Abstract
Background: Peroxisome proliferator-activated receptor gamma (PPARG), a nuclear hormone receptor, plays a critical role in the lipid and glucose homeostasis, adipocyte differentiation, as well as intracellular insulin-signaling events. Several studies have been conducted to explore the associations of PPARG polymorphisms with breast cancer (BC), yet the findings are inconsistent. Methods: Databases of Pubmed and Embase were searched until October 5, 2014. The association between PPARG polymorphisms and BC risk was determined by crude odds ratios (ORs) with their 95% confidence intervals (CIs). Results: Finally, there are nine publications involving 3,931 BC cases and 5,382 controls included in this meta-analysis. No significant association was observed between PPARG rs1801282 C>G variants and overall BC risk in all genetic comparison models. However, in a subgroup analysis by ethnicity, significant association was observed between PPARG rs1801282 C>G variants and decreased BC risk in three genetic models: GG+CG vs. CC (OR, 0.83; 95% CI, 0.71-0.96; P = 0.011), CG vs. CC (OR, 0.82; 95% CI, 0.71-0.96; P = 0.011) and G vs. C (OR, 0.85; 95% CI, 0.75-0.97; P = 0.016) in Caucasians and in a subgroup analysis by menopausal status, significantly decreased BC risk was also found in two genetic models: GG+CG vs. CC (OR, 0.79; 95% CI, 0.67-0.95; P = 0.011) and CG vs. CC (OR, 0.77; 95% CI, 0.64-0.92; P = 0.005) in post-menopause subgroup. For PPARG rs3856806 C>T, we found no significant association between PPARG rs3856806 C>T polymorphism and breast cancer. Conclusions: In summary, despite some limitations, the results suggest that PPARG rs1801282 C>G polymorphism may be a protective factor for BC in Caucasians and in post-menopause women.
Keywords: Breast cancer, polymorphism, peroxisome proliferator-activated receptor gamma, meta-analysis
Introduction
Breast cancer (BC) is the most frequently diagnosed cancer in female with an estimated 1,676,600 new BC cases and 521,900 BC related deaths in 2012 worldwide [1]. BC is a common disease that is attributed to multiple genetic and environmental risk factors. Recently, a number of candidate genes, such as BRCA1/2, TP53, BRIP1 and PALB2, have been confirmed to contribute to the risk of breast cancer [2-6]. These mutations may play a critical role in the development of BC. BRCA1/2 is a high penetrance gene [4] and 80% of individuals who carry this mutation may develop to BC. Compared to the normal population, the medium penetrance genes PALB2, CHEK2 and BRIP1 can increase 2.3-, 2.2-, and 2.0-fold risks of BC, respectively [2,3]. Some low penetrance genes FGFR2, ESR1, MAP3K1 and TOX3 have been investigated in several genome-wide association studies (GWAS) and were confirmed as candidates of BC [7-11]. Thus, in all probability, there are a crowd of low penetrance mutations in genes contributing to the remaining unexplained susceptibility of BC, which have not yet been verified.
Accumulating epidemiological evidence highlights that impaired glucose tolerance and type 2 diabetes are associated with the risk of cancer [12-15]. Peroxisome proliferator-activated receptor gamma (PPARG), a nuclear hormone receptor, acts as a critical regulator of lipid and glucose homeostasis, adipocyte differentiation, and intracellular insulin-signaling events. A number of investigations have therefore explored the hypothesis that the mutation of PPARG gene influences the development and progression of malignancy [16-21]. The PPARG single nucleotide polymorphisms (SNPs) are deemed to alter the activity of PPARG. This gene is polymorphic, and a number of SNPs have been studied, such as the rs1801282, rs3856806, rs4135247, rs1175543, rs709158 and rs2938395 polymorphisms, etc. Among them, the rs1801282 C>G and rs3856806 C>T are the most widely explored for correlation with cancer susceptibility. In a previous review, PPARG rs1801282 C>G polymorphism were correlated with protection from colorectal cancer, but with an increased susceptibility of gastric carcinoma and rs3856806 C>T polymorphism was marginally associated with the risk of cancer [22].
Recently, more studies have focused on the association of PPARG SNPs with BC [23-30]. Some of them identified the potential correlation between PPARG SNPs with BC risk [26,28]. A meta-analyses including three investigations confirmed that PPARG rs1801282 C>G was associated with the decreased risk of BC [31]; however, the other meta-analysis suggested that PPARG SNPs were not associated with BC [30]. At present, more studies have demonstrated that PPARG SNPs may clarify the causes and events correlated with BC; nevertheless, the results remain inconclusive. Therefore, in this study, we performed an updated meta-analysis to further explore the role of the PPARG polymorphisms in BC risk.
Materials and methods
Our study is reported on the basis of the PRISMA (Preferred Reporting Items for Meta-analyses) guideline (Table S1. PRISMA checklist) [32].
Search strategy
We searched literatures from PubMed and Embase databases (published up to October 5, 2014) using the following terms ‘Peroxisome proliferator-activated receptor gamma’, ‘PPARG’ ‘PPARγ’, ‘polymorphism’, ‘mutation’, ‘variant’, ‘cancer’, ‘carcinoma’, ‘malignance’ and ‘breast’. In order to minimize potential publication bias, additional relevant studies in the citations were also manually scanned. Only the latest study with the largest samples was recruited in our study to avoid overlapping data.
Inclusion and exclusion criteria
In our meta-analysis, all studies included had to meet all the following criteria: (a) case-control or cohort studies which assessed the association of PPARG SNPs with BC risk; (b) the available frequencies of genotypes or alleles must be provided and the genotype distribution among controls complied with the Hardy-Weinberg equilibrium (HWE). The major reasons for exclusion of studies were: (a) incomplete data; (b) duplicated studies or overlapping data; (c) only relevant to BC treatment; (d) meta-analysis, review, editorial, comment or letter.
Data extraction
In a standardized form, three reviewers (W. Tang, Y. Chen and Y. Wang) independently checked and extracted the data from all included publications. The following characteristic terms were extracted: the surname of first author, the year of publication, the country of origin, the ethnicity of participants, the allele and genotype frequencies, the genotyping method, the sample size, and the evidence of HWE in controls. If different results generated, disputes were settled by consulting the third reviewer (H. Gu).
Methodological quality assessment
The quality assessment was carefully performed by three authors (W. Tang, Y. Chen and Y. Wang) according to the ‘methodological quality assessment scale’ described previously [33,34]. Scores range from 0 to 10, and if the quality scores were ≥ 6, publications were defined as ‘high quality’; otherwise, they were classified as ‘low quality’.
