Abstract
Background: Published data regarding the associations between glutathione S-transferase (GST) T1, M1 and P1 polymorphisms and breast cancer risk are inconclusive. The aim of this study is to comprehensively evaluate the genetic risk of GST genes for breast cancer. Materials and Methods: A systematic literature search was carried out in Pubmed, Medline (Ovid), Embase, CBM, CNKI, Weipu, and Wanfang database, covering all publications (last search was performed on May 20, 2015). Statistical analysis was performed using Revman 5.2 and STATA 12.0 softwares. Results: A total of 12,035 cases and 13,911 controls in 34 case-control studies were included in this meta-analysis. The results suggested that the GSTM1 and GSTP1 polymorphisms can obviously increase the risk of breast cancer in Asian population (odds ratio (OR) = 1.18, 95% confidence interval (CI) = 1.04-1.33, P = 0.008 and OR = 1.23, 95% CI = 1.07-1.41, P = 0.003, respectively), especially in East Asian (OR = 1.14, 95% CI = 1.01-1.27, P = 0.03 and OR = 1.15, 95% CI = 1.03-1.28, P = 0.01, respectively) and hospital-based case-control study (HCC) group (OR = 1.32, 95% CI = 1.11-1.56, P = 0.001 and OR = 1.38, 95% CI = 1.03-1.84, P = 0.03, respectively), while the association between GSTT1 null genotype and breast cancer risk is not significant (OR = 1.08, 95% CI = 0.93-1.25, P = 0.3). Conclusions: This meta-analysis indicated that the GSTM1 and GSTP1 polymorphisms might significantly contribute to breast cancer susceptibility in Asian population, especially in East Asian, while the GSTT1 polymorphism might not be associated with breast cancer.
Keywords: GSTT1, GSTM1, GSTP1, polymorphism, breast cancer, susceptibility, meta-analysis
Introduction
Breast cancer was reported to be the most frequently diagnosed cancer and one of the leading causes of cancer-related death in females worldwide, which has become a major public health challenge [1,2]. Some studies suggested that Asian women were highly susceptible to breast cancer, and it was reported that the number of women with incident breast cancer in Asia was estimated at 651,000 in 2012, comprising 38.8% of all cases globally, followed by Europe (27.7% of all cases) and North America (15.3% of all cases) [3,4]. Now, the mechanism of breast cancer is still not fully understood. It has been suggested that susceptibility genes combining with environmental factors may be important in the development of breast cancer [5,6].
In recent years, several common genes have been identified as potential breast cancer susceptibility genes. An important one is glutathione S-transferase (GST), which plays a key role in the detoxification of a broad range of toxic and potentially carcinogenic compounds [7]. In humans, five common classes of GST enzymes have been identified (GST classes α, μ, π, ω and θ) and each class is encoded by a separate gene or gene family (respectively are GSTA, GSTM, GSTP, GSTO and GSTT genes). Allelic variants for each of these genes may result in less effective or absent enzymatic detoxification and thus increase susceptibility to cancer, although the exact biochemical processes are not yet fully understood. Among these genes, the deletion mutations in GSTT1 and GSTM1 and the amino acid transition (A313G→Ile105Val) in GSTP1 to breast cancer risk have been a research focus in scientific community and have drawn increasing attention. Despite the fact that lots of the epidemiologic investigations studying the association of these three polymorphisms with breast cancer risk were conducted in the past decades, the available evidences are still weak at present, due to the possible small effect of each individual polymorphism on breast cancer risk and the relatively small sample size in each of published studies. Therefore, we performed the present meta-analysis aimed at utilizing the acquirable data of GST polymorphisms with breast cancer risk in Asian population to derive a more precise estimation of these associations and evaluating the trends in occurrence of breast cancer in this population.
Materials and methods
Selection of studies
A comprehensive literature search was carried out in Pubmed, Medline (Ovid), Embase, Chinese biomedical database (CBM), China national knowledge infrastructure (CNKI), Weipu and Wanfang database to identify studies involving association between the GSTT1, GSTM1 and GSTP1 polymorphisms and breast cancer risk in Asian population (last search was updated on May 20, 2015). The search terms were used as follows: (glutathione S-transferase T1) OR (glutathione S-transferase M1) OR (glutathione S-transferase P1) OR (GSTT1) OR (GSTM1) OR (GSTP1) in combination with (polymorphism) OR (variant) OR (mutation), (breast cancer) OR (breast carcinoma) OR (breast neoplasm) AND (Asia) OR (Asian). The search results were limited to English and Chinese languages. Studies included in our meta-analysis met the following inclusion criteria: (1) evaluation of the glutathione S-transferase T1, M1 and P1 polymorphisms and breast cancer risk in Asian population, (2) the design had to be a case-control design published in a journal, (3) genotype distributions in both cases and controls were available for estimating an odds ratio with 95% confidence interval (CI) and P value, and (4) genotype distributions in control group should be consistent with Hardy-Weinberg equilibrium (HWE). Studies were excluded if one of the following existed: (1) no controls, (2) genotype frequencies or numbers not reported, and (3) abstracts, reviews. For duplications or overlapping publications, the studies with larger number of cases and controls or been published latest were included.
