29:4818-25; PMID:11726691; http://dx.doi.org/ 10.1093/nar/29.23.4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. Embo J 2006; 25:565-74; PMID:16424902; http://dx.doi.org/ 10.1038/sj.emboj.7600952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhao Y, Abreu E, Kim J, Stadler G, Eskiocak U, Terns MP, Terns RM, Shay JW, Wright WE. Processive and distributive extension of human telomeres by telomerase under homeostatic and nonequilibrium conditions. Mol Cell 2011; 42:297-307; PMID:21549308; http://dx.doi.org/ 10.1016/j.molcel.2011.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Romero OA, Setien F, John S, Gimenez-Xavier P, Gomez-Lopez G, Pisano D, Condom E, Villanueva A, Hager GL, Sanchez-Cespedes M. The tumour suppressor and chromatin-remodelling factor BRG1 antagonizes Myc activity and promotes cell differentiation in human cancer. EMBO Mol Med 2012; 4:603-16; PMID:22407764; http://dx.doi.org/ 10.1002/emmm.201200236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ouellette MM, Liao M, Herbert BS, Johnson M, Holt SE, Liss HS, Shay JW, Wright WE. Subsenescent telomere lengths in fibroblasts immortalized by limiting amounts of telomerase. J Biol Chem 2000; 275:10072-6; PMID:10744686; http://dx.doi.org/ 10.1074/jbc.275.14.10072 [DOI] [PubMed] [Google Scholar]
- 52. Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer 1997; 33:787-91; PMID:9282118; http://dx.doi.org/ 10.1016/S0959-8049(97)00062-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.