Statistical analysis
In our study, the pooled odds ratios (ORs) with 95% confidence intervals (CIs) were assessed for dominant model, recessive model, heterozygote comparison, homozygote comparison and allelic comparison. Stratified analyses were conducted by ethnicity, menopausal status, source of controls and sample sizes. Heterogeneity among the studies was assessed by using a χ2-test-based Q statistic test. The value of P < 0.1 showed substantial heterogeneity across the publications, then the data were pooled by using the random-effects model (DerSimonian and Laird) [35]; otherwise, the fixed-effects model was used (Mantel-Haenszel) [36]. Both one-way sensitivity analysis and “trim-and-fill” method were conducted to evaluate the stability of this meta-analysis. Potential publication bias across the studies was assessed by a funnel plots and Egger’s linear regression test. The distribution of the genotypes in control subjects was checked for HWE using a web-based χ2 test program (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). All data analysis was conducted with STATA 12.0 software package (Stata Corp LP, College Station, Texas).
Results
Study characteristics
As shown in Figure 1, a total of 142 potentially relevant publications were obtained based on the search terms from PubMed and Embase databases. Finally, there were nine publications involving 3,931 BC cases and 5,382 controls included in this meta-analysis. All subjects were female. For PPARG rs1801282 C>G polymorphism, eight publications focusing on the association of this SNP with BC risk remained in the pooled analysis [23-29]. As for subjects in these studies, four were Caucasians [25,26,28,29]; two were mixed populations [23,27] and two were Asians [24,30]. As for menopausal status, three studies investigated post-menopause women [25,26,28], while five studies investigated overall adult women [23,24,27,29,30]. For PPARG rs3856806 C>T polymorphism, three studies were included [24,30,37]. Among them, all were Asians and investigated overall adult women. The detailed characteristics of the eligible studies and distribution of the PPARG polymorphisms as well as alleles are summarized in Tables 1 and 2, respectively.
Figure 1.
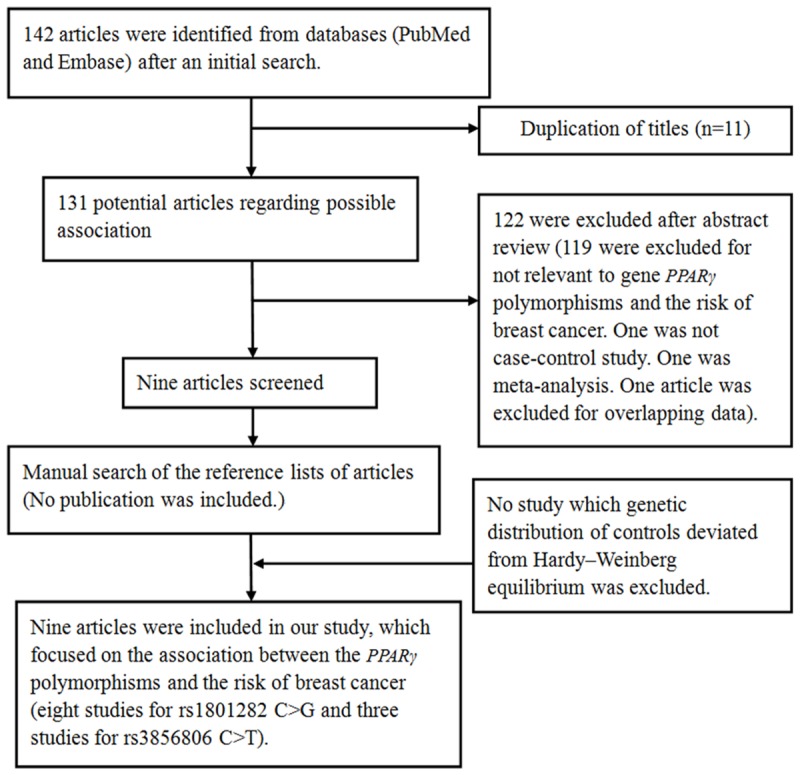
Flow chart shows studies included procedure for meta-analysis.
Table 1.
Characteristics of the included studies and the results of the methodological quality assessment scale
| Study | Publication year | Ethnicity | Country | Source of controls | Menopausal status | Sample size (case/control) | PPARG polymorphisms | Genotype method |
|---|---|---|---|---|---|---|---|---|
| Park et al. | 2014 | Asians | Korea | HB | Mixed status | 456/461 | rs1801282 and rs3856806 | MALDI-TOF MS |
| Martinez-Nava et al. | 2013 | Mixed populations | Mexico | PB | Mixed status | 208/220 | rs1801282 | RT-PCR |
| Wei | 2013 | Asians | China | HB | Mixed status | 216/216 | rs3856806 | MALDI-TOF MS |
| Petersen et al. | 2012 | Caucasians | Denmark | PB | Post-menopaused | 798/798 | rs1801282 | TaqMan |
| Wu et al. | 2011 | Asians | China | HB | Mixed status | 291/589 | rs1801282 and rs3856806 | RT-PCR |
| Justenhoven et al. | 2008 | Caucasians | German | PB | Mixed status | 688/724 | rs1801282 | MALDI-TOF MS |
| Gallicchio et al. | 2007 | Caucasians | USA | PB | Post-menopaused | 61/933 | rs1801282 | TaqMan |
| Wang et al. | 2007 | Caucasians | USA | PB | Post-menopaused | 488/488 | rs1801282 | TaqMan |
| Memisoglu et al. | 2002 | Mixed populations | USA | PB | Mixed status | 725/953 | rs1801282 | PCR-RFLP |
RT-PCR: real-time PCR; PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism; MALDI-TOF MS: Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry.
Table 2.