Data extraction
Two independent reviewers (QXZ and JQT) collected the data and reached a consensus on all items. In case of disagreement, a third author (FZ) would assess these articles. A standardized data form was used and included: first author’s name, year of publication, original country, subregion of Asia, case age, study design, total number of cases and controls and genotyping method.
Quality assessment
We evaluated the methodological quality of the included studies according to the Newcastle-Ottawa Scale (NOS) criteria [8]. The NOS criteria is scored based on three aspects: (1) subject selection, 0~4; (2) comparability of subject, 0~2; and (3) clinical outcome, 0~3. Total NOS scores range from 0 to 9, with scores ≥ 7 indicating good quality.
Statistical analysis
Odds ratios (OR) with 95% CI were used to assess the strength of association between the glutathione S-transferase T1, M1 and P1 polymorphisms and breast cancer risk in Asian population. We first examined GSTT1 and GSTM1 genotypes using (Null vs Present) model. Then, the relationship between the GSTP1 polymorphism and susceptibility to breast cancer was estimated with the dominant (GG+AG vs AA) and allelic (G vs A) models. The pooled OR was calculated by a fixed-effect model or a random-effect model according to the heterogeneity. Heterogeneity was checked by a χ2-based Q statistic and P < 0.10 was considered statistically significant. A P-value ≥ 0.10 for the Q-test indicated the lack of heterogeneity among the studies, and so the summary OR estimate of each study was calculated by the fixed-effect model [9]. Otherwise, the random-effect model was used [10]. The statistical significance of OR was analyzed by Z test, and P < 0.05 was considered statistically significant. To evaluate the subregion-specific, menopausal status-specific and study design-specific effects, we performed stratification analyses on subregion, menopausal status and study design. For the subgroup analysis by subregion, the study populations were stratified into four groups: East Asia, Southeast Asia, South Asia and West Asia. And for stratification analysis by menopausal status, the available study populations were stratified into two groups: premenopausal and postmenopausal. In addition, subjects were categorized into different classifications according to study design: population-based case-control study (PCC) and hospital-based case-control study (HCC). Sensitivity analysis was also performed by sequentially excluding individual study to check the robustness of the result [11]. The possible publication bias was examined visually in a Begg’s funnel plot and the degree of asymmetry was tested by Egger’s test (P < 0.05 was considered representative of statistically significant publication bias). HWE was tested by Pearson’s x2 test [12]. Statistical analysis was performed using Revman 5.2 and Stata 12.0 softwares.
Results
Study inclusion and characteristics
As shown in Figure 1, the initial search identified 591 results from the selected electronic databases. After reading the titles and abstracts, 122 potential articles were included for full-text view. After reading full texts, 86 studies were excluded for being irrelevant to the glutathione S-transferase T1, M1 and P1 polymorphisms and breast cancer risk. Therefore, 36 full-text articles remained for data extraction. 1 article was excluded for repeating or overlapping [13]. In addition, the control group genotype for GSTP1 in 1 case-control study was not consistent with HWE and this study was excluded [14]. Finally, a total of 34 case-control studies published in 34 articles which met our inclusion criteria were identified, including 12,035 cases and 13,911 controls. The characteristics and methodological quality of each case-control study were listed in Table 1. GST genotypes and allele distributions for each case-control study are shown in Table 2. GST genotypes distributions for each case-control study in subgroup by menopausal status are shown in Table 3. There was 1 case-control study of GSTT1 polymorphism [15], 3 of GSTM1 polymorphism [16-18], 6 of GSTP1 polymorphism [19-24], 10 of GSTT1 and GSTM1 polymorphisms [25-34], 2 of GSTM1 and GSTP1 polymorphisms [35,36], 12 of GSTT1, GSTM1 and GSTP1 polymorphisms [37-48]. All the included 34 eligible reports were written in English or Chinese.
Figure 1.
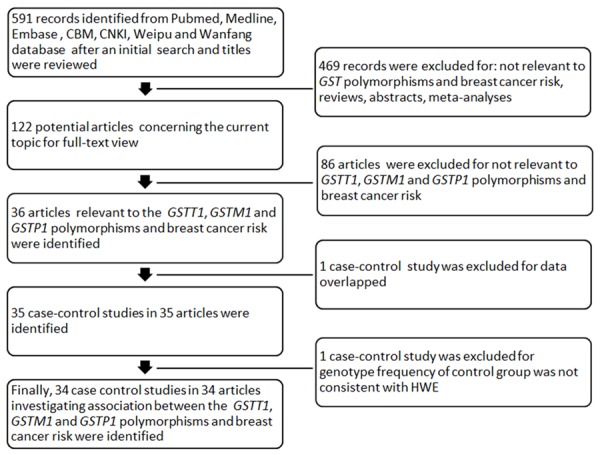
Flow diagram of included/excluded studies in this meta-analysis.
Table 1.
