Abstract
Cyclin-dependent kinases (CDKs) are central regulators of eukaryotic cell cycle progression. In contrast to interphase CDKs, the mitotic phase CDK1 is the only CDK capable of driving the entire cell cycle and it can do so from yeast to mammals. Interestingly, plants and the marine chordate, Oikopleura dioica, possess paralogs of the highly conserved CDK1 regulator. However, whereas in plants the 2 CDK1 paralogs replace interphase CDK functions, O. dioica has a full complement of interphase CDKs in addition to its 5 odCDK1 paralogs. Here we show specific sub-functionalization of odCDK1 paralogs during oogenesis. Differential spatiotemporal dynamics of the odCDK1a, d and e paralogs and the meiotic polo-like kinase 1 (Plk1) and aurora kinase determine the subset of meiotic nuclei in prophase I arrest that will seed growing oocytes and complete meiosis. Whereas we find odCDK1e to be non-essential, knockdown of the odCDK1a paralog resulted in the spawning of non-viable oocytes of reduced size. Knockdown of odCDK1d also resulted in the spawning of non-viable oocytes. In this case, the oocytes were of normal size, but were unable to extrude polar bodies upon exposure to sperm, because they were unable to resume meiosis from prophase I arrest, a classical function of the sole CDK1 during meiosis in other organisms. Thus, we reveal specific sub-functionalization of CDK1 paralogs, during the meiotic oogenic program.
Keywords: aurora kinase, centrosome, endocycle, MTOC, polo-like kinase, syncytium, urochordate
Abbreviations
- CDK
Cyclin Dependent Kinase
- cmRNA
capped messenger RNA
- dsRNA
double-stranded RNA
- MAPK
Mitogen-Activated Protein Kinase
- MTOC
microtubule organizing center
- DMYPT
Drosophila myosin phosphatase
- NEBD
nuclear envelope breakdown
- NPC
Nuclear Pore Complex
- OC
Organizing Center
- Plk1
Polo-like kinase 1
- GVBD
germinal vesicle breakdown
Introduction
The cyclinB-CDK1 (Maturation Promoting Factor; MPF) complex is a focal regulatory module during both mitosis and meiosis.1 Release from metazoan, meiotic, prophase I primary arrest in immature oocytes can be viewed as similar to the G2-M phase transition in mitosis,2 in that both require activation of the cyclinB-CDK1 kinase. The activities of MAPK and polo-like kinase (Plk1) are also integral to the meiotic cycle.3,4 While the oocytes of some animals, such as nematodes, only undergo the primary prophase I arrest, additional intricacy in the meiotic regulation of CDK1 activity can be imposed by a secondary arrest at metaphase I (many invertebrates), metaphase II (most vertebrates) or the post-meiotic G1 phase (sea urchins and starfish).2 Further constraints on the spatiotemporal regulation of CDK1 activity are required in cases where meiotic nuclei in immature oocytes share a common cytoplasm with endocycling nurse nuclei, dedicated to generating the maternal components stocked in the oocyte. In Drosophila, 15 endocycling nurse nuclei share the ovariole with a single meiotic nucleus,5 whereas in the urochordate, Oikopleura dioica, thousands of nurse and meiotic nuclei share a common cytoplasm, in a 1:1 ratio, in one giant cell, the coenocyst.6,7 Absence of CDK1 activation during entry into, and maintenance of, endocycles in vertebrates and Drosophila is required to prevent M-phase entry.8-11 Inactivation of CDK1 activity is also required in meiotic prophase I arrest.12-14 On the other hand, CDK1 is required for resumption of meiosis from prophase I arrest and once activated,2,15 maintains meiotic progression until completion of anaphase II.2,16 Thus, though both meiotic arrest and endocycles depend on the inhibition of CDK1 activity, additional spatiotemporal regulatory finesse may be necessary to coordinate events around the increase in CDK1 activity required to drive resumption of meiosis, in cases where such different cell cycles share a common cytoplasm.
Centrosomal, microtubule organizing centers (MTOCs) at the mitotic spindle poles are essential in enabling correct sister chromatid separation during mitosis and meiosis and a number of mitotic kinases, including CDK1, and Plk1, localize to spindle poles prior to nuclear envelope breakdown (NEBD). Daughter cells then inherit one MTOC each, which duplicates during interphase in a process known as the centrosome cycle.17 Orthogonally paired centrioles are a usual component of centrosomes, but in vertebrate oocytes the centrioles are lost, a mechanism that ensures inheritance of a pair of centrosomes from sperm. In this case, acentriolar MTOCs coordinate asymmetric cell divisions producing oocyte polar bodies.18-20 To our knowledge, the presence of CDK1 on meiotic centrosomes has not been directly demonstrated. Nonetheless, protein kinase B, implicated in the initial activation of CDK1 in mouse oocytes, has been found to localize to acentriolar meiotic centrosomes.21 Furthermore, though not specifically identified as centrosomes at the time, cyclin B1 localizes to centrosome-like structures adjacent to the nuclear envelope just prior to germinal vesicle breakdown (GVBD) in mouse oocytes.22
Given that endocycles involve inactivation of CDK1, and that O. dioica extensively employs endocycles in development, patterning, rapid growth and the female germline,6,23-25 it was surprising to find that it is the only metazoan known to possess more than one CDK1 paralog.26 An unusual feature is that none of the 5 odCDK1 paralogs possesses a perfect PSTAIRE motif, which is invariant in known CDK1s of other metazoans. Several odCDK1 paralogs possess a proline substitution that mimics plant CDK1 homologs (CDKBs),27 and all odCDK1 paralogs share an alanine to serine substitution at position 4 of the PSTAIRE motif (e.g. odCDK1a: PSTSIRE) that mimics the PLSTIRE motif of vertebrate CDK6. Transcripts for the odCDK1a, b and c paralogs are present throughout the life cycle, including stages predominated by endocycles, whereas odCDK1d and e paralogs are essentially limited to expression in maturing females and early embryos.26 The strong enrichment of odCDK1d and e paralog expression in maturing females, contrasts a modest enrichment of odCDK1b and c paralogs in maturing males. The odCDK1a paralog exhibits a slight enrichment in expression in maturing females as opposed to maturing males.
Here we have investigated the roles of the odCDK1a, d and e paralogs during coenocystic oogenesis in O. dioica. Previous studies have shown that the female germline initially undergoes syncytial mitotic nuclear divisions from metamorphosis to day 3 of the life cycle yielding between 103–104 nuclei.6,7,28 At this point (phase P1), female germline nuclei undergo fate differentiation to generate a 1:1 ratio of asynchronously endocycling nurse nuclei and meiotic nuclei in prophase I. Each meiotic nucleus seeds a small pro-oocyte, connected to the general coenocyst cytoplasm by a ring canal. During phase P2, meiotic nuclei progress to zygotene and in P3, the coenocyst grows considerably in size. Upon the transition to P4, a subset of pro-oocytes, with their resident meiotic nuclei, is selected to increase in size via transfer of cytoplasmic material through the ring canal and all meiotic chromatin condenses into the π−configuration. Finally, during P5, oocyte maturation is completed, nurse and excess meiotic nuclei undergo apoptosis in a minimal cytoplasmic volume, and spawning occurs on day 6 via rupture of the ovarian wall. In this study we characterized kinase dynamics focused on a meiotic MTOC-like structure in the O. dioica ovary as the central organizing node. We found that odCDK1 paralogs, Plk1 and Aurora kinase exhibit different temporal spatial dynamics at meiotic nuclei selected to populate growing oocytes, versus those that were not selected and simply collected in the general coenocyst cytoplasm to eventually undergo apoptosis. We then revealed differential essential functions for the odCDK1a and d paralogs, in the regulation of oocyte size (vitellogenesis) and resumption of meiosis from prophase I arrest. Whereas no discernable effects of the knockdown of odCDK1e were observed on the developmental potential of spawned oocytes, impairment of the activity of either of the odCDK1a or d paralogs resulted in the spawning of infertile oocytes.