Distribution of PPARG polymorphisms genotype and allele
| PPARG polymorphisms | Study | Case genotype | Control genotype | Case allele | Control allele | HWE | Quality scores | ||||||
|
|
|
|
|
|
|||||||||
| rs1801282 C>G | CC | CG | GG | CC | CG | GG | G | C | G | C | |||
|
| |||||||||||||
| Martinez-Nava et al. | 165 | 43 | 0 | 169 | 49 | 2 | 43 | 373 | 53 | 387 | 0.448105 | 8.0 | |
| Park et al. | 413 | 40 | 2 | 412 | 42 | 1 | 44 | 866 | 44 | 866 | 0.948224 | 6.5 | |
| Wu et al. | 260 | 29 | 0 | 546 | 40 | 0 | 29 | 549 | 40 | 1132 | 0.392337 | 7.0 | |
| Gallicchio et al. | 48 | 7 | 1 | 689 | 188 | 18 | 9 | 103 | 224 | 1566 | 0.223793 | 8.5 | |
| Wang et al. | 376 | 87 | 15 | 375 | 98 | 5 | 117 | 839 | 108 | 848 | 0.615475 | 6.5 | |
| Memisoglu et al. | 563 | 148 | 14 | 752 | 190 | 11 | 176 | 1274 | 212 | 1694 | 0.795703 | 7.0 | |
| Petersen et al. | 616 | 167 | 15 | 569 | 209 | 20 | 197 | 1399 | 249 | 1347 | 0.876910 | 7.5 | |
| Justenhoven et al. | 452 | 135 | 6 | 462 | 145 | 15 | 147 | 1039 | 175 | 1069 | 0.372101 | 7.5 | |
|
| |||||||||||||
| rs3856806 C>T | CC | CT | TT | CC | CT | TT | C | T | C | T | |||
|
| |||||||||||||
| Park et al. | 320 | 128 | 8 | 311 | 117 | 15 | 768 | 144 | 739 | 147 | 0.335483 | 6.5 | |
| Wei | 115 | 69 | 15 | 122 | 69 | 9 | 299 | 99 | 313 | 87 | 0.848027 | 6.5 | |
| Wu et al. | 162 | 110 | 19 | 328 | 219 | 40 | 434 | 148 | 875 | 299 | 0.675591 | 7.0 | |
HWE: Hardy-Weinberg equilibrium.
Association of PPARG rs1801282 C>G polymorphism with BC risk
In total, 3,715 BC cases and 5,166 controls from eight eligible studies were relevant to the association between PPARG rs1801282 C>G polymorphism and BC. In overall meta-analysis, we found no association between PPARG rs1801282 C>G polymorphism and BC risk: GG+CG vs. CC (OR, 0.92; 95% CI, 0.82-1.03; P = 0.132), GG vs. CG+CC (OR, 1.05; 95% CI, 0.56-1.96; P = 0.884), GG vs. CC (OR, 1.02; 95% CI, 0.54-1.93; P = 0.963), CG vs. CC (OR, 0.91; 95% CI, 0.81-1.02; P = 0.107) and G vs. C (OR, 0.95; 95% CI, 0.81-1.11; P = 0.503) (Table 3). In a subgroup analysis by ethnicity, significantly decreased BC risk was confirmed in three genetic models: GG+CG vs. CC (OR, 0.83; 95% CI, 0.71-0.96; P = 0.011), CG vs. CC (OR, 0.82; 95% CI, 0.71-0.96; P = 0.011) and G vs. C (OR, 0.85; 95% CI, 0.75-0.97; P = 0.016) in Caucasians, but not in non-Caucasians (Table 3; Figure 2). In a subgroup analysis by menopausal status, significant decreased BC risk was also found in two genetic models: GG+CG vs. CC (OR, 0.79; 95% CI, 0.67-0.95; P = 0.011) and CG vs. CC (OR, 0.77; 95% CI, 0.64-0.92; P = 0.005) in post-menopause subgroup, but not mixed status (Table 3; Figure 3).
Table 3.
Meta-Analysis of PPARG rs1801282 C>G polymorphism with the breast cancer risk
| No. of study | G vs. C | GG vs. CC | GG+CG vs. CC | GG vs. CG+CC | CG vs. CC | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||||||||
| OR (95% CI) | P | P (Q-test) | OR (95% CI) | P | P (Q-test) | OR (95% CI) | P | P (Q-test) | OR (95% CI) | P | P (Q-test) | OR (95% CI) | P | P (Q-test) | ||
| Total | 8 | 0.95 (0.81-1.11) | 0.503 | 0.065 | 1.02 (0.54-1.93) | 0.963 | 0.062 | 0.92 (0.82-1.03) | 0.132 | 0.119 | 1.05 (0.56-1.96) | 0.884 | 0.071 | 0.91 (0.81-1.02) | 0.107 | 0.178 |
| Ethnicity | ||||||||||||||||
| Asians | 2 | 1.19 (0.86-1.64) | 0.302 | 0.225 | 2.00 (0.18-22.09) | 0.573 | NA | 1.18 (0.85-1.65) | 0.327 | 0.192 | 2.00 (0.18-22.18) | 0.571 | NA | 1.17 (0.83-1.64) | 0.363 | 0.171 |
| Caucasians | 4 | 0.85 (0.75-0.97) | 0.016 | 0.158 | 0.90 (0.37-2.16) | 0.809 | 0.038 | 0.83 (0.71-0.96) | 0.011 | 0.287 | 0.94 (0.39-2.25) | 0.890 | 0.038 | 0.82 (0.71-0.96) | 0.011 | 0.353 |
| Mixed populations | 2 | 1.05 (0.86-1.26) | 0.647 | 0.265 | 1.39 (0.66-2.92) | 0.389 | 0.184 | 1.03 (0.83-1.27) | 0.792 | 0.402 | 1.38 (0.66-2.90) | 0.394 | 0.190 | 1.01 (0.81-1.25) | 0.939 | 0.582 |
| Menopausal status | ||||||||||||||||
| Post-menopaused | 3 | 0.85 (0.63-1.15) | 0.298 | 0.075 | 1.21 (0.41-3.57) | 0.723 | 0.062 | 0.79 (0.67-0.95) | 0.011 | 0.202 | 1.29 (0.46-3.64) | 0.631 | 0.074 | 0.77 (0.64-0.92) | 0.005 | 0.442 |
| Mixed status | 5 | 1.00 (0.88-1.15) | 0.958 | 0.237 | 0.85 (0.30-2.38) | 0.750 | 0.093 | 1.01 (0.87-1.01) | 0.865 | 0.395 | 0.85 (0.31-2.35) | 0.754 | 0.099 | 1.02 (0.88-1.18) | 0.801 | 0.537 |
| Source of controls | ||||||||||||||||
| PB | 6 | 0.91 (0.77-1.06) | 0.234 | 0.093 | 0.97 (0.49-1.93) | 0.930 | 0.039 | 0.89 (0.79-1.00) | 0.051 | 0.203 | 1.00 (0.51-1.98) | 0.991 | 0.045 | 0.88 (0.78-0.99) | 0.041 | 0.322 |
| HB | 2 | 1.19 (0.86-1.64) | 0.302 | 0.225 | 2.00 (0.18-22.09) | 0.573 | NA | 1.18 (0.85-1.65) | 0.327 | 0.192 | 2.00 (0.18-22.09) | 0.571 | NA | 1.17 (0.83-1.64) | 0.363 | 0.171 |
| Sample sizes | ||||||||||||||||
| < 1000 | 5 | 1.02 (0.85-1.22) | 0.829 | 0.242 | 1.78 (0.85-3.73) | 0.125 | 0.316 | 0.98 (0.81-1.19) | 0.837 | 0.252 | 1.86 (0.89-3.89) | 0.102 | 0.332 | 0.94 (0.77-1.15) | 0.563 | 0.231 |
| ≥ 1000 | 3 | 0.90 (0.72-1.12) | 0.341 | 0.043 | 0.80 (0.37-1.73) | 0.578 | 0.064 | 0.89 (0.71-1.12) | 0.318 | 0.069 | 0.83 (0.39-1.74) | 0.619 | 0.075 | 0.89 (0.77-1.03) | 0.118 | 0.115 |
PB: Population-based; HB: Hospital-based.