Baseline characteristics and methodological quality of all included studies in the meta-analysis
| First author | Year | Country | Subregion | Case age (year) | Study design | Sample size (Cases/Controls) | Genotyping method | NOS score |
|---|---|---|---|---|---|---|---|---|
| Ceschi et al. | 2005 | Singapore | Southeast Asia | 55.6 ± 7.4† | PCC | 257/668 | TaqMan & PCR | 7 |
| Chacko et al. | 2005 | India | South Asia | 49 ± 10.3† | HCC | 112/112 | multiplex-PCR | 6 |
| Chang et al. | 2006 | China | East Asia | NM | HCC | 189/420 | PCR | 7 |
| Cheng et al. | 2005 | China | East Asia | NM | PCC | 465/736 | multiplex-PCR | 7 |
| Egan et al. | 2004 | China | East Asia | 47 | PCC | 1143/1221 | multiplex-PCR & RFLP-PCR | 8 |
| Gago-Dominguez et al. | 2004 | Singapore | Southeast Asia | NM | PCC | 180/466 | TaqMan | 7 |
| Ge et al. | 2013 | China | East Asia | 54.3 | HCC | 920/783 | TaqMan | 7 |
| Geng et al. | 2010 | China | East Asia | 46.8 | HCC | 50/15 | PCR | 5 |
| Hashemi et al. | 2012 | Iran | West Asia | 47.9 ± 13.3† | HCC | 134/152 | multiplex-PCR & PCR | 7 |
| Kadouri et al. | 2008 | Israel | West Asia | NM | HCC | 211/109 | PCR | 6 |
| Kaushal et al. | 2010 | India | South Asia | 45.5 ± 12.86† | PCC | 117/174 | RFLP-PCR | 7 |
| Khabaz et al. | 2014 | Jordan | West Asia | 44.66 | PCC | 100/48 | RFLP-PCR | 5 |
| Khabaz et al. | 2015 | Saudi Arabia | West Asia | 54.6 | HCC | 86/35 | PCR | 5 |
| Kim et al. | 2004 | Korea | East Asia | NM | HCC | 171/171 | RFLP-PCR | 6 |
| Lee et al. | 2008 | China | East Asia | 49.6 ± 8.3† | PCC | 3026/3037 | RFLP-PCR & TaqMan | 8 |
| Li et al. | 2008 | China | East Asia | 46.7 ± 8.75† | HCC | 78/78 | multiplex-PCR | 8 |
| Luo et al. | 2012 | China | East Asia | 52.8 ± 8.8† | PCC | 353/701 | PCR | 7 |
| Ma et al. | 2007 | China | East Asia | 46 ± 9† | HCC | 105/100 | PCR | 7 |
| Masoudi et al. | 2010 | Iran | West Asia | 45.9 | HCC | 181/181 | PCR | 7 |
| Nosheen et al. | 2011 | Pakistan | South Asia | 48 | PCC | 150/150 | PCR | 7 |
| Park et al. | 2000 | Korea | East Asia | NM | HCC | 188/181 | PCR | 7 |
| Park et al. | 2004 | Korea | East Asia | 47.9 ± 11.2† | HCC | 200/289 | multiplex-PCR | 7 |
| Pongtheerat et al. | 2009 | Thailand | Southeast Asia | NM | HCC | 43/56 | mutiplex-PCR & PCR | 5 |
| Rajkumar et al. | 2008 | India | South Asia | 46 | PCC | 250/500 | PCR | 7 |
| Sakoda et al. | 2008 | China | East Asia | 45 | PCC | 615/878 | multiplex-PCR & PCR | 8 |
| Samson et al. | 2007 | India | South Asia | 46 | PCC | 250/500 | TaqMan & PCR | 7 |
| Saxena et al. | 2009 | India | South Asia | NM | HCC | 406/403 | multiplex-PCR & RFLP-PCR | 7 |
| Sohail et al. | 2013 | Pakistan | South Asia | NM | HCC | 100/102 | multiplex-PCR & PCR | 7 |
| Syamala et al. | 2008 | India | South Asia | NM | HCC | 347/250 | multiplex-PCR & RFLP-PCR | 6 |
| Wang et al. | 2002 | China | East Asia | 49 | PCC | 42/108 | PCR | 5 |
| Wu et al. | 2002 | China | East Asia | 46.7 ± 10.2† | HCC | 60/60 | PCR | 7 |
| Wu et al. | 2006 | China | East Asia | 49.11 | HCC | 262/225 | PCR | 7 |
| Yu et al. | 2009 | China | East Asia | 47.6 ± 10.6† | HCC | 1017/903 | RFLP-PCR | 7 |
| Zgheib et al. | 2013 | Lebanon | West Asia | 48.9 ± 11.6† | HCC | 227/99 | PCR | 7 |
HCC: hospital-based case-control study; PCC: population-based case-control study; NM: not mentioned; PCR: polymerase chain reaction; RFLP-PCR: polymerase chain reaction-restriction fragment length polymorphism; NOS: Newcastle-Ottawa Scale;
Mean ± SD.
Table 2.