Results
MTOC-like organizing center (OC) dynamics during oogenesis
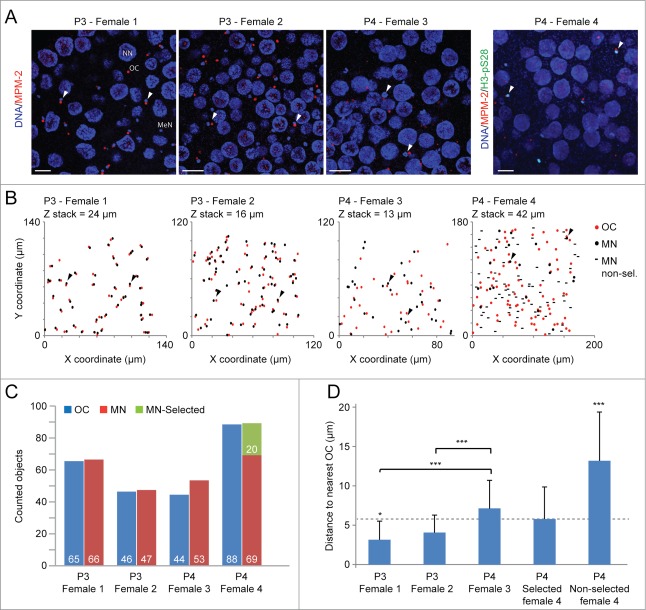
In order to observe meiotic kinase activity within the coenocyst, animals were collected and fixed at days 4 and 5 and assessed by immunostaining with the MPM-2 primary antibody targeting mitotic phospho epitopes,29 also phosphorylated during meiosis. Kinase activity was revealed within cytoplasmic bodies found in proximity to each of the meiotic prophase I nuclei during the P3 stage of oogenesis (Fig. 1A, B). The presence of pERK1/2 on microtubule organizing centers (MTOC) in O. dioica embryos28, co-localization of MPM-2 with gamma-tubulin, and MPM-2 staining of MTOCs in O. dioica testes (Fig. S1, see also Fig. 2A), all indicate the cytoplasmic ovarian MPM-2 stained structures to have centrosomal organizing characteristics. We subsequently refer to these structures as organizing centers (OCs). MPM-2 staining also appeared on smaller foci within nurse nuclei, an observation similar to that with pERK1/2 localization following activation of the MAPK pathway.28 OCs were not associated with nurse nuclei but were observed at a 1:1 ratio to meiotic nuclei (Fig. 1C). Most OC lost association with meiotic nuclei upon entry into the P4 stage (Fig. 1D). On the other hand, OCs remained juxtaposed to meiotic nuclei that were selected to populate maturing oocytes, as judged by persistence of H3-pS28 staining (Fig. 1D).28 Since they populate growing oocytes, selected meiotic nuclei are also identified in images where H3-pS28 staining is not used, by the cytoplasmic space that separates them from nurse nuclei. In contrast, non-selected meiotic nuclei often juxtapose to nurse nuclei or exhibit a minimal cytoplasmic distance from them.
Figure 1.
Organizing Center dynamics within the O. dioica coenocyst. Stages pre- (P3) and post- (P4) selection of meiotic nuclei (MeN) to populate growing pro-oocytes (P3/4, phase 3/4; (see refs. 6,7,28) were assessed. A) MPM-2 immunofluorescence reveals cytoplasmic organizing centers (OC) within the coenocyst. Figure represents confocal stack Z-projections with a depth of 3 μm. Arrowheads indicate points of reference in B. Scale bars = 25 μm. B) Scatter plot of OCs and meiotic nuclei XY coordinates (in μm) through confocal Z stack (depths displayed for each stack) reveal that OCs (stained by MPM-2 antibody) were juxtaposed to meiotic nuclei in the coenocyst prior to oocyte selection. In P4, the close juxtaposition of OCs to meiotic nuclei was reduced, though it was preferentially retained in meiotic nuclei selected to seed growing oocytes (H3-pS28 positive). OC, organizing center; MN, meiotic nuclei, MN non-sel, meiotic nuclei not selected to populate growing oocytes. C) The total number of OCs coincided with the number of meiotic nuclei, indicating that terminally differentiated endocycling nurse nuclei (NN) did not retain a juxtaposed OC. MN-selected, meiotic nuclei selected to populate growing oocytes. D) Graphical representation of measured distances between meiotic nuclei and proximal OCs confirms that OC proximity is reduced in non-selected nuclei.
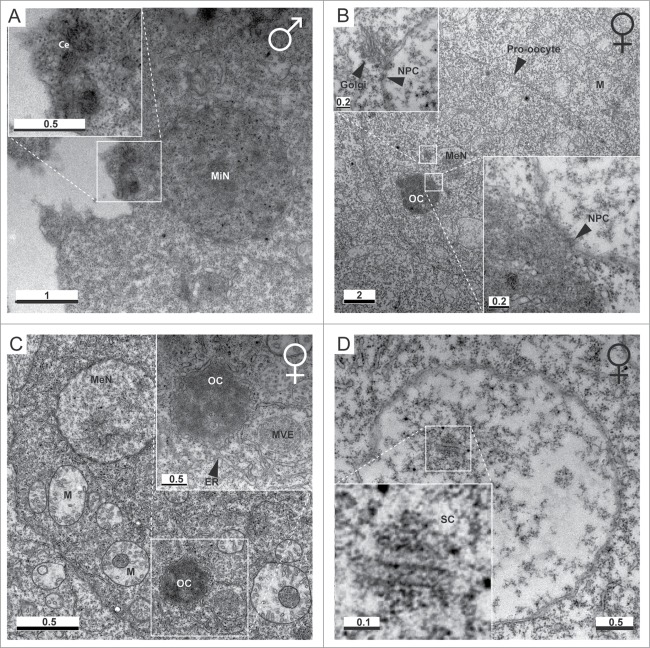
Figure 2.
Ultrastructure of the O. dioica germline. (A) Day 5 male mitotic germline nuclei (MiN) are each associated with an MTOC that possesses a pair of centrioles (Ce). (B) Centrioles were not observed within female P3 ovaries. Meiotic nuclei (MeN) possessed nuclear membranes containing nuclear pore complexes (NPC). Golgi apparati were observed in proximity to the nuclear membrane. An electron dense structure of similar size to meiotic nuclei was observed in contact with the nuclear envelope, consistent with the immunofluorescent staining (Fig. 3) of acentriolar OCs. NPCs were observed at the OC/MN interface. (C) OCs were surrounded by endoplasmatic reticulum (ER). M, mitochondria; MVE, multi-vesicular endosome-like structure. (D) Electron dense regions resembling 3 parallel lines, with 100 nm gaps between the central and flanking lines, were observed, consistent with the structure of synaptonemal complexes (SCs). Scale bars in μm.
Transmission electron microscopy of ultra-thin sections of the male testes (Fig. 2A) vs. the female coenocyst in P3 (Fig. 2B, C) revealed ultrastructural differences in the respective OCs. Paired centrioles were clearly detected in MTOCs associated with each of the mitotic germline nuclei in male testes (Fig. 2A). On the other hand, OCs in the P3 ovary appeared as electron dense structures that did not contain centrioles (Fig. 2B, C). Ovarian OC were surrounded by endoplasmic reticulum (ER) and resided within pro-oocytes in close proximity to meiotic nuclei. The OC was sometimes found in close contact with the nuclear membrane of its associated meiotic nucleus, where nuclear pore complexes (NPC) were present. Synaptonemal complexes, characteristic of zygotene/pachytene were also observed in meiotic nuclei during P3 (Fig. 2D). Since primary prophase I meiotic arrest usually occurs at diplotene or diakinesis, and meiotic nuclei in the P3 stage display synaptonemal complexes, we conclude that O. dioica oocytes have not yet entered primary arrest during the P3 stage of oogenesis. O. dioica meiotic nuclei enter primary arrest in diakinesis, consistent with DNA condensation in π−configuration, during P4.7 Immunofluorescence also revealed the co-localization of pERK1/2 and MPM-2 on OC (Fig. 3A), consistent with pERK1/2 being a meiotic regulator.1,30
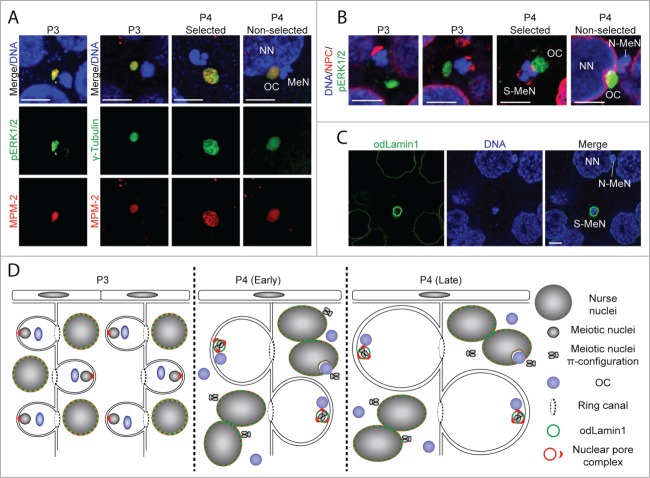
Figure 3.
Kinase activity at OCs and rearrangement of the nuclear envelope of meiotic nuclei. (A) Activation of the MAPK pathway and phosphorylation of meiotic substrates occurs at OCs, analogous to MTOCs (gamma-tubulin staining), as indicated by pERK1/2 and MPM-2 stainings, respectively. MeN, meiotic nucleus; NN, nurse nucleus. (B) OCs are positioned toward the vegetal pole of pro-oocytes, opposite clustered nuclear pore complexes (NPCs) at the animal pole in P3.7 In selected pro-oocyte meiotic nuclei (S-MeN), NPCs form clusters at the OC-nucleus interface at P4. Non-selected meiotic nuclei (N-MeN) associate with nurse nuclei, along with OCs which localize within a large invagination of the nurse nuclear envelope. (C) Nurse nuclei and selected meiotic nuclei of P4 ovaries are bounded by O. dioica lamin1,44 whereas no odLamin1 was observed surrounding non-selected meiotic nuclei. D) Schematic representation of events in the coenocyst during P3 and P4, illustrating NPC, OC, and lamin dynamics from pre- to post-selection of meiotic nuclei. (A-C) Scale bars = 5 μm.