Figure 2.
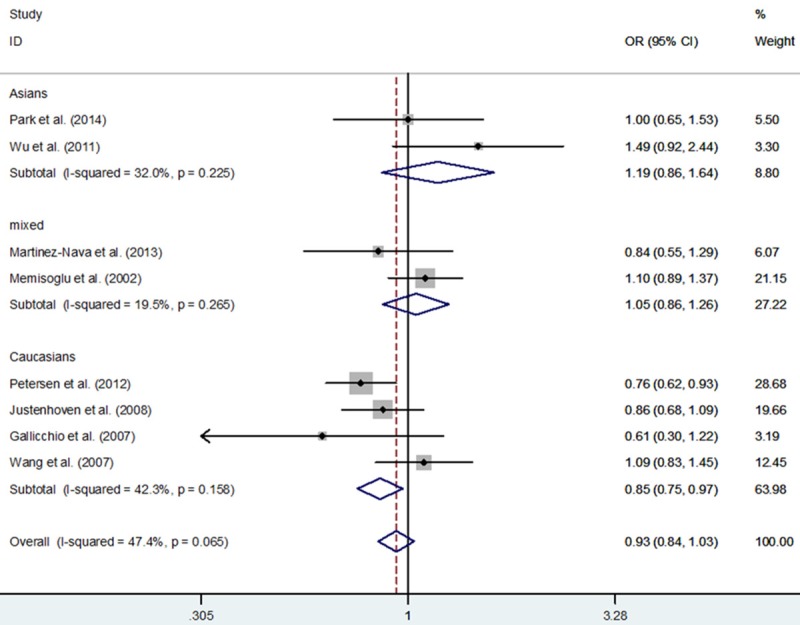
Forest plot of breast cancer risk associated with PPARG rs1801282 C>G polymorphism for the G vs. C (fixed effects model).
Figure 3.
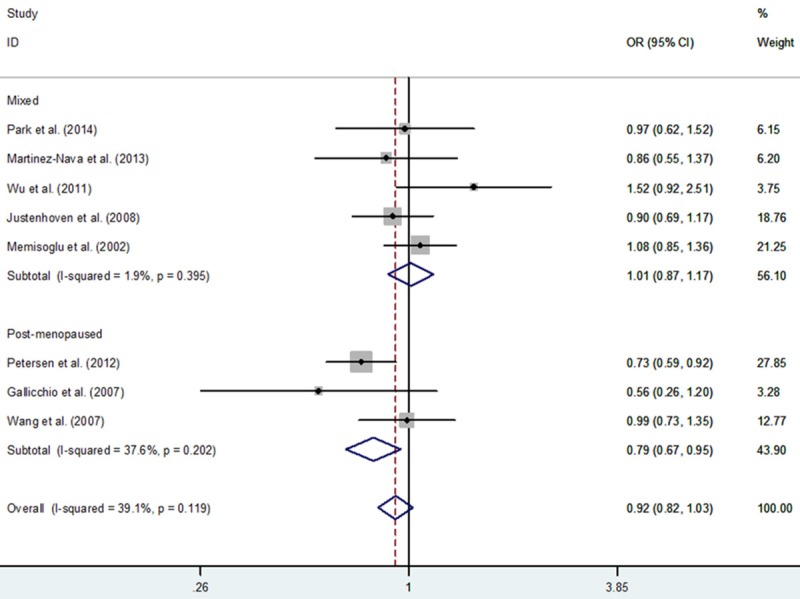
Forest plot of breast cancer risk associated with PPARG rs1801282 C>G polymorphism for the GG+CG vs. CC (fixed effects model).
Association of PPARG rs3856806 C>T polymorphism with BC risk
A total of 963 BC cases and 1,266 controls from three publications focused on the association of PPARG rs3856806 C>T with BC were enrolled for the current study. In pooled analysis, we found no significant association between them: TT+CT vs. CC (OR, 1.03; 95% CI, 0.86-1.23; P = 0.737), TT vs. CT+CC (OR, 0.95; 95% CI, 0.63-1.43; P = 0.806), TT vs. CC (OR, 0.96; 95% CI, 0.63-1.45; P = 0.843), CT vs. CC (OR, 1.04; 95% CI, 0.87-1.26; P = 0.652) and T vs. C (OR, 1.01; 95% CI, 0.87-1.18; P = 0.849) (Table 4).
Table 4.
Meta-Analysis of PPARG rs3856806 C>T polymorphism with the breast cancer risk
| Genetic comparison | OR (95% CI) | P | Test of heterogeneity | |
|---|---|---|---|---|
|
| ||||
| p-Value | Model | |||
| TT+CT vs. CC | 1.03 (0.86-1.23) | 0.737 | 0.853 | F |
| TT vs. CT+CC | 0.95 (0.63-1.43) | 0.806 | 0.143 | F |
| TT vs. CC | 0.96 (0.63-1.45) | 0.843 | 0.147 | F |
| CT vs. CC | 1.04 (0.87-1.26) | 0.652 | 0.975 | F |
| T vs. C | 1.01 (0.87-1.18) | 0.849 | 0.531 | F |
Publication bias for PPARG rs1801282 C>G polymorphism
Funnel plots and the Egger’s linear regression test were conducted to check potential publication bias across literatures. The shape of the funnel plot appeared to be symmetrical in all comparison models supported by Egger’s test (G vs. C: Begg’s test P = 0.711, Egger’s test P = 0.780; GG vs. CC: Begg’s test P = 1.000, Egger’s test P = 0.929; GG+CG vs. CC: Begg’s test P = 1.000, Egger’s test P = 0.826; GG vs. CG+CC: Begg’s test P = 1.000, Egger’s test P = 0.925; CG vs. CC: Begg’s test P = 1.000, Egger’s test P = 0.865; Figure 4).
Figure 4.