Distribution of GST genotypes and allele among breast cancers and controls
| Author | GSTT1 | GSTM1 | GSTP1 | HWEa for control P | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||||
| Cases (n) | Controls (n) | Cases (n) | Controls (n) | Cases (n) | Controls (n) | Cases (n) | Controls (n) | ||||||||||||
|
| |||||||||||||||||||
| Null | Present | Null | Present | Null | Present | Null | Present | GG | AG | AA | GG | AG | AA | G | A | G | A | ||
| Ceschi et al. | 87 | 169 | 282 | 385 | 119 | 137 | 298 | 369 | 9 | 87 | 161 | 27 | 199 | 442 | 105 | 409 | 253 | 1083 | 0.4429 |
| Chang et al. | 111 | 78 | 210 | 210 | 107 | 82 | 227 | 193 | NA | 66* | 123 | NA | 133* | 288 | — | — | — | — | |
| Egan et al. | 557 | 579 | 596 | 614 | 628 | 497 | 683 | 523 | 53 | 363 | 723 | 31 | 371 | 809 | 469 | 1809 | 433 | 1989 | 0.1315 |
| Gago-Dominguez et al. | 66 | 114 | 204 | 262 | 82 | 98 | 218 | 248 | NA | 65* | 115 | NA | 162* | 304 | — | — | — | — | |
| Hashemi et al. | 18 | 116 | 12 | 140 | 86 | 48 | 71 | 81 | 26 | 72 | 36 | 3 | 52 | 97 | 124 | 144 | 58 | 246 | 0.1833 |
| Kadouri et al. | 53 | 158 | 24 | 84 | 105 | 106 | 63 | 46 | 16 | 74 | 121 | 3 | 29 | 76 | 106 | 316 | 35 | 181 | 0.9073 |
| Kaushal et al. | 33 | 84 | 69 | 105 | 23 | 94 | 52 | 122 | 7 | 48 | 62 | 4 | 62 | 108 | 62 | 172 | 70 | 278 | 0.1515 |
| Pongtheer at et al. | 18 | 25 | 25 | 28 | 14 | 26 | 24 | 32 | NA | 13* | 30 | NA | 21* | 32 | — | — | — | — | |
| Saxena et al. | 96 | 310 | 88 | 315 | 215 | 191 | 134 | 269 | 66 | 193 | 147 | 32 | 171 | 200 | 325 | 487 | 235 | 571 | 0.5860 |
| Sohail et al. | 27 | 73 | 32 | 70 | 43 | 57 | 45 | 57 | 90 | 10 | 0 | 67 | 28 | 7 | 190 | 10 | 162 | 42 | 0.1050 |
| Syamala et al. | 56 | 291 | 23 | 227 | 119 | 228 | 63 | 187 | 21 | 140 | 186 | 16 | 109 | 125 | 182 | 512 | 141 | 359 | 0.2254 |
| Zgheib et al. | 43 | 183 | 20 | 78 | 111 | 115 | 47 | 51 | NA | 110* | 117 | NA | 49* | 49 | — | — | — | — | |
| Sakoda et al. | — | — | — | — | 321 | 294 | 428 | 450 | 20 | 215 | 378 | 30 | 277 | 569 | 255 | 971 | 337 | 1415 | 0.6000 |
| Samson et al. | — | — | — | — | 65 | 185 | 110 | 390 | 29 | 103 | 118 | 51 | 219 | 230 | 161 | 339 | 321 | 679 | 0.9150 |
| Chacko et al. | 29 | 83 | 10 | 102 | 40 | 72 | 28 | 84 | — | — | — | — | — | — | — | — | — | — | |
| Cheng et al. | 223 | 238 | 336 | 400 | 234 | 231 | 362 | 371 | — | — | — | — | — | — | — | — | — | — | |
| li et al. | 35 | 43 | 44 | 34 | 31 | 47 | 37 | 41 | — | — | — | — | — | — | — | — | — | — | |
| Luo et al. | 186 | 167 | 364 | 337 | 207 | 146 | 414 | 286 | — | — | — | — | — | — | — | — | — | — | |
| Ma et al. | 49 | 56 | 22 | 78 | 52 | 53 | 25 | 75 | — | — | — | — | — | — | — | — | — | — | |
| Masoudi et al. | 47 | 134 | 45 | 136 | 111 | 70 | 91 | 90 | — | — | — | — | — | — | — | — | — | — | |
| Nosheen et al. | 13 | 137 | 28 | 122 | 3 | 147 | 12 | 138 | — | — | — | — | — | — | — | — | — | — | |
| Park et al. -2000 | 94 | 94 | 76 | 105 | 110 | 78 | 95 | 86 | — | — | — | — | — | — | — | — | — | — | |
| Park et al. -2004 | 101 | 99 | 121 | 168 | 116 | 84 | 152 | 137 | — | — | — | — | — | — | — | — | — | — | |
| Wu et al. -2002 | 27 | 33 | 26 | 34 | 34 | 26 | 25 | 35 | — | — | — | — | — | — | — | — | — | — | |
| Rajkumar et al. | 44 | 206 | 84 | 416 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | |
| Wang et al. | — | — | — | — | 24 | 18 | 52 | 56 | — | — | — | — | — | — | — | — | — | — | |
| Wu et al. -2006 | — | — | — | — | 123 | 139 | 103 | 122 | — | — | — | — | — | — | — | — | — | — | |
| Yu et al. | — | — | — | — | 622 | 395 | 510 | 393 | — | — | — | — | — | — | — | — | — | — | |
| Ge et al. | — | — | — | — | — | — | — | — | 55 | 325 | 540 | 34 | 230 | 519 | 435 | 1405 | 298 | 1268 | 0.1903 |
| Geng et al. | — | — | — | — | — | — | — | — | NA | 12* | 38 | NA | 1* | 14 | — | — | — | — | |
| Khabaz et al. -2014 | — | — | — | — | — | — | — | — | 2 | 40 | 58 | 2 | 18 | 28 | 44 | 156 | 22 | 74 | 0.6704 |
| Khabaz et al. -2015 | — | — | — | — | — | — | — | — | 1 | 45 | 40 | 2 | 14 | 19 | 47 | 125 | 18 | 52 | 0.7809 |
| Kim et al. | — | — | — | — | — | — | — | — | 5 | 44 | 122 | 6 | 52 | 113 | 54 | 288 | 64 | 278 | 0.9953 |
| Lee et al. | — | — | — | — | — | — | — | — | 123 | 953 | 1950 | 85 | 949 | 2003 | 1199 | 4853 | 1119 | 4955 | 0.2910 |
HWE: Hardy-Weinberg equilibrium for controls of GSTP1 gene;
NA: not available;
Numbers of GG+AG.