Disposition of the Organizing Center with respect to nuclear envelope components of selected and non-selected meiotic nuclei during oogenesis
During P3 in the coenocyst, NPCs form clusters in the meiotic nuclear envelope, juxtaposed to an actin scaffold at the future animal pole.7 The individual OCs associated with each meiotic nucleus were oriented opposite the NPC clusters in P3 ovaries (Fig. 3B). During the P4 stage, OCs contacted the selected meiotic nuclei. This coincided with further fragmentation of the NPC clusters such that one cluster became localized at the nuclear-OC interface (Fig. 3B). NPC clusters did not appear at the nuclear-OC interface of non-selected nuclei, and these nuclei became less closely associated with OCs. Instead, these “released” OCs increasingly associated with nurse nuclei, often found in pockets formed by large invaginations of the nurse nuclear envelope (Fig. 3B).
The nuclear lamina plays roles in localization and function of NPCs.31 In concert with NPC re-localization in the nuclear envelope during P4, we also observed the appearance of the odlamin1 splice variant in the selected meiotic nuclei, which persisted through P4 and P5 (spawning), whereas non-selected nuclei did not display Lamin1 envelopment during P4 (Fig. 3C). The progression of these events is summarized schematically in Fig. 3D.
CDK1 paralogs translocate from Organizing Centers to meiotic nuclei during the progression of coenocystic oogenesis
During P3, Plk1 and Aurora kinases were observed at OCs which co-stained with MPM-2 phospho-epitopes (Fig. 4). Aurora was also present within meiotic nuclei during P3 whereas Plk1 was not (Fig. 4A). Active Plk1 translocated exclusively to selected nuclei in P4, whereas active Aurora was retained in selected, but lost from non-selected meiotic nuclei. The loss of active Aurora kinase from non-selected meiotic nuclei paralleled the loss of H3-pS28 staining in these same nuclei (Fig. S2A and 28). The presence of Plk1 in selected meiotic nuclei in growing oocytes, would be consistent with the previously reported role of Plk1 in disassembly of synaptonemal complexes required for exit from pachytene.32
Figure 4.
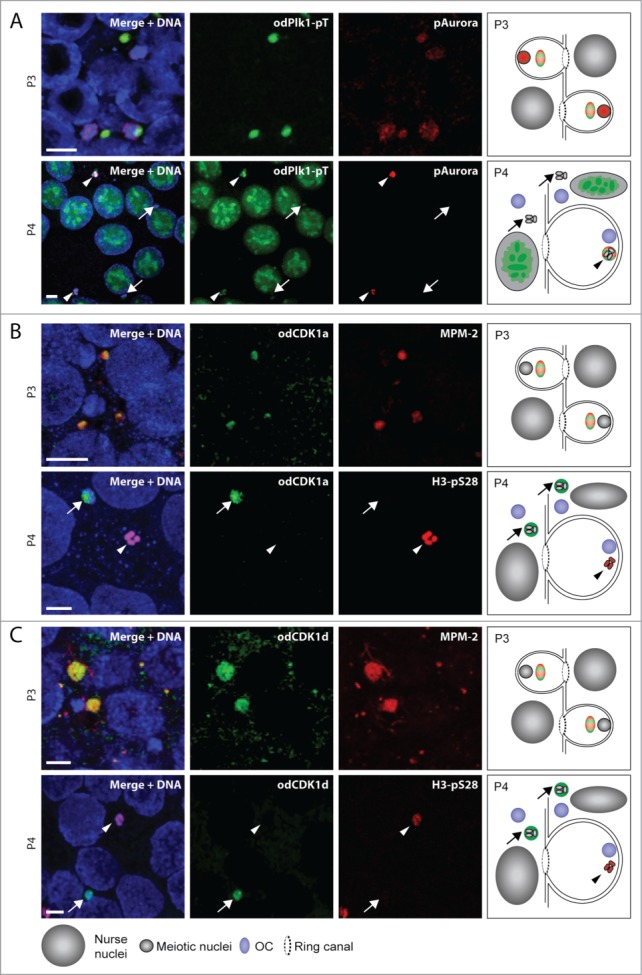
Localization of cell cycle kinases within the P3 and P4 coenocyst. (A) Phospho-Plk1 and phospho-Aurora localized to OCs juxtaposed to meiotic nuclei in P3. Active Aurora kinase was also present in meiotic nuclei at this stage. In P4, active Plk1 translocated only to meiotic nuclei (arrowheads) that had been selected (H3-pS28 positive; see P4 panels in (B and C) and Fig. S2A) to populate growing oocytes. It was not present on non-selected meiotic nuclei (arrows) and was no longer present on OCs. During this stage, active Aurora kinase was retained only in meiotic nuclei that had been selected to populate growing oocytes, and was no longer present on OCs or on unselected meiotic nuclei. At the P4 stage, chromatin in all meiotic nuclei had attained the more condensed π−configuration.6,7,28 B) O. dioica CDK1a localized to MPM-2 stained OCs juxtaposed to meiotic nuclei in P3. In P4, odCDK1a was no longer observed at OCs. Instead, it was present on meiotic nuclei which did not retain H3-pS28 staining (arrows) and had not been selected to populate growing oocytes. (C) O. dioica CDK1d exhibited the same spatiotemporal P3 to P4 dynamics as od CDK1a. Schemas in the right column of panels summarize the results in the corresponding rows. Coloring of epitopes in the schemas corresponds to that given in the labels at the left side of each row of images. Scale bars = 5 μm.
Of the 5 O. dioica CDK1 paralogs, odCDK1a shows the closest phylogenetic affinity to CDK1 homologs in other species.26 It possesses the least derived PSTAIRE motif (PSTSIRE) among the 5 paralogs and is expressed throughout development, at similar levels in males and females. odCDK1b and c are also expressed throughout somatic development but in addition exhibit preferential expression in males compared to females. On the other hand, odCDK1d and e expression is restricted to maturing females and early embryonic development.26 Capped mRNAs (cmRNAs) encoding eGFP fusions with the 3 odCDK1 paralogs (a, d and e) expressed during oogenesis, were injected into gonads of day 4 animals which were subsequently cultured until day 5, and then assessed for CDK1 paralog localization. During P3, the 3 odCDK1 paralogs, were observed at OCs, where they co-stained with MPM-2 phospho-epitopes (Fig. 4B, C; Fig. S2B). The three odCDK1 paralogs were never observed within nurse nuclei, consistent with the down regulation of CDK1 activity during endocycling in the Drosophila and mouse models.8,9,33 During P4, odCDK1a, d and e were observed within non-selected meiotic nuclei (Fig. 4B, C; Fig. S2B), but were not observed within selected nuclei.
The spatiotemporal localizations of the evaluated cell cycle kinases and markers are summarized schematically in Fig. 5. During P3, cell cycle kinases important in the progression of mitosis and meiosis were present on the OC. In P4, concomitant with the selection of meiotic nuclei that will populate growing oocytes, there was a selective release of kinases into the selected (Plk1 and Aurora) or non-selected meiotic nuclei (CDK1 a, d and e). In selected meiotic nuclei in P4, both the persistence of lamin staining, and the absence of translocated CDK1, indicate that these nuclei are still in prophase I of meiosis, consistent with EM data indicating synaptonemal complexes at this stage (Fig. 2D).
Figure 5.