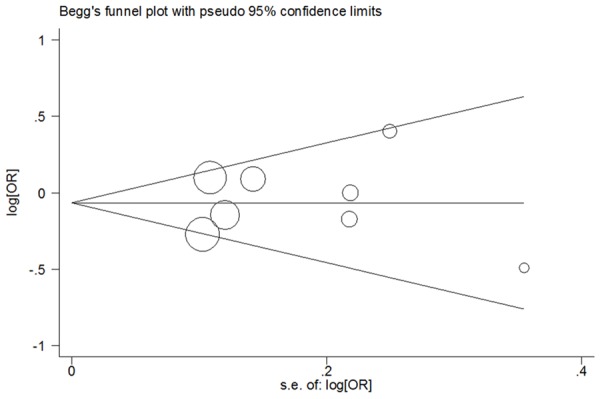
Begg’s funnel plot analysis of PPARG rs1801282 C>G polymorphism with breast cancer risk for the G vs. C (random-effects model).
Sensitivity analyses for PPARG rs1801282 C>G polymorphism
Both one-way sensitivity analysis and “trim-and-fill” method were carried out to verify the stability of this meta-analysis. The adjusted ORs and CIs of nonparametric “trim-and-fill” method were not substantially altered (G vs. C: adjusted pooled OR = 0.95, 95% CI: 0.81-1.11, P = 0.503; GG vs. CC: adjusted pooled OR = 1.02, 95% CI: 0.54-1.93, P = 0.963; GG+CG vs. CC: adjusted pooled OR = 0.92, 95% CI: 0.82-1.03, P = 0.148; GG vs. CG+CC: adjusted pooled OR = 1.05, 95% CI: 0.56-1.96, P = 0.884; CG vs. CC: adjusted pooled OR = 0.91, 95% CI: 0.81-1.03, P = 0.122; Figure 5), verifying the stability of our findings. Results of one-way sensitivity analysis were not significantly changed when any study was removed in turn, attesting the robustness of our findings (Figure 6).
Figure 5.
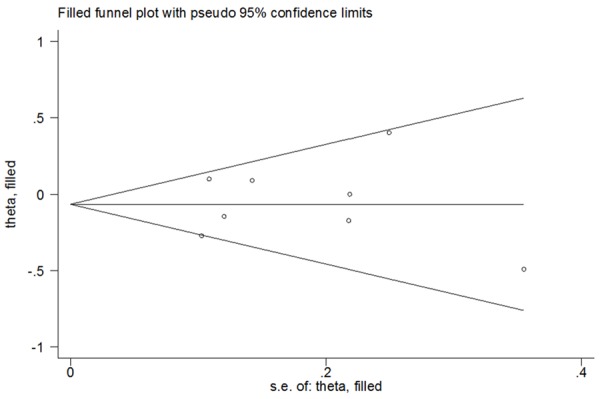
Filled funnel plot of PPARG rs1801282 C>G polymorphism with breast cancer risk for the G vs. C (random-effects model).
Figure 6.
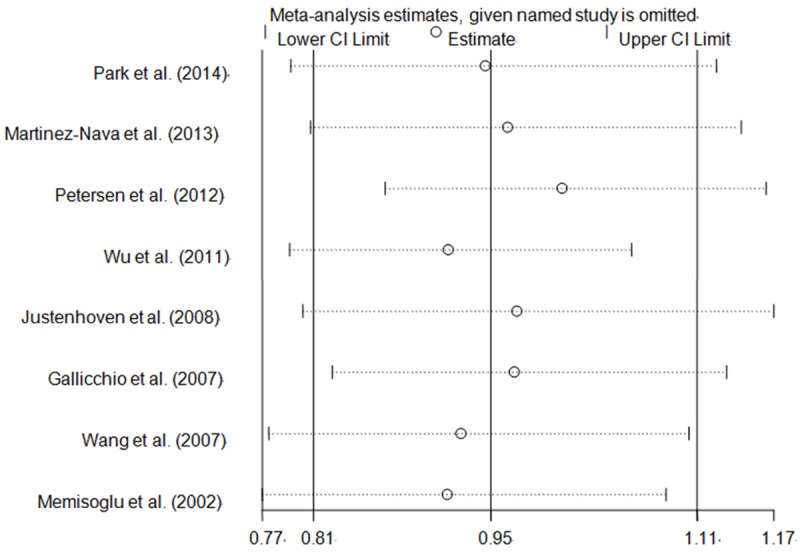
One-way sensitivity analysis of PPARG rs1801282 C>G polymorphism with breast cancer risk for the G vs. C (random-effects model).
Tests for heterogeneity for PPARG rs1801282 C>G polymorphism
Heterogeneity between studies was summarized in Table 3. Results of subgroup analysis indicated that the investigations conducted in Caucasians, post-menopause, population-based and large sample sizes (≥ 1000) subgroups might contribute to the major heterogeneity.
Results of quality assessment
According to the ‘methodological quality assessment scale’ [33,34], quality assessment was performed in all included publications. The results indicated that all studies were “high quality” (quality scores ≥ 6; Table 2), suggesting the reliability of our findings.
Discussion
PPARG, a member of the nuclear hormone receptor superfamily, could recognize and bind to PPARG response elements, subsequently regulate and potentially affect the transcription of target genes in the promoter region. Given the PPARG has shown pro-apoptotic, pro-differentiation and anti-proliferative properties after activation, it is deemed to have overall anti-carcinogenic effects in a number of cell types [38]. In view of these findings, the PPARG SNPs have been intensively studied for the association with BC recently. Results of one pooled analysis highlighted that the PPARG rs1801282 G allele modestly modified the susceptibility of breast cancer [31]. In contrast, the other previous meta-analysis indicated that both PPARG rs1801282 G allele and rs3856806 T allele did not affect the BC risk [30]. These seemingly conflicting findings have inspired more studies on correlation of PPARG SNPs with BC risk. In the light of these results, we summarized data for 3,931 BC cases and 5,382 controls from nine recruited publications and attempted to evaluate the risk of PPARG SNPs to BC by a comprehensive meta-analysis. Our results indicated that PPARG rs1801282 G allele might modify the susceptibility of breast cancer in Caucasians and in post-menopause women [31].
PPARG is an important transcription factor which acts as a controller in inflammatory cytokine production, insulin sensitization, lipogenesis, glucose homeostasis and cell differentiation [39]. The PPARG rs1801282 C>G polymorphism, a most common SNP in exon B of PPARG, encodes a Pro→Ala substitution at amino acid residue 12 (Pro 12 Ala) [40]. As a previous study has shown this missense substitution of rs1801282 C>G could cause less transcriptional activation of target genes in vitro [41], it may presumably affect cell differentiation and then alters the risk of BC. In combination with our results, these findings suggested that the PPARG rs1801282 C>G variants might be a protective factor for BC, probably through increasing binding capacity for certain PPARG response elements and promoting the ability of pro-apoptotic, pro-differentiation and anti-proliferative properties.