Table 3.
Distribution of GST genotypes among breast cancers and controls in subgroup by menopausal status
| Author | Premenopausal | Postmenopausal | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Cases (n) | Controls (n) | Cases (n) | Controls (n) | |||||||||
|
| ||||||||||||
| Null | Present | Null | Present | Null | Present | Null | Present | |||||
| GSTT1 | ||||||||||||
| Chacko et al. | 9 | 45 | 3 | 51 | 6 | 52 | 7 | 51 | ||||
| Hashemi et al. | 9 | 54 | 8 | 111 | 9 | 62 | 4 | 29 | ||||
| Park et al. -2000 | 57 | 57 | 42 | 55 | 37 | 37 | 32 | 48 | ||||
| Park et al. -2004 | 61 | 59 | 75 | 92 | 40 | 40 | 46 | 76 | ||||
| Saxena et al. | 34 | 146 | 24 | 150 | 62 | 164 | 64 | 165 | ||||
| GSTM1 | ||||||||||||
| Chacko et al. | 9 | 45 | 3 | 51 | 6 | 52 | 7 | 51 | ||||
| Hashemi et al. | 9 | 54 | 8 | 111 | 9 | 62 | 4 | 29 | ||||
| Park et al. -2000 | 57 | 57 | 42 | 55 | 37 | 37 | 32 | 48 | ||||
| Park et al. -2004 | 61 | 59 | 75 | 92 | 40 | 40 | 46 | 76 | ||||
| Saxena et al. | 34 | 146 | 24 | 150 | 62 | 164 | 64 | 165 | ||||
| Chacko et al. | 9 | 45 | 3 | 51 | 6 | 52 | 7 | 51 | ||||
| Hashemi et al. | 9 | 54 | 8 | 111 | 9 | 62 | 4 | 29 | ||||
| GSTM1 | ||||||||||||
| GG | AG | AA | GG | AG | AA | GG | AG | AA | GG | AG | AA | |
| Kim et al. | 2 | 32 | 67 | 4 | 27 | 70 | 3 | 12 | 55 | 2 | 25 | 43 |
| Lee et al. | 86 | 579 | 1161 | 48 | 553 | 1096 | 37 | 374 | 789 | 37 | 396 | 907 |
| Sakoda et al. | 11 | 100 | 181 | 18 | 156 | 353 | 9 | 115 | 197 | 12 | 121 | 216 |
| Saxena et al. | 18 | 92 | 70 | 14 | 106 | 51 | 48 | 101 | 77 | 18 | 65 | 149 |