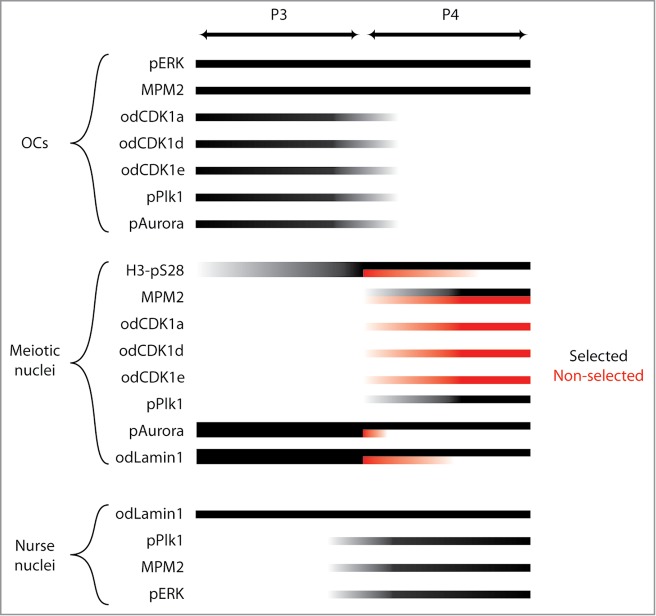
Summary of localizations of cell cycle regulators and markers during P3 and P4 in the O. dioica coenocyst, with respect to organizing centers (OC), meiotic nuclei in prophase I, and endocycling nurse nuclei. In P4, a proportion of the meiotic nuclei had been selected to populate growing oocytes (black bars), whereas the remaining meiotic nuclei had not been selected (red bars) and were present in the general coenocyst cytoplasm. The degree of presence of each parameter is indicated by degree of shading of the lines from light (weakly present) to dark (strongly present). M-phase MPM-2 phosphoepitopes were observed on OCs throughout P3 and P4, as was pERK. Cell cycle regulators odCDK1a,d,e, phospho-Plk1 (pPLK1) and phospho-Aurora (pAurora) were present on OCs during P3 but lost from OCs during P4. Toward the transition from P3 to P4 H3-pS28 staining increased on meiotic nuclei. This staining was retained throughout P4 on selected meiotic nuclei but was progressively lost on non-selected meiotic nuclei. At P4, odCDK1 paralogs translocated from OCs to non-selected meiotic nuclei whereas pPlk1, translocated from OCs to selected meiotic nuclei. The pAurora kinase, present on all meiotic nuclei in P3, was retained on selected nuclei in P4 but lost from non-selected meiotic nuclei. MPM-2 phosphoepitopes, absent from meiotic nuclei in P3, became progressively enriched on both selected and non-selected meiotic nuclei during P4. MPM-2 phophoepitopes also appeared as small foci within nurse nuclei during P4, as did foci of pERK, the latter consistent with observations in previous work.28 Nurse nuclei also exhibited nucleolar pPlk1 staining during P4. In P3, odLamin1 surrounded both meiotic and nurse nuclei. During P4, odLamin was retained on nurse and selected meiotic nuclei but was lost from non-selected meiotic nuclei.
Differential functions of odCDK1 paralogs during meiosis I of oogenesis
To determine whether the odCDK1 paralogs exhibited any sub-functionalization during oogenesis, CDK1a, d and e paralogs were each specifically targeted for knockdown using dsRNA constructs. In each case, gonads of day 4 animals were injected with the respective dsRNA construct along with a cmRNA coding histone H2B-eGFP as a marker for successful injection.34 Following vitellogenesis and spawning, unfertilized oocytes were collected and processed to verify successful knockdown by qRT-PCR. Knockdown rescue experiments were conducted as above, replacing H2B-eGFP cmRNA with odCDK1 paralog-eGFP cmRNA, possessing silent mutations within the dsRNA target regions.
Injection of dsRNA efficiently knocked-down CDK1a, with no significant off-target effects on other CDK1 paralogs detected (Fig. 6A). Oocytes produced from successfully injected females were infertile when exposed to wild type sperm (Fig. 6B) and displayed significant reduction in size (Fig. 6C). This mutant phenotype was rescued when the dsRNA targeting odCDK1a was co-injected with an odCDK1a-eGFP cmRNA that was engineered to be resistant to the dsRNA targeting (Figs. S4A, B, and S5A–C). Previous reports on Drosophila oocytes showed reduction of oocyte size in response to knockdown of myosin phosphatase (DMYPT) and protein phosphatase 1 (Flw/PP1B),35,36 causing constriction of ring canals. As a consequence oocyte growth was limited due to reduced influx of cytoplasm. Similar to odCDK1a knock-down, inhibition of the PP1 phosphatase in O. dioica females by incubation with calyculin A, also resulted in significant reduction in oocyte size (Fig. 6C and Fig. S3A, B) and impaired developmental potential upon exposure to wild type sperm. Finally, inhibition of CDK1 activity by roscovitine also resulted in spawning of oocytes with a reduced size and did so in a dose-dependent manner (Fig. 6C and Fig. S3C, D). Our results suggest that odCDK1a is involved in the regulation of oocyte size prior to spawning. To assess whether this might involve pre-mature constriction of ring canals, we measured ring canal diameters in ovaries where odCDK1a had been knocked down by dsRNA, or where protein phosphatase 1 was inhibited by calyculin. We observed no significant difference in ring canal diameters under either of these 2 conditions when compared to ovaries in wild type females or those that had been incubated in the presence of DMSO (Fig. 6D).
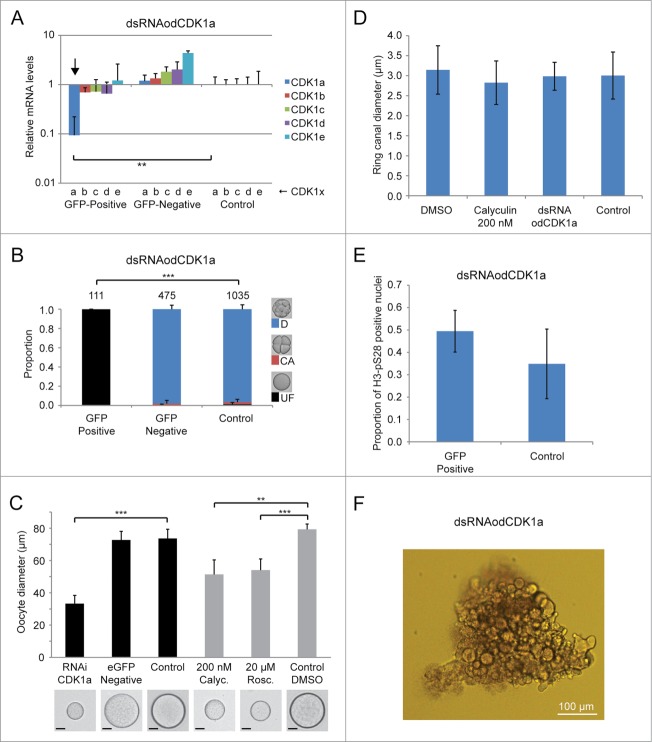
Figure 6.
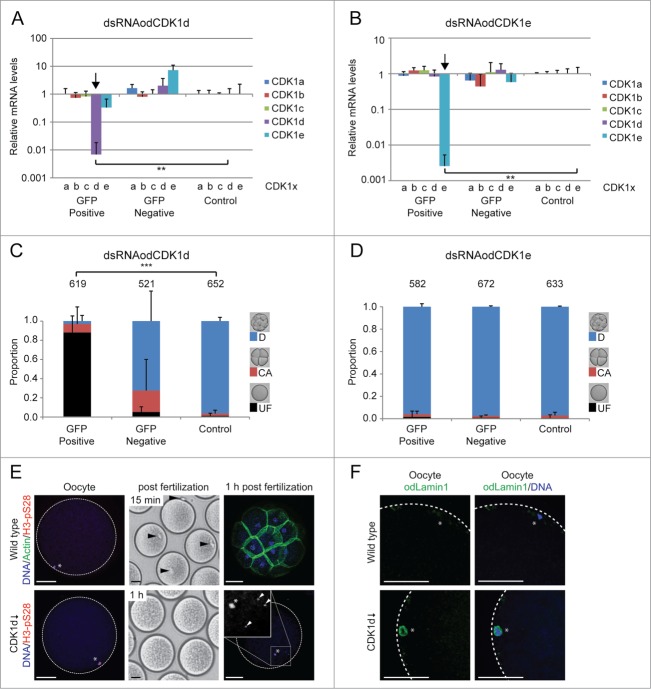
Phenotypes resulting from dsRNA knockdown of odCDK1a. A) Significant (**P < 0.01) knockdown of the targeted odCDK1a was verified by qRT-PCR. No significant off-target effects on other odCDK1 paralogs were detected. B) Oocytes spawned from females where ovaries had been co-injected with dsRNA (GFP-positive) against odCDK1a and capped mRNA coding for histone H2B-eGFP (selection marker for successful injection), failed to develop after exposure to sperm from wild type males. Oocytes spawned from wild-type non-injected females (Control) exposed to the same pool of wild type sperm developed normally. Oocytes derived from females whose ovaries had been co-injected as above, but failed to exhibit histone H2B-eGFP fluorescence (GFP-negative) also developed normally. The number of oocytes/embryos assessed is given across the top of the histogram bars. Legend: UF, UnFertilized; CA, early embryo Cleavage Arrest; D, Developed normally. C) Oocytes that were spawned from ovaries where odCDK1a had been knocked down were considerably smaller (***P < 0.001) compared to oocytes produced from wild-type and H2B-eGFP negative females. A similar effect in reducing spawned oocyte size was observed upon treatment of day 6 females with 200 nM calyculin A (Calyc.; **P < 0.01) or 20 μM roscovitine (Rosc.; ***P < 0.001) as compared to control females incubated in the presence of the DMSO solute alone. Scale bars = 20 μm. D) Ring canal diameters of growing oocytes were not affected by dsRNA knockdown of odCDK1a or by inhibiting PP1 phosphatase activity with 200 nM calyculin. E) Knockdown of odCDK1a did not significantly alter the proportions of selected vs. non-selected meiotic nuclei as determined by H3-pS28 staining (P = 0.19). F) Females injected with dsRNA targeting odCDK1a often released clumps of unresolved oocytes embedded in excess cytoplasm. This was never observed in wild type females, or in females injected with dsRNA targeting odCDK1d or odCDK1e.