PPARG rs3856806 C>T polymorphism, another important SNP, has been suggested to have inverse associations with body weight compared to PPARG rs1801282 C>G polymorphism and relate to inflammation response [42]. It has been reported that PPARG rs3856806 C>T polymorphism is correlated with several cancer risk including colorectal cancer [43-45], follicular lymphoma [46] and ovarian carcinoma [47]. This pooled study is to explore possible association of this functional mutation (rs3856806 C>T) in the PPARG gene with BC risk. Our results indicated that PPARG rs3856806 C>T polymorphism was not associated with the risk of BC, which was consistent with the previous study [30]. This conclusion, however, should be elucidated with caution as only three moderate sample sizes studies were included, which may have insufficient power to detect a reliable correlation. In the future, more studies with large sample sizes are warranted to verify our findings.
There are some merits in our study. First of all, the sample sizes were larger as compared with previous studies. Secondly, we confirmed for the first time PPARG rs1801282 G allele was correlated with the susceptibility of breast cancer in Caucasians and in post-menopause women. Finally, the quality scores of all recruited studies were ≥ 6.5 (‘high quality’, Table 2), suggesting the reliability of our results. However, certain potential limitations that may lead to bias are also acknowledged and addressed. This meta-analysis only used published studies; certain publication bias may inevitably exist. Moreover, the included publications were performed mainly in Caucasians, only two Asians and two mixed populations were recruited, which restricted the power to detect a real influence. Hence, more large-scale studies in more diverse populations are needed. Furthermore, due to lack of genotype frequency information, we did not conduct further evaluation of potential interactions, such as age, family history, hormone replacement therapy use, oral contraceptives use, body mass index, other environmental factors and lifestyle. In consideration of the complexity of cancer etiology, these gene-environment interactions should not be ignored. Finally, the association between other important polymorphisms (e.g., PPARG rs4135247, rs1175543, rs709158 and rs2938395) and BC was seldom explored, these polymorphisms were not considered in our study.
In summary, this updated meta-analysis suggests that PPARG rs1801282 C>G variants are associated with a significantly decreased risk of BC in Caucasians and in post-menopause women. As only nine publications were included in our analysis and the evidence was relatively limited, more large and well-designed epidemiological studies with the consideration of gene-gene and gene-environment interactions are definitely demanded.
Acknowledgements
This study was supported in part by Jiangsu University Clinical Medicine Science and Technology Development Fund (JLY20140012), National Natural Science Foundation of China (81472332, 81341006), Fujian Province Natural Science Foundation (2013J01126, 2013J05116), Fujian Medical University professor fund (JS12008). The Fund of Union Hospital (2015TC-1-048 and 2015TC-2-004), Fujian Province Science and Technology Programmed Fund (2012Y0030), Fujian Medical Innovation Fund (2014-CX-15) and Union Hospital Fund (2015TC-1-048 and 2015TC-2-004).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T, Jayatilake H, McGuffog L, Hanks S, Evans DG, Eccles D Breast Cancer Susceptibility Collaboration (UK) Easton DF, Stratton MR. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, Chagtai T, Jayatilake H, Ahmed M, Spanova K, North B, McGuffog L, Evans DG, Eccles D Breast Cancer Susceptibility Collaboration (UK) Easton DF, Stratton MR, Rahman N. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 4.Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, Bishop DT, Weber B, Lenoir G, Chang-Claude J, Sobol H, Teare MD, Struewing J, Arason A, Scherneck S, Peto J, Rebbeck TR, Tonin P, Neuhausen S, Barkardottir R, Eyfjord J, Lynch H, Ponder BA, Gayther SA, Zelada-Hedman M, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vargas AC, Reis-Filho JS, Lakhani SR. Phenotype-genotype correlation in familial breast cancer. J Mammary Gland Biol Neoplasia. 2011;16:27–40. doi: 10.1007/s10911-011-9204-6. [DOI] [PubMed] [Google Scholar]
- 6.Hemel D, Domchek SM. Breast cancer predisposition syndromes. Hematol Oncol Clin North Am. 2010;24:799–814. doi: 10.1016/j.hoc.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Kim HC, Lee JY, Sung H, Choi JY, Park SK, Lee KM, Kim YJ, Go MJ, Li L, Cho YS, Park M, Kim DJ, Oh JH, Kim JW, Jeon JP, Jeon SY, Min H, Kim HM, Park J, Yoo KY, Noh DY, Ahn SH, Lee MH, Kim SW, Lee JW, Park BW, Park WY, Kim EH, Kim MK, Han W, Lee SA, Matsuo K, Shen CY, Wu PE, Hsiung CN, Lee JY, Kim HL, Han BG, Kang D. A genome-wide association study identifies a breast cancer risk variant in ERBB4 at 2q34: results from the Seoul Breast Cancer Study. Breast Cancer Res. 2012;14:R56. doi: 10.1186/bcr3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rinella ES, Shao Y, Yackowski L, Pramanik S, Oratz R, Schnabel F, Guha S, LeDuc C, Campbell CL, Klugman SD, Terry MB, Senie RT, Andrulis IL, Daly M, John EM, Roses D, Chung WK, Ostrer H. Genetic variants associated with breast cancer risk for Ashkenazi Jewish women with strong family histories but no identifiable BRCA1/2 mutation. Hum Genet. 2013;132:523–536. doi: 10.1007/s00439-013-1269-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barzan D, Veldwijk MR, Herskind C, Li Y, Zhang B, Sperk E, Du WD, Zhang XJ, Wenz F. Comparison of genetic variation of breast cancer susceptibility genes in Chinese and German populations. Eur J Hum Genet. 2013;21:1286–1292. doi: 10.1038/ejhg.