Quantitative data synthesis
GSTT1 polymorphism with breast cancer risk
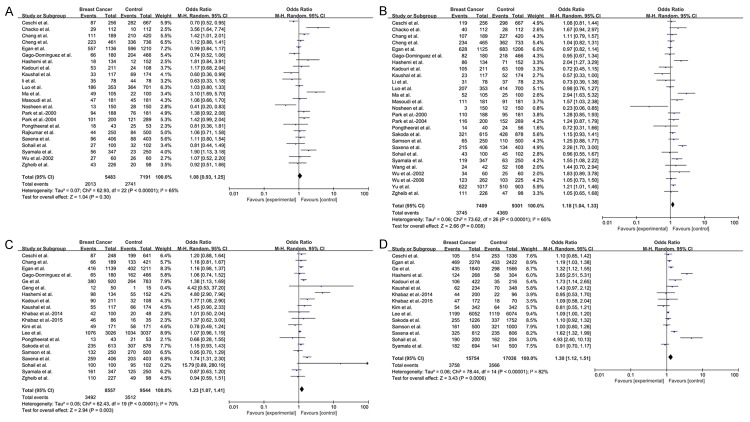
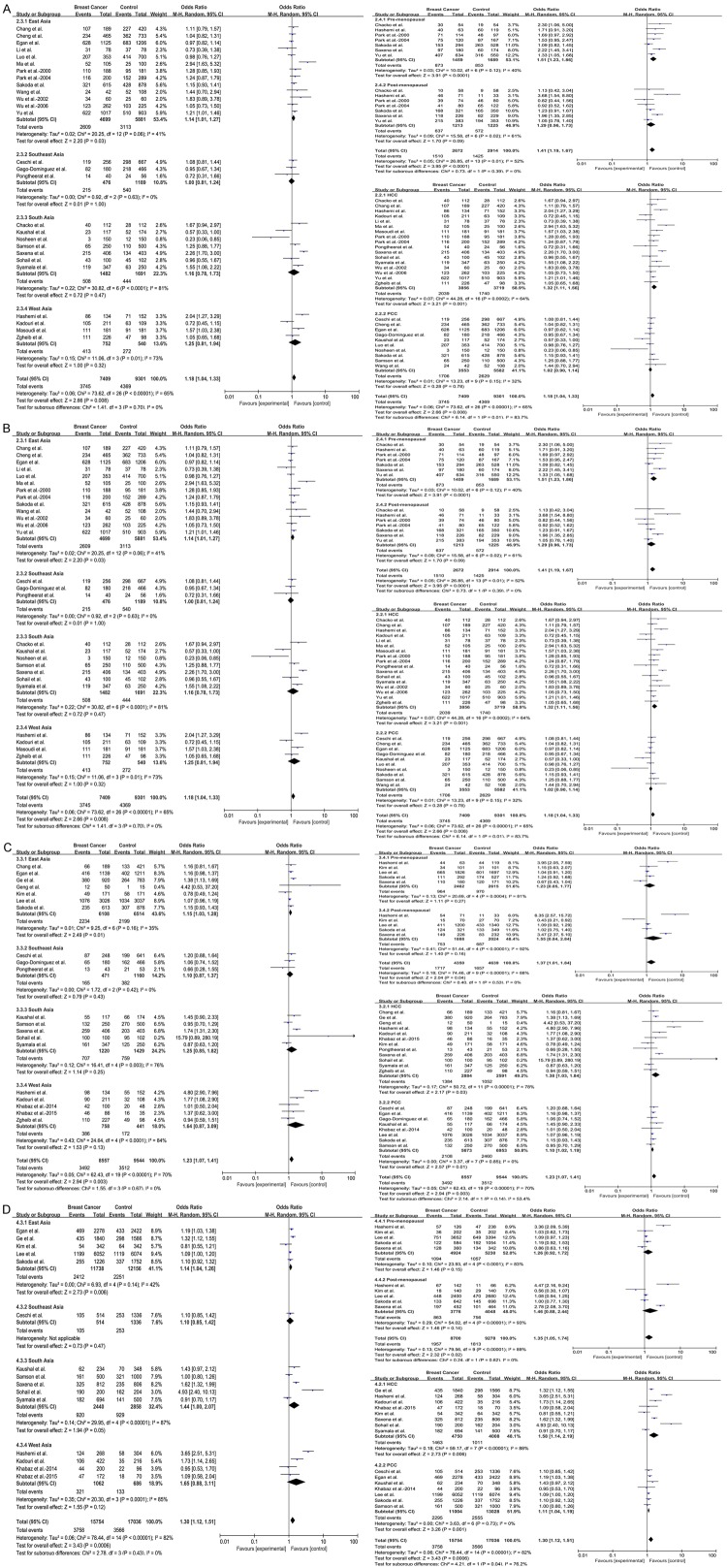
In this meta-analysis, we found that GSTT1 polymorphism was not associated with breast cancer risk in Asian population (OR = 1.08, 95% CI = 0.93-1.25, P = 0.30) (Figure 2A). However, in the subgroup analyses, this meta-analysis indicated that null/present polymorphism of GSTT1 significantly increased breast cancer risk in East Asian (OR = 1.20, 95% CI = 1.00-1.45, P = 0.05), premenopausal (OR = 1.45, 95% CI = 1.10-1.93, P = 0.009) and HCC (OR = 1.30, 95% CI = 1.07-1.59, P = 0.009) groups. Interestingly, GSTT1 polymorphism may have a lowered risk for breast cancer in Southeast Asian (OR = 0.73, 95% CI = 0.58-0.90, P = 0.004) (Figure 3A). The detailed data were listed in Table 4.
Figure 2.
Forest plots for the association between GST polymorphisms and breast cancer risk. Boxes represent the ORs of individual studies, and diamonds represent the overall OR. Horizontal lines represent the 95% CI. A. GSTT1 polymorphism. B. GSTM1 polymorphism. C. GSTP1 polymorphism under dominant model (GG+AG vs AA). D. GSTP1 polymorphism under allelic model (G vs A).
Figure 3.
Subgroup analyses for the association between GST polymorphisms and breast cancer risk. Boxes represent the OR of individual studies, and diamonds represent the overall OR. Horizontal lines represent the 95% CI. A. GSTT1 polymorphism. B. GSTM1 polymorphism. C. GSTP1 polymorphism under dominant model (GG+AG vs AA). D. GSTP1 polymorphism under allelic model (G vs A).
Table 4.