We then asked whether the knockdown of odCDK1a resulted in an increased number of smaller oocytes. This might arise, for example, if the translocation of odCDK1a to non-selected meiotic nuclei during P4 was required to maintain their non-selected status. The mean diameter of wild type oocytes was 79 μm compared to 33 μm from females where odCDK1a had been knocked down (Fig. 6C). This would mean then that a similar coenocyst cytoplasmic volume would be apportioned into ∼13.7-fold more, smaller, spawned oocytes in the case of the dsRNA odCDK1a females. There is commonly a 2-3 fold variation in the number of oocytes spawned by wild type females under the standard nutrient conditions used in these experiments. Similar variability in oocyte numbers was observed in the spawns from dsRNA odCDK1a females and the predicted, more than an order of magnitude increase in number of oocytes, calculated on reduced oocyte diameter, was never observed. To examine this further, in ovaries prior to spawning, we examined the ratios of H3-pS28 stained, selected meiotic nuclei, to unstained, non-selected meiotic nuclei, in both wild type and dsRNA odCDK1a females. No significant differences were observed in this ratio, indicating that there was not an increase in the relative proportion of selected nuclei to populate more growing oocytes when odCDK1a was knocked down (Fig. 6E). Finally, in spawns from dsRNA odCDK1a females, we often observed clumps of small oocytes that were not fully resolved and embedded in excess cytoplasm (Fig. 6F). This was never observed in wild type females, or in females injected with dsRNA targeting odCDK1d or odCDK1e. Taken together, our data show that knockdown of odCDK1a results in small, infertile oocytes, through impaired and incomplete transfer of coenocyst cytoplasm into growing oocytes, as opposed to increasing the proportion of selected meiotic nuclei and, therefore, oocyte number. The mechanism through which odCDK1a exerts this effect on cytoplasmic flow remains unclear but does not appear to involve pre-mature constriction of ring canals.
The most recently duplicated odCDK1 paralogs, odCDK1d and odCDK1e were also targeted for dsRNA knockdown as above (Fig. 7).26 Each of the dsRNAs efficiently knocked-down their respective paralog target, with no significant off-target effects detected (Fig. 7A, B). Females exhibiting successful knockdown of odCDK1d or odCDK1e spawned oocytes of normal size, but of quite different developmental potential (Fig. 7C–E). Following fertilization with wild-type sperm, oocytes in which odCDK1e had been knocked down developed normally (Fig. 7D). In contrast, oocytes in which odCDK1d had been knocked down were infertile, a phenotype that was rescued by dsRNA-insensitive odCDK1d-eGFP cmRNA (Figs. S4A, C and S5D, E). An analysis of early events following fertilization showed that these oocytes were unable to emit polar bodies or cleave when exposed to wild type sperm (Fig. 7C, E). Some of these oocytes displayed non-cortical nuclear positioning and the appearance of 2–4 small nuclei (Fig. 7E, bottom panels), indicative of polyspermy. Interestingly, when odCDK1d was knocked down, oocyte chromatin retained a nuclear envelope as shown by persistent lamin staining (Fig. 7F) as opposed to wild type oocytes that are spawned in metaphase I arrest.7 Therefore, when CDK1d activity was reduced via dsRNA knockdown, resumption of meiosis from prophase I arrest to metaphase I was impaired and NEBD did not occur.
Figure 7.
Phenotypes resulting from dsRNA knockdown of odCDK1d and e. (A and B) Significant (**P < 0.01) knockdown of the targeted odCDK1d (A) and odCDK1e (B) was verified by qRT-PCR. No significant off-target effects on other odCDK1 paralogs were detected in either case. (C) Oocytes spawned from females where ovaries had been co-injected with dsRNA (GFP-Positive) against odCDK1d and capped mRNA coding for histone H2B-eGFP (selection marker for successful injection), failed to develop after exposure to sperm from wild type males. Oocytes spawned from wild-type non-injected females (Control) exposed to the same pool of wild type sperm developed normally. Oocytes derived from females whose ovaries had been co-injected as above, but failed to exhibit histone H2B-eGFP fluorescence (GFP-Negative) also developed normally in most cases though there was an elevated degree of embryonic arrest following initial cleavages. The number of oocytes/embryos assessed is given across the top of the histogram bars. (D) Similar experiments generating the knockdown of odCDK1e did not affect the developmental potential of oocytes spawned from females positive for H2B-eGFP. E) The majority of oocytes that were spawned from ovaries where odCDK1d had been knocked down appeared normal, with chromatin (*) at the oocyte cortex (left panels), as in wild type oocytes. However, whereas polar body extrusion (arrowheads, upper mid-panel) was observed within 15 minutes in wild type embryos, no polar body was extrusion was observed after 1 h post fertilization in odCDK1d deficient embryos (lower mid-panel). Control embryos reached the 32-cell stage by 1 h, whereas odCDK1d deficient embryos did not develop and contained one H3-pS28 positive prophase-I arrested female nucleus and up to 4 sperm nuclei (arrowheads, right panel insert). Scale bars = 50 μm. F) Wild type oocytes were spawned in Metaphase I arrest whereas those produced from females where CDK1d had been knocked down, retained a nuclear lamin envelope, indicating that they had not resumed meiosis from prophase I arrest. Scale bars = 20 μm.
Thus, whereas the odCDK1e paralog does not appear to execute essential non-redundant functions, the odCDK1a and d paralogs clearly play different and essential roles in the successful maturation of viable oocytes from the coenocystic ovary.
Discussion
In yeast, a single CDK1 ortholog (cdc28) interacts with multiple different cyclins to control cell cycle progression. The classical view of the evolution of cell cycle control mechanisms was that in higher, multicellular, eukaryotes, each cell cycle phase came to be driven by specific CDKs, notably CDK4, CDK6 and CDK2, with CDK1 retaining a primary control of M-phase. Genetic experiments in mice challenged this view,37 revealing that CDK1 alone was still able to drive the complete mammalian cell cycle, leading to an alternative view that primary CDK1 control of the “essential” cell cycle was conserved and that interphase CDKs were required for cell type specialization.38 The chordate, Oikopleura dioica model provides an additional new element to this scenario. It not only possesses the normal higher eukaryote complement of specialized interphase CDKs, but is the first known example of a metazoan to have more than one CDK1, expressing 5 distinct CDK1 paralogs over the course of its life cycle.26 Here we show specific sub-functionalization of 2 of these paralogs, CDK1a and CDK1d, during the O. dioica meiotic oogenesis program.
Centrosome-like Organizing Centers associate differentially with coenocyst nuclei as oogenesis progresses
The OCs in the coenocyst showed a clear spatial and compositional evolution as oogenesis progressed. Following the asymmetric germline nuclear division in P1,28 which gives rise to endocycling nurse nuclei and meiotic nuclei in a 1:1 ratio, OCs were only retained in close proximity to each of the meiotic nuclei through to P3 and were no longer associated with the terminally differentiated nurse nuclei, analogous to the loss of centrosomes from endocycling nurse nuclei in the Drosophila ovariole.39 Within each pro-oocyte, there was a clear polarity: the meiotic nucleus was anchored to the future animal pole, opposite the ring canal connection to the general coenocyst cytoplasm, and the OC was positioned intermediately along this axis, in closer proximity to the meiotic nucleus than to the ring canal. In the syncytial environment of the Drosophila ovariole there is a clear axis of polarity with the growing oocyte positioned at the posterior end while the 15 nurse nuclei occupy the anterior region. In the syncytial O. dioica coenocyst there is no such linear polarity. Nurse nuclei, and pro-oocytes harboring meiotic nuclei, are distributed throughout this unicellular cytoplasm. Instead, each growing oocyte must establish its own localized polar environment and the large OC may play an important role in such an organizational context.
During the transition to P4, all meiotic nuclei exhibited further chromatin compaction to the π−configuration, but only a subset were retained in oocytes selected to mature. The unselected meiotic nuclei were present in the general coenocyst cytoplasm and lost their close association with OC, and were often found embedded in the envelope of nurse nuclei. In contrast the association of OC with selected meiotic nuclei was enhanced, with OC abutting the selected nuclei adjacent to a region of clustered nuclear pore complexes. At this stage, the odLamin 1 splice variant (present on all of the nurse nuclei) appeared in the nuclear envelope of selected meiotic nuclei but remained absent from the nuclear envelope of unselected meiotic nuclei. As P4 progressed, OC that had been released from association with unselected meiotic nuclei were increasingly found in large pocket invaginations in nurse nuclear envelopes and appeared to be a focal point for increased collection of coenocyst nurse and unselected meiotic nuclei in a reduced local cytoplasmic volume.