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Brien KM, Cole SR, Poole C, Bensen JT, Herring AH, Engel LS, Millikan RC. Replication of breast cancer susceptibility loci in whites and African Americans using a Bayesian approach. Am J Epidemiol. 2014;179:382–394. doi: 10.1093/aje/kwt258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han MR, Deming-Halverson S, Cai Q, Wen W, Shrubsole MJ, Shu XO, Zheng W, Long J. Evaluating 17 breast cancer susceptibility loci in the Nashville breast health study. Breast Cancer. 2015;22:544–51. doi: 10.1007/s12282-014-0518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oh SW, Park CY, Lee ES, Yoon YS, Lee ES, Park SS, Kim Y, Sung NJ, Yun YH, Lee KS, Kang HS, Kwon Y, Ro J. Adipokines, insulin resistance, metabolic syndrome, and breast cancer recurrence: a cohort study. Breast Cancer Res. 2011;13:R34. doi: 10.1186/bcr2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Healy LA, Ryan AM, Carroll P, Ennis D, Crowley V, Boyle T, Kennedy MJ, Connolly E, Reynolds JV. Metabolic syndrome, central obesity and insulin resistance are associated with adverse pathological features in postmenopausal breast cancer. Clin Oncol (R Coll Radiol) 2010;22:281–288. doi: 10.1016/j.clon.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Amir E, Cecchini RS, Ganz PA, Costantino JP, Beddows S, Hood N, Goodwin PJ. 25-Hydroxy vitamin-D, obesity, and associated variables as predictors of breast cancer risk and tamoxifen benefit in NSABP-P1. Breast Cancer Res Treat. 2012;133:1077–1088. doi: 10.1007/s10549-012-2012-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rose DP, Komninou D, Stephenson GD. Obesity, adipocytokines, and insulin resistance in breast cancer. Obes Rev. 2004;5:153–165. doi: 10.1111/j.1467-789X.2004.00142.x. [DOI] [PubMed] [Google Scholar]
- 16.Kopp TI, Friis S, Christensen J, Tjonneland A, Vogel U. Polymorphisms in genes related to inflammation, NSAID use, and the risk of prostate cancer among Danish men. Cancer Genet. 2013;206:266–278. doi: 10.1016/j.cancergen.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Canbay E, Kurnaz O, Canbay B, Bugra D, Cakmakoglu B, Bulut T, Yamaner S, Sokucu N, Buyukuncu Y, Yilmaz-Aydogan H. PPAR-gamma Pro12Ala polymorphism and gastric cancer risk in a Turkish population. Asian Pac J Cancer Prev. 2012;13:5875–5878. doi: 10.7314/apjcp.2012.13.11.5875. [DOI] [PubMed] [Google Scholar]
- 18.Crous-Bou M, Rennert G, Salazar R, Rodriguez-Moranta F, Rennert HS, Lejbkowicz F, Kopelovich L, Lipkin SM, Gruber SB, Moreno V. Genetic polymorphisms in fatty acid metabolism genes and colorectal cancer. Mutagenesis. 2012;27:169–176. doi: 10.1093/mutage/ger066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abuli A, Fernandez-Rozadilla C, Alonso-Espinaco V, Munoz J, Gonzalo V, Bessa X, Gonzalez D, Clofent J, Cubiella J, Morillas JD, Rigau J, Latorre M, Fernandez-Banares F, Pena E, Riestra S, Paya A, Jover R, Xicola RM, Llor X, Carvajal-Carmona L, Villanueva CM, Moreno V, Pique JM, Carracedo A, Castells A, Andreu M, Ruiz-Ponte C, Castellví-Bel S Gastrointestinal Oncology Group of the Spanish Gastroenterological Association. Case-control study for colorectal cancer genetic susceptibility in EPICOLON: previously identified variants and mucins. BMC Cancer. 2011;11:339. doi: 10.1186/1471-2407-11-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang H, Dong X, Hassan M, Abbruzzese JL, Li D. Body mass index and obesity- and diabetes-associated genotypes and risk for pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2011;20:779–792. doi: 10.1158/1055-9965.EPI-10-0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim WY, Chen Y, Ali SM, Chuah KL, Eng P, Leong SS, Lim E, Lim TK, Ng AW, Poh WT, Tee A, Teh M, Salim A, Seow A. Polymorphisms in inflammatory pathway genes, host factors and lung cancer risk in Chinese female never-smokers. Carcinogenesis. 2011;32:522–529. doi: 10.1093/carcin/bgr006. [DOI] [PubMed] [Google Scholar]
- 22.Xu W, Li Y, Wang X, Chen B, Liu S, Wang Y, Zhao W, Wu J. PPARgamma polymorphisms and cancer risk: a meta-analysis involving 32,138 subjects. Oncol Rep. 2010;24:579–585. [PubMed] [Google Scholar]
- 23.Martinez-Nava GA, Burguete-Garcia AI, Lopez-Carrillo L, Hernandez-Ramirez RU, Madrid-Marina V, Cebrian ME. PPARgamma and PPARGC1B polymorphisms modify the association between phthalate metabolites and breast cancer risk. Biomarkers. 2013;18:493–501. doi: 10.3109/1354750X.2013.816776. [DOI] [PubMed] [Google Scholar]
- 24.Wu MH, Chu CH, Chou YC, Chou WY, Yang T, Hsu GC, Yu CP, Yu JC, Sun CA. Joint effect of peroxisome proliferator-activated receptor gamma genetic polymorphisms and estrogen-related risk factors on breast cancer risk: results from a case-control study in Taiwan. Breast Cancer Res Treat. 2011;127:777–784. doi: 10.1007/s10549-010-1282-4. [DOI] [PubMed] [Google Scholar]
- 25.Gallicchio L, McSorley MA, Newschaffer CJ, Huang HY, Thuita LW, Hoffman SC, Helzlsouer KJ. Body mass, polymorphisms in obesity-related genes, and the risk of developing breast cancer among women with benign breast disease. Cancer Detect Prev. 2007;31:95–101. doi: 10.1016/j.cdp.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, McCullough ML, Stevens VL, Rodriguez C, Jacobs EJ, Teras LR, Pavluck AL, Thun MJ, Calle EE. Nested case-control study of energy regulation candidate gene single nucleotide polymorphisms and breast cancer. Anticancer Res. 2007;27:589–593. [PubMed] [Google Scholar]
- 27.Memisoglu A, Hankinson SE, Manson JE, Colditz GA, Hunter DJ. Lack of association of the codon 12 polymorphism of the peroxisome proliferator-activated receptor gamma gene with breast cancer and body mass. Pharmacogenetics. 2002;12:597–603. doi: 10.1097/00008571-200211000-00003. [DOI] [PubMed] [Google Scholar]