Meta-analysis of the GST polymorphisms on breast cancer risk in Asian population
| Description (No. of studies) | Subgroup (No. of studies) | Sample size | Analysis model | Test of association☆ | P value for Egger’s test | Test for heterogeneity | |||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| Cases | Controls | OR (95% CI) | P | P | I2 % | ||||
| GSTT1 (Null vs Present) | |||||||||
| Total [23] | 5483 | 7191 | R | 1.08 [0.93, 1.25] | 0.3000 | 0.493 | <0.00001 | 65 | |
| Subregion [23] | East Asia [9] | 2770 | 3775 | R | 1.20 [1.00, 1.45] | 0.0500 | 0.0070 | 62 | |
| Southeast Asia [3] | 479 | 1186 | R | 0.73 [0.58, 0.90] | 0.0040 | 0.9400 | 0 | ||
| South Asia [7] | 1482 | 1691 | R | 1.05 [0.70, 1.59] | 0.8000 | 0.0001 | 78 | ||
| West Asia [4] | 752 | 539 | R | 1.13 [0.85, 1.51] | 0.3900 | 0.5700 | 0 | ||
| Menopausal status [5] | Premenopausal [5] | 531 | 611 | F | 1.45 [1.10, 1.93] | 0.0090 | 0.6000 | 0 | |
| Postmenopausal [5] | 509 | 522 | F | 1.19 [0.90, 1.58] | 0.2100 | 0.5500 | 0 | ||
| Study design [23] | HCC [15] | 2580 | 2587 | R | 1.30 [1.07, 1.59] | 0.0090 | 0.0080 | 53 | |
| PCC [8] | 2903 | 4604 | R | 0.87 [0.73, 1.03] | 0.1100 | 0.0200 | 59 | ||
| GSTM1 (Null vs Present) | |||||||||
| Total [27] | 7409 | 9301 | R | 1.18 [1.04, 1.33] | 0.0080 | 0.836 | <0.00001 | 65 | |
| Subregion (27) | East Asia [13] | 4699 | 5881 | R | 1.14 [1.01, 1.27] | 0.0300 | 0.0600 | 41 | |
| Southeast Asia [3] | 476 | 1189 | R | 1.00 [0.81, 1.24] | 1.0000 | 0.6300 | 0 | ||
| South Asia [7] | 1482 | 1691 | R | 1.16 [0.78, 1.73] | 0.4700 | <0.0001 | 81 | ||
| West Asia [4] | 752 | 540 | R | 1.25 [0.81, 1.94] | 0.3200 | 0.0100 | 73 | ||
| Menopausal status [7] | Premenopausal [7] | 1459 | 1689 | R | 1.51 [1.23, 1.86] | <0.0001 | 0.1200 | 40 | |
| Postmenopausal [7] | 1213 | 1225 | R | 1.29 [0.96, 1.73] | 0.0900 | 0.0200 | 61 | ||
| Study design [27] | HCC [17] | 3856 | 3719 | R | 1.32 [1.11, 1.56] | 0.0010 | 0.0002 | 64 | |
| PCC [10] | 3553 | 5582 | R | 1.02 [0.90, 1.14] | 0.7800 | 0.1500 | 32 | ||
| GSTP1 (GG+AG vs AA) | |||||||||
| Total [20] | 8557 | 9544 | R | 1.23 [1.07, 1.41] | 0.0030 | 0.204 | <0.00001 | 70 | |
| Subregion [20] | East Asia [7] | 6108 | 6514 | R | 1.15 [1.03, 1.28] | 0.0100 | 0.1600 | 35 | |
| Southeast Asia [3] | 471 | 1160 | R | 1.10 [0.87, 1.37] | 0.4300 | 0.4200 | 0 | ||
| South Asia [5] | 1220 | 1429 | R | 1.25 [0.85, 1.82] | 0.2500 | 0.0030 | 76 | ||
| West Asia [5] | 758 | 441 | R | 1.64 [0.87, 3.09] | 0.1300 | <0.0001 | 84 | ||
| Menopausal status [5] | Premenopausal [5] | 2462 | 2615 | R | 1.23 [0.85, 1.77] | 0.2700 | 0.0004 | 81 | |
| Postmenopausal [5] | 1888 | 2024 | R | 1.55 [0.84, 2.84] | 0.1600 | <0.00001 | 92 | ||
| Study design [20] | HCC [12] | 2884 | 2591 | R | 1.38 [1.03, 1.84] | 0.0300 | <0.00001 | 78 | |
| PCC [8] | 5673 | 6953 | R | 1.10 [1.02, 1.19] | 0.0100 | 0.8500 | 0 | ||
| GSTP1 (G vs A) | |||||||||
| Total [15] | 15754 | 17036 | R | 1.30 [1.12, 1.51] | 0.0006 | 0.170 | <0.00001 | 82 | |
| Subregion [15] | East Asia [5] | 11738 | 12156 | R | 1.14 [1.04, 1.26] | 0.0060 | 0.1400 | 42 | |
| Southeast Asia [1] | 514 | 1336 | R | 1.10 [0.85, 1.42] | 0.4700 | — | — | ||
| South Asia [5] | 2440 | 2858 | R | 1.44 [1.00, 2.07] | 0.0500 | <0.00001 | 87 | ||
| West Asia [4] | 1062 | 686 | R | 1.65 [0.88, 3.11] | 0.1200 | 0.0001 | 85 | ||
| Menopausal status [15] | Premenopausal [5] | 4924 | 5230 | R | 1.26 [0.92, 1.72] | 0.1500 | <0.0001 | 83 | |
| Postmenopausal [5] | 3776 | 4048 | R | 1.46 [0.88, 2.44] | 0.1400 | <0.00001 | 93 | ||
| Study design [15] | HCC [8] | 4750 | 4008 | R | 1.58 [1.14, 2.19] | 0.0060 | <0.00001 | 88 | |
| PCC [7] | 11004 | 13028 | R | 1.11 [1.04, 1.19] | 0.0010 | 0.7300 | 0 | ||
vs: versus; OR: odds ratio; CI: confidence interval; F: fixed-effect mode; R: random-effect model; HCC: hospital-based case-control study; PCC: population-based case-control study;
The data of positive results are represented in bold type.