Differential translocation of meiotic kinases from Organizing Centers to prophase I nuclei during P4 define selected versus non-selected meiotic nuclei
Activated Aurora kinase was present in OC and all meiotic nuclei during P3 but was lost from OC and became restricted only to selected meiotic nuclei in P4. On the other hand Plk1, only observed on OC during P3, translocated to selected meiotic nuclei and nurse nuclei in P4, and was no longer retained on OC at this stage. The odCDK1a, d and e paralogs, like Plk1, were present only on OC during P3, but during P4, they translocated exclusively to non-selected meiotic nuclei, a complementary, opposite, behavior to that of Plk1. The absence of Plk1 in non-selected nuclei indicates that they will be unable to proceed properly with synaptonemal complex disassembly during meiotic progression.32 The purpose of the P4 translocation of odCDK1a, d and e paralogs to non-selected meiotic nuclei is at present unclear. In the absence of synaptonemal complex disassembly regulated by Plk1, they might drive non-selected nuclei into a meiotic crisis, culminating in their eventual apoptosis in P5.28 Alternatively it may be that retention of these CDK1 paralogs on OC during P4 is incompatible with changes in OC localization/function in the transition from P3 to P4. Thus, the non-selected meiotic nuclei could serve as a site of sequestration of CDK1 activity in P4, so as to prevent interference with continued nurse nuclear endocycles or premature resumption of meiosis in vitellogenic oocytes. Taken together, our results show that Plk1 and Aurora kinase concentrations designate selected meiotic nuclei, whereas premature odCDK1 translocation, combined with absence of Plk1 and Aurora kinase, define non-selected meiotic nuclei.
Sub-functionalization of odCDK1a and d paralogs during O. dioica oogenesis
Knockout experiments in mice have demonstrated that CDK2 and CDK4 are required for fertility in females.40 This approach has not been able to assess specific requirement for CDK1 in meiosis because of the early embryonic lethality of CDK1 knockout mice. In general, the loss of individual cyclins or CDKs generates more pronounced effects in the meiotic as opposed to mitotic cell cycle, suggesting more specialized roles during meiotic progression. Thus we were interested as to whether the odCDK1 paralogs exhibited specific or redundant functions during coenocystic oogenesis.
Knockdown of the odCDK1a paralog in females produced small infertile oocytes. In Drosophila egg chambers, similar small, non-viable, oocytes were produced in lines mutant for the DMYPT1 (myosin targeting) or Flw/PP1β (phosphatase) subunits of myosin phosphatase.35,36 Mutant DMYPT1 or Flw/PP1β subunits prevent inactivation of myosin II and cause over-constriction of ring canals, restricting cytoplasmic transfer into developing Drosophila oocytes. CDK1 is known to generate phosphoepitopes that regulate MYPT1 activity and local inhibition of CDK1 activity drives cleavage furrow formation in Drosophila oocytes.41,42 Indeed, inhibition of CDK1 activity by roscovitine, or PP1 activity by calyculin A, also produced small, non-viable oocytes from the O. dioica coenocyst. However, we did not observe any direct alterations in ring canal diameter under these conditions. The knockdown of odCDK1a by dsRNA did not increase the proportion of meiotic nuclei selected to populate growing oocytes as compared to those meiotic nuclei that remained non-selected in the general coenocyst cytoplasm. Consistent with this, an increase in oocyte number, proportionate to the decrease in oocyte diameter, was not observed. Instead, our data show that knockdown of odCDK1a results in small, infertile oocytes, through impaired and incomplete transfer of coenocyst cytoplasm into growing oocytes. The mechanism through which odCDK1a exerts this regulatory control on cytoplasmic flow, and ultimately, oocyte size, remains unclear.
The odCDK1d paralog was also required to produce viable oocytes. Knockdown of odCDK1 yielded normal size oocytes, but they were unable to extrude polar bodies upon exposure to wild type sperm. This resulted from loss of the classical function of CDK1 in meiotic resumption from prophase I arrest. Thus, both odCDK1a and d paralogs are required to generate viable oocytes, through different specialized functions in regulating oocyte growth and meiotic progression.
Conclusions
The capacity of O. dioica to adjust its reproductive output over 3 orders of magnitude in a short time frame is important in its ability to rapidly adjust population size in response to algal blooms.6,24 The coenocystic oogenesis strategy is central to this adaptation, in which an excess of meiotic nuclei in prophase I is generated, with a variable subset subsequently selected to seed maturing oocytes as a function of nutritional resources. Here we have characterized the spatiotemporal dynamics of cytoplasmic OC during oogenesis and shown that differential translocation of meiotic kinases from OC to meiotic nuclei defines the selection of this subset. Finally, we demonstrate specific roles of CDK1 paralogs in carrying out this meiotic oogenic program.
Materials and Methods
Animal culture and collection
O. dioica were maintained in culture at 15°C.43 Day 4–6 animals were placed in filtered seawater, removed from their houses and anesthetized in cold ethyl 3-aminobenzoate methanesulfonate salt (MS-222, 0.125 mg/ml; Sigma), before collection.
In vitro fertilization (IVF) and developmental analysis
Sperm solutions were prepared by transferring 3–4 mature males to a petri dish containing artificial seawater, on ice, and sperm quality was assessed using a Nikon Eclipse E400 microscope. Mature females were transferred to artificial sea water in 6-well plates coated with 0.1% gelatin. Aliquots of spawned oocytes were collected for qRT-PCR and remaining oocytes fertilized with 100 μl sperm solution. Development was documented using a Spot RT-KE camera on a Nikon Eclipse TE2000-S inverted microscope. Unfertilized (UF) (1-cell), cleavage arrested (CA) (2–4 cell) and normally developing (D) embryos were counted in ImageJ using the Cell Counter plugin (http://rsbweb.nih.gov/ij/plugins/cell-counter.html). Oocyte diameters were measured in ImageJ and treatments compared using Student's T-test.
Quantitative reverse transcription-PCR
Oocytes were washed with 100 μl PBS. PBS was replaced by 100 μl of lysis solution and samples were snap-frozen in liquid nitrogen and stored at −80°C until further processing. RNA was isolated from 20–30 oocytes following the manufacturer's protocol using RNAqueous®-Micro Kit (Ambion). Total RNA (200 ng) was reverse transcribed using M-MLV reverse transcriptase (Life Technologies), with oligo-dT primers (Promega). Quantitative reverse transcription–polymerase chain reactions (qRT-PCR) of 20 μl were assembled using cDNA equivalent to 2 ng total RNA, 500 nM gene specific forward and reverse primers26 and 10 μl iQ™ SYBR® Green Supermix (Bio-Rad). qRT-PCR reactions were run on a CFX-96 (Bio-Rad). Relative expression levels were normalized to RPL23 and EF-1β mRNA expression. Statistical significance of knockdown experiments was assessed by Student's T-test.
Microinjection
Injection solutions were prepared by mixing 400 ng/μl capped mRNA (cmRNA) and 100 ng/μl Alexa Fluor® 568 dye (Molecular Probes), with 400 ng/μl double stranded RNA (dsRNA) in RNase free water. Anesthetized day 4 animals in filtered seawater were transferred to Petri dishes coated with 3% agarose. Gonads were injected on a Nikon Eclipse TE2000-S inverted microscope equipped with Narishige micromanipulators and transjector 5246 micro injector (Eppendorf). Holding pipettes were crafted from borosilicate capillaries with an outer diameter of 1 mm and inner diameter of 0.5 mm (Sutter instruments). Injection needles (quartz, Sutter instruments) were pulled with outer diameter of 1 mm and inner diameter of 0.7 mm on a Flaming/Brown P87 Micropipette puller (Sutter instruments). Injected day 4 animals were transferred to watch glasses containing filtered seawater for recovery and then transferred to 3 L beakers for subsequent culture.43 Animals were collected from day 4–6 for immunofluorescent assessment, or raised until spawning for IVF and immunofluorescence. Injected embryos were transferred to 6-well plates coated with 0.1% gelatin and collected shortly after hatching.
eGFP fusion constructs
O. dioica CDK1 paralog a, d and e protein coding sequences and 3’UTRs were amplified separately by polymerase chain reaction (PCR) on cDNA (primers, Table S1). PCR products were cloned into TOPO 2.1 vectors (Life Technologies). CDK1 eGFP fusion constructs for cmRNA expression were driven by a T7 promoter. Constructs were purified by maxi prep (QIAGEN). dsRNA insensitive odCDK1 paralog-eGFP fusion constructs, for RNAi rescue, were constructed by replacing the dsRNA target sequence with gBlocks® gene fragments (Integrated DNA Technologies), possessing numerous silent mutations within the mRNA coding regions (Fig. S5). An H2B-eGFP construct was used to produce cmRNA,34 as a successful injection control in dsRNA experiments. The odLamin 1-eGFP construct was as previously described.44
Capped mRNA and dsRNA synthesis
O. dioica CDK1-eGFP fusion-constructs including endogenous 3’UTRs were linearized using XmaI (odCDK1a and d), AfeI (odCDK1e), or Xba1 (H2B-eGFP) and purified by phenol-chloroform extraction followed by ethanol precipitation. cmRNAs were synthesized from these templates using mMessage mMachine (Ambion) followed by poly(A) tailing (Ambion) according to the manufacturer's protocol and purified by Lithium chloride precipitation.