- 28.Petersen RK, Larsen SB, Jensen DM, Christensen J, Olsen A, Loft S, Nellemann C, Overvad K, Kristiansen K, Tjonneland A, Vogel U. PPARgamma-PGC-1alpha activity is determinant of alcohol related breast cancer. Cancer Lett. 2012;315:59–68. doi: 10.1016/j.canlet.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 29.Justenhoven C, Hamann U, Schubert F, Zapatka M, Pierl CB, Rabstein S, Selinski S, Mueller T, Ickstadt K, Gilbert M, Ko YD, Baisch C, Pesch B, Harth V, Bolt HM, Vollmert C, Illig T, Eils R, Dippon J, Brauch H. Breast cancer: a candidate gene approach across the estrogen metabolic pathway. Breast Cancer Res Treat. 2008;108:137–149. doi: 10.1007/s10549-007-9586-8. [DOI] [PubMed] [Google Scholar]
- 30.Park B, Shin A, Kim KZ, Lee YS, Hwang JA, Kim Y, Sung J, Yoo KY, Lee ES. Lack of effects of peroxisome proliferator-activated receptor gamma genetic polymorphisms on breast cancer risk: a case-control study and pooled analysis. Asian Pac J Cancer Prev. 2014;15:9093–9099. doi: 10.7314/apjcp.2014.15.21.9093. [DOI] [PubMed] [Google Scholar]
- 31.Mao Q, Guo H, Gao L, Wang H, Ma X. Peroxisome proliferator-activated receptor gamma2 Pro12Ala (rs1801282) polymorphism and breast cancer susceptibility: a meta-analysis. Mol Med Rep. 2013;8:1773–1778. doi: 10.3892/mmr.2013.1735. [DOI] [PubMed] [Google Scholar]
- 32.Mills E, Slotkin TA, Sampson SR. Letter: Carotid body chemoreceptors. Nature. 1975;258:268–269. doi: 10.1038/258268b0. [DOI] [PubMed] [Google Scholar]
- 33.Guo J, Jin M, Zhang M, Chen K. A genetic variant in miR-196a2 increased digestive system cancer risks: a meta-analysis of 15 case-control studies. PLoS One. 2012;7:e30585. doi: 10.1371/journal.pone.0030585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu MT, Hu JW, Ding XX, Yang X, Zhang Z, Yin R, Xu L. Hsa-miR-499 rs3746444 polymorphism contributes to cancer risk: a meta-analysis of 12 studies. PLoS One. 2012;7:e50887. doi: 10.1371/journal.pone.0050887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hua Z, Li D, Xiang G, Xu F, Jie G, Fu Z, Jie Z, Da P, Li D. PD-1 polymorphisms are associated with sporadic breast cancer in Chinese Han population of Northeast China. Breast Cancer Res Treat. 2011;129:195–201. doi: 10.1007/s10549-011-1440-3. [DOI] [PubMed] [Google Scholar]
- 36.Bayram S, Akkiz H, Ulger Y, Bekar A, Akgollu E, Yildirim S. Lack of an association of programmed cell death-1 PD1.3 polymorphism with risk of hepatocellular carcinoma susceptibility in Turkish population: a case-control study. Gene. 2012;511:308–313. doi: 10.1016/j.gene.2012.09.119. [DOI] [PubMed] [Google Scholar]
- 37.Wei W, Jiang M, Luo L, Li Z, Wang P, Dong WQ. Colorectal cancer susceptibility variants alter risk of breast cancer in a Chinese Han population. Genet Mol Res. 2013;12:6268–6274. doi: 10.4238/2013.December.4.14. [DOI] [PubMed] [Google Scholar]
- 38.Michalik L, Desvergne B, Wahli W. Peroxisome-proliferator-activated receptors and cancers: complex stories. Nat Rev Cancer. 2004;4:61–70. doi: 10.1038/nrc1254. [DOI] [PubMed] [Google Scholar]
- 39.He W. PPARgamma2 polymorphism and human health. PPAR Res. 2009;2009:849538. doi: 10.1155/2009/849538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yen CJ, Beamer BA, Negri C, Silver K, Brown KA, Yarnall DP, Burns DK, Roth J, Shuldiner AR. Molecular scanning of the human peroxisome proliferator activated receptor gamma (hPPAR gamma) gene in diabetic Caucasians: identification of a Pro12Ala PPAR gamma 2 missense mutation. Biochem Biophys Res Commun. 1997;241:270–274. doi: 10.1006/bbrc.1997.7798. [DOI] [PubMed] [Google Scholar]
- 41.Masugi J, Tamori Y, Mori H, Koike T, Kasuga M. Inhibitory effect of a proline-to-alanine substitution at codon 12 of peroxisome proliferator-activated receptor-gamma 2 on thiazolidinedione-induced adipogenesis. Biochem Biophys Res Commun. 2000;268:178–182. doi: 10.1006/bbrc.2000.2096. [DOI] [PubMed] [Google Scholar]
- 42.Doney A, Fischer B, Frew D, Cumming A, Flavell DM, World M, Montgomery HE, Boyle D, Morris A, Palmer CN. Haplotype analysis of the PPARgamma Pro12Ala and C1431T variants reveals opposing associations with body weight. BMC Genet. 2002;3:21. doi: 10.1186/1471-2156-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kury S, Buecher B, Robiou-du-Pont S, Scoul C, Colman H, Le Neel T, Le Houerou C, Faroux R, Ollivry J, Lafraise B, Chupin LD, Sebille V, Bezieau S. Low-penetrance alleles predisposing to sporadic colorectal cancers: a French case-controlled genetic association study. BMC Cancer. 2008;8:326. doi: 10.1186/1471-2407-8-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang J, Gajalakshmi V, Wang J, Kuriki K, Suzuki S, Nakamura S, Akasaka S, Ishikawa H, Tokudome S. Influence of the C161T but not Pro12Ala polymorphism in the peroxisome proliferator-activated receptor-gamma on colorectal cancer in an Indian population. Cancer Sci. 2005;96:507–512. doi: 10.1111/j.1349-7006.2005.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vogel U, Christensen J, Dybdahl M, Friis S, Hansen RD, Wallin H, Nexo BA, Raaschou-Nielsen O, Andersen PS, Overvad K, Tjonneland A. Prospective study of interaction between alcohol, NSAID use and polymorphisms in genes involved in the inflammatory response in relation to risk of colorectal cancer. Mutat Res. 2007;624:88–100. doi: 10.1016/j.mrfmmm.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 46.Wang SS, Davis S, Cerhan JR, Hartge P, Severson RK, Cozen W, Lan Q, Welch R, Chanock SJ, Rothman N. Polymorphisms in oxidative stress genes and risk for non-Hodgkin lymphoma. Carcinogenesis. 2006;27:1828–1834. doi: 10.1093/carcin/bgl013. [DOI] [PubMed] [Google Scholar]
- 47.Smith WM, Zhou XP, Kurose K, Gao X, Latif F, Kroll T, Sugano K, Cannistra SA, Clinton SK, Maher ER, Prior TW, Eng C. Opposite association of two PPARG variants with cancer: overrepresentation of H449H in endometrial carcinoma cases and underrepresentation of P12A in renal cell carcinoma cases. Hum Genet. 2001;109:146–151. doi: 10.1007/s004390100563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