GSTM1 polymorphism with breast cancer risk
Using the random-effect model, significantly elevated breast cancer risk was associated with the GSTM1 null/present polymorphism when all 27 studies were pooled into the current study (OR = 1.18, 95% CI = 1.04-1.33, P = 0.008) (Figure 2B). In the subgroup analysis by subregion, obviously increased risk was found in East Asian (OR = 1.14, 95% CI = 1.01-1.27, P = 0.03), but no significant associations were found in other subregions. When stratified by menopausal status, statistically significantly increased risk was detected in premenopausal group (OR = 1.51, 95% CI = 1.23-1.86, P < 0.0001) but not in postmenopausal group (OR = 1.29, 95% CI = 0.96-1.73, P = 0.09). In the subgroup analysis by study design, the data suggested that GSTM1 was significantly associated with breast cancer risk in HCC group (OR = 1.32, 95% CI = 1.11-1.56, P = 0.001) (Figure 3B). The detailed data were listed in Table 4.
GSTP1 polymorphism with breast cancer risk
Analysis using available data of GSTP1 genotypes revealed statistical noteworthy association in Asian population (GG+AG vs AA: OR = 1.23, 95% CI = 1.07-1.41, P = 0.003; G vs A: OR = 1.30, 95% CI = 1.12-1.51, P = 0.0006) (Figure 2C, 2D). Furthermore, the GSTP1 A/G polymorphism might play an effective role in the risk of breast cancer in East Asian (GG+AG vs AA: OR = 1.15, 95% CI = 1.03-1.28, P = 0.01 and G vs A: OR = 1.14, 95% CI = 1.04-1.26, P = 0.006), HCC (GG+AG vs AA: OR = 1.38, 95% CI = 1.03-1.84, P = 0.03 and G vs A: OR = 1.58, 95% CI = 1.14-2.19, P = 0.006) and PCC (GG+AG vs AA: OR = 1.10, 95% CI = 1.02-1.19, P = 0.01 and G vs A: OR = 1.11, 95% CI = 1.04-1.19, P = 0.001) groups. In the subgroup analysis by menopausal status, no associations were detected in premenopausal or postmenopausal groups not only under dominant model (GG+AG vs AA: OR = 1.23, 95% CI = 0.85-1.77, P = 0.27 and OR = 1.55, 95% CI = 0.84-2.84, P = 0.16, respectively) but also under allelic model (G vs A: OR = 1.26, 95% CI = 0.92-1.72, P = 0.15 and OR = 1.46, 95% CI = 0.88-2.44, P = 0.14, respectively) (Figure 3C, 3D). The detailed data were listed in Table 4.
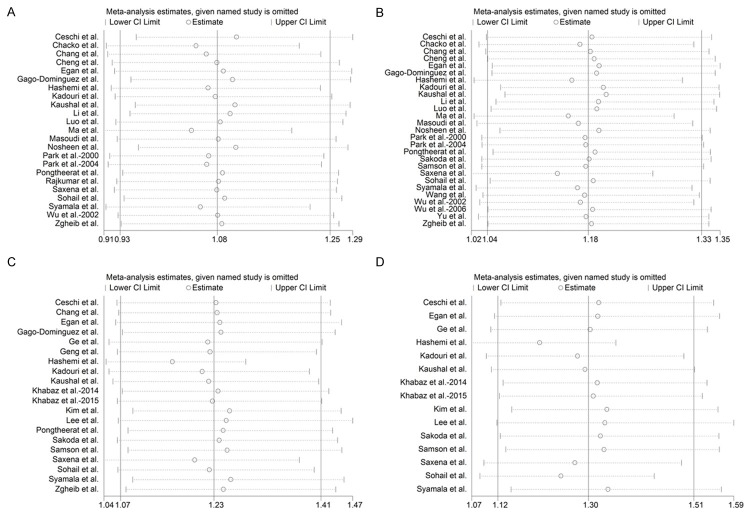
Sensitivity analysis
The one-way sensitivity analyses were performed to assess the stability of the results, namely, a single study in the meta-analysis was deleted each time to reflect the influence of the individual data set to the pooled OR. After sequentially excluding each case-control study, the corresponding pooled ORs were not materially altered (Figure 4), confirming that our meta-analysis was statistically robust.
Figure 