DNA templates for dsRNA synthesis were prepared by PCR using gene-specific primers with T7 overhang (Table S1), and purified by phenol-chloroform extraction. Sense and antisense RNA were synthesized in a single reaction following the recommended protocol using T7 RiboMAXTM Express RNAi system (Promega).
Chemical Inhibitors
Day 6 females were transferred to artificial seawater containing 50–200 nM calyculin A (PP1 inhibitor; LC Laboratories) or 5–20 mM roscovitine (CDK1 inhibitor; Cell Signaling Technology) in DMSO and incubated until spawning (2–6 h). Controls were treated with DMSO diluted 1:2000 in artificial seawater. Diameters of successfully spawned oocytes and development post fertilization were analyzed as above. We were unable to use RO-3306 as an inhibitor of CDK1 activity because of difficulties in sustaining its solubility in seawater.
Immunofluorescence
Samples were processed for immunofluorescence and analyzed by confocal microscopy as described,26 with the following modifications. Gonads were separated from the trunk and tail using 2 needles after 1 h fixation at room temperature. Gonads were transferred back to fixative O/N at 4°C. After washing, gonads were permeabilized with 20 μg/ml proteinase K (New England BioLabs) in washing solution for 3 min. Gonads were washed 6 × 3 min in washing solution and blocked O/N at 4°C. For gamma-tubulin staining, live animals were exposed to 1 μM Paclitaxel (Sigma) in filtered seawater for 15 min before transfer to fixative containing 1 μM Paclitaxel. For actin detection, 0.6 U/ml Alexa488-conjugated phalloidin (Molecular Probes) was added to the secondary antibody incubation step. Images were processed in Image J.
Organizing Center/meiotic nuclei distance analysis
X, Y and Z coordinates of organizing centers (OCs) and meiotic nuclei were obtained from confocal Z-stack images using ImageJ with the Cell Counter plugin. Distances between OCs and meiotic nuclei were calculated based on the distance from each meiotic nucleus to the closest OC. Distance variations were assessed using the Students T-test.
Transmission electron microscopy
Sample preparation procedures were performed at room temperature. Day 5 and 6 male and female animals were fixed in 2.5% glutaraldehyde/0.1 M sodium cacodylate/0.24 M NaCl (pH 7.4) for 1 h. After rinsing in 0.1 M sodium cacodylate/0.24 M NaCl (pH 7.4), dissected gonads were post-fixed in 1% OsO4/0.1 M sodium cacodylate/0.24 M NaCl (pH 7.4) for 1 h and rinsed in distilled water. Specimens were en bloc stained for 30 min in 2% OsO4/1.5% potassium ferricyanide/0.1 M sodium cacodylate (pH 7.4). Following rinsing in distilled water; they were incubated in 2% aqueous uranyl acetate for 30 min and washed in distilled water. Samples were dehydrated stepwise in 50%, 70%, 80%, 90%, 96% and 100% ethanol for 10 min at each step with the incubation at 100% repeated 3 times before twice exchanging with propylene oxide. Embedding was performed using the TAAB Araldite 502/812 kit (TAAB Laboratories Equipment Ltd), in which mixture A consisted of Embed-812/Araldite 502/DDSA (Dodecenyl Succinic Anhydride). To embed, propylene oxide was replaced by successive 4 h incubations of 1:3, 1:1 and 3:1 propylene oxide:mixture A, before O/N incubation in pure mixture A. Samples were transferred to fresh medium consisting of Embed-812/Araldite 502/DDSA/BDMA (Benzyl Dimethyl Amine) and polymerized at 60°C for 48 h. Ultrathin sections (70 nm) were cut with a Leica EM UC7 and counterstained with 2% uranyl acetate and lead citrate. Images were acquired on a Jeol JEM-1230 transmission electron microscope equipped with a 1k × 1k CCD camera (Gatan multiscan).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
J.I.Ø. C.C. and E.M.T conceived and designed the experiments. J.I.Ø, J.K. and M.R. performed experiments. J.I.O., C.C., J.K., H.H. and E.M.T. analyzed the data. J.I.Ø. and E.M.T. wrote the manuscript. All authors edited and approved the manuscript.
Acknowledgments
We thank J.-M. Bouquet, M. Reeve and A. Aasjord from Appendic Park for providing animals from the Oikopleura culture.
Funding
This work was supported by a PhD fellowship from the Department of Biology, University of Bergen (J.I.Ø) and grants 183690/S10 NFR-FUGE and 133335/V40 from the Norwegian Research Council (E.M.T.).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Kishimoto T. Cell-cycle control during meiotic maturation. Curr Opin Cell Biol 2003; 15:654-63; PMID:14644189; http://dx.doi.org/ 10.1016/j.ceb.2003.10.010 [DOI] [PubMed] [Google Scholar]
- 2. Sagata N. Meiotic metaphase arrest in animal oocytes: its mechanisms and biological significance. Trends Cell Biol 1996; 6:22-28; PMID:15157528; http://dx.doi.org/ 10.1016/0962-8924(96)81034-8 [DOI] [PubMed] [Google Scholar]
- 3. Tachibana K, Machida T, Nomura Y, Kishimoto T. MAP kinase links the fertilization signal transduction pathway to the G1/S-phase transition in starfish eggs. EMBO J 1997; 16:4333-39; PMID:9250677; http://dx.doi.org/ 10.1093/emboj/16.14.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Okano-Uchida T, Okumura E, Iwashita M, Yoshida H, Tachibana K, Kishimoto T. Distinct regulators for Plk1 activation in starfish meiotic and early embryonic cycles. EMBO J 2003; 22:5633-42; PMID:14532135; http://dx.doi.org/ 10.1093/emboj/cdg535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Spradling AC. Germline Cysts - Communes That Work. Cell 1993; 72:649-51; PMID:8453660; http://dx.doi.org/ 10.1016/0092-8674(93)90393-5 [DOI] [PubMed] [Google Scholar]
- 6. Ganot P, Bouquet JM, Kallesoe T, Thompson EM. The Oikopleura coenocyst, a unique chordate germ cell permitting rapid, extensive modulation of oocyte production. Dev Biol 2007; 302:591-600; PMID:17126826; http://dx.doi.org/ 10.1016/j.ydbio.2006.10.021 [DOI] [PubMed] [Google Scholar]
- 7. Ganot P, Kallesoe T, Thompson EM. The cytoskeleton organizes germ nuclei with divergent fates and asynchronous cycles in a common cytoplasm during oogenesis in the chordate Oikopleura. Dev Biol 2007; 302:577-90; PMID:17123503; http://dx.doi.org/ 10.1016/j.ydbio.2006.10.022 [DOI] [PubMed] [Google Scholar]
- 8. Narbonne-Reveau K, Senger S, Pal M, Herr A, Richardson HE, Asano M, Deak P, Lilly MA. APC/CFzr/Cdh1 promotes cell cycle progression during the Drosophila endocycle. Development 2008; 135:1451-61; PMID:18321983; http://dx.doi.org/ 10.1242/dev.016295 [DOI] [PubMed] [Google Scholar]
- 9. Ullah Z, Kohn MJ, Yagi R, Vassilev LT, DePamphilis ML. Differentiation of trophoblast stem cells into giant cells is triggered by p57/Kip2 inhibition of CDK1 activity. Genes Dev 2008; 22:3024-36; PMID:18981479; http://dx.doi.org/ 10.1101/gad.1718108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ullah Z, Lee CY, Lilly MA, DePamphilis ML. Developmentally programmed endoreduplication in animals. Cell Cycle 2009; 8:1501-09; PMID:19372757; http://dx.doi.org/ 10.4161/cc.8.10.8325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zielke N, Edgar BA, DePamphilis ML. Endoreplication. CSH Perspect Biol 2013; 5:a012948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hayashi S. A Cdc2 dependent checkpoint maintains diploidy in Drosophila. Development 1996; 122:1051-58; PMID:8620832 [DOI] [PubMed] [Google Scholar]
- 13. Sauer K, Knoblich JA, Richardson H, Lehner CF. Distinct modes of cyclin E/Cdc2c kinase regulation and S-phase control in mitotic endoreduplication cycles of Drosophila embryogenesis. Genes Dev 1995; 9:1327-39; PMID:7797073; http://dx.doi.org/ 10.1101/gad.9.11.1327 [DOI] [PubMed] [Google Scholar]
- 14. Sigrist SJ, Lehner CF. Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell 1997; 90:671-81; PMID:9288747; http://dx.doi.org/ 10.1016/S0092-8674(00)80528-0 [DOI] [PubMed] [Google Scholar]
- 15. Von Stetina JR, Tranguch S, Dey SK, Lee LA, Cha B, Drummond-Barbosa D. α-Endosulfine is a conserved protein required for oocyte meiotic maturation in Drosophila. Development 2008; 135:3697-06; PMID:18927152; http://dx.doi.org/ 10.1242/dev.025114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Swan A, Schupbach T. The Cdc20 (Fzy)/Cdh1-related protein, Cort, cooperates with Fzy in cyclin destruction and anaphase progression in meiosis I and II in Drosophila. Development 2007; 134:891-99; PMID:17251266; http://dx.doi.org/ 10.1242/dev.02784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Avidor-Reiss T, Gopalakrishnan J. Building a centriole. Curr Opin Cell Biol 2013; 25:72-77; PMID:23199753; http://dx.doi.org/ 10.1016/j.ceb.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luksza M, Queguigner I, Verlhac MH, Brunet S. Rebuilding MTOCs upon centriole loss during mouse oogenesis. Dev Biol 2013; 382:48-56; PMID:23954884; http://dx.doi.org/ 10.1016/j.ydbio.2013.07.029 [DOI] [PubMed] [Google Scholar]
- 19. Szollosi D, Calarco P, Donahue RP. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J Cell Sci 1972; 11:521-41; PMID:5076360 [DOI] [PubMed] [Google Scholar]
- 20. Mikeladze-Dvali T, von Tobel L, Strnad P, Knott G, Leonhardt H, Schermelleh L, Gönczy P. Analysis of centriole elimination during C. elegans oogenesis. Development 2012; 139:1670-79; PMID:22492357; http://dx.doi.org/ 10.1242/dev.075440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kalous J, Solc P, Baran V, Kubelka M, Schultz RM, Motlik J. PKB/AKT is involved in resumption of meiosis in mouse oocytes. Biol Cell 2006; 98:111-23; PMID:15842198; http://dx.doi.org/ 10.1042/BC20050020 [DOI] [PubMed] [Google Scholar]
- 22. Marangos P, Carroll J. The dynamics of cyclin B1 distribution during meiosis I in mouse oocytes. Reproduction 2004; 128:153-62; PMID:15280554; http://dx.doi.org/ 10.1530/rep.1.00192 [DOI] [PubMed] [Google Scholar]
- 23. Ganot P, Thompson EM. Patterning through differential endoreduplication in epithelial organogenesis of the chordate, Oikopleura dioica. Dev Biol 2002; 252:59-71; PMID:12453460; http://dx.doi.org/ 10.1006/dbio.2002.0834 [DOI] [PubMed] [Google Scholar]
- 24. Troedsson C, Bouquet JM, Aksnes DL, Thompson EM. Resource allocation between somatic growth and reproductive output in the pelagic chordate Oikopleura dioica allows opportunistic response to nutritional variation. Mar Ecol Prog Ser 2002; 243:83-91; http://dx.doi.org/ 10.3354/meps243083 [DOI] [Google Scholar]
- 25. Troedsson C, Ganot P, Bouquet JM, Aksnes DL, Thompson EM. Endostyle cell recruitment as a frame of reference for development and growth in the Urochordate Oikopleura dioica. Biol Bull USA 2007; 213:325-34; PMID:18083972; http://dx.doi.org/ 10.2307/25066650 [DOI] [PubMed] [Google Scholar]
- 26. Campsteijn C, Ovrebo JI, Karlsen BO, Thompson EM. Expansion of cyclin D and CDK1 paralogs in Oikopleura dioica, a chordate employing diverse cell cycle variants. Mol Biol Evol 2012; 29:487-502; PMID:21734012; http://dx.doi.org/ 10.1093/molbev/msr136 [DOI] [PubMed] [Google Scholar]
- 27. Dewitte W, Murray JA. The plant cell cycle. Annu Rev Plant Biol 2003; 54:235-64; PMID:14502991; http://dx.doi.org/ 10.1146/annurev.arplant.54.031902.134836 [DOI] [PubMed] [Google Scholar]
- 28. Ganot P, Moosmann-Schulmeister A, Thompson EM. Oocyte selection is concurrent with meiosis resumption in the coenocystic oogenesis of Oikopleura. Dev Biol 2008; 324:266-76; PMID:18845138; http://dx.doi.org/ 10.1016/j.ydbio.2008.09.016 [DOI] [PubMed] [Google Scholar]
- 29. Davis FM, Tsao TY, Fowler SK, Rao PN. Monoclonal antibodies to mitotic cells. Proc Natl Acad Sci USA 1983; 80:2926-30; PMID:6574461; http://dx.doi.org/ 10.1073/pnas.80.10.2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hajnal A, Berset T. The C.elegans MAPK phosphatase LIP-1 is required for the G(2)/M meiotic arrest of developing oocytes. EMBO J 2002; 21:4317-26; PMID:12169634; http://dx.doi.org/ 10.1093/emboj/cdf430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Capell BC, Collins FS. Human laminopathies: nuclei gone genetically awry. Nat Rev Genet 2006; 7:940-52; PMID:17139325; http://dx.doi.org/ 10.1038/nrg1906 [DOI] [PubMed] [Google Scholar]
- 32. Jordan PW, Karppinen J, Handel MA. Polo-like kinase is required for synaptonemal complex disassembly and phosphorylation in mouse spermatocytes. J Cell Sci 2012; 125:5061-72; PMID:22854038; http://dx.doi.org/ 10.1242/jcs.105015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee HO, Davidson JM, Duronio RJ. Endoreplication: polyploidy with purpose. Genes Dev 2009; 23:2461-77; PMID:19884253; http://dx.doi.org/ 10.1101/gad.1829209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Omotezako T, Nishino A, Onuma TA, Nishida H. RNA interference in the appendicularian Oikopleura dioica reveals the function of the Brachyury gene. Dev Genes Evol 2013; 223:261-67; PMID:23494664; http://dx.doi.org/ 10.1007/s00427-013-0438-8 [DOI] [PubMed] [Google Scholar]
- 35. Ong S, Foote C, Tan CG. Mutations of DMYPT cause over constriction of contractile rings and ring canals during Drosophila germline cyst formation. Dev Biol 2010; 346:161-69; PMID:20542024; http://dx.doi.org/ 10.1016/j.ydbio.2010.06.008 [DOI] [PubMed] [Google Scholar]
- 36. Yamamoto S, Bayat V, Bellen HJ, Tan C. Protein phosphatase 1ss limits ring canal constriction during Drosophila germline cyst formation. Plos One 2013; 8:e70502; PMID:23936219; http://dx.doi.org/ 10.1371/journal.pone.0070502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Santamaria D, Barriere C, Cerqueira A, Hunt S, Tardy C, Newton K, Gönczy P. Cdk1 is sufficient to drive the mammalian cell cycle. Nature 2007; 448:811-15; PMID:17700700; http://dx.doi.org/ 10.1038/nature06046 [DOI] [PubMed] [Google Scholar]
- 38. Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 2009; 9:153-66; PMID:19238148; http://dx.doi.org/ 10.1038/nrc2602 [DOI] [PubMed] [Google Scholar]
- 39. Mahowald AP, Strassheim JM. Intercellular migration of centrioles in the germarium of Drosophila melanogaster: an electron microscopic study. J Cell Biol 1970; 45:306-20; PMID:4327572; http://dx.doi.org/ 10.1083/jcb.45.2.306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Satyanarayana A, Kaldis P. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene 2009; 28:2925-39; PMID:19561645; http://dx.doi.org/ 10.1038/onc.2009.170 [DOI] [PubMed] [Google Scholar]
- 41. Yamashiro S, Yamakita Y, Totsukawa G, Goto H, Kaibuchi K, Ito M, Hartshorne DJ, Matsumura F. Myosin phosphatase-targeting subunit 1 regulates mitosis by antagonizing polo-like kinase 1. Dev Cell 2008; 14:787-97; PMID:18477460; http://dx.doi.org/ 10.1016/j.devcel.2008.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Menant A, Karess RE. Inducing "cytokinesis" without mitosis in unfertilized Drosophila eggs. Cell Cycle 2012; 11:2856-63; PMID:22801541; http://dx.doi.org/ 10.4161/cc.21190 [DOI] [PubMed] [Google Scholar]
- 43. Bouquet JM, Spriet E, Troedsson C, Ottera H, Chourrout D, Thompson EM. Culture optimization for the emergent zooplanktonic model organism Oikopleura dioica. J Plankton Res. 2009; 31:359-70; PMID:19461862; http://dx.doi.org/ 10.1093/plankt/fbn132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Clarke T, Bouquet JM, Fu X, Kallesoe T, Schmid M, Thompson EM. Rapidly evolving lamins in a chordate, Oikopleura dioica, with unusual nuclear architecture. Gene 2007; 396:159-69; PMID:17449201; http://dx.doi.org/ 10.1016/j.gene.2007.03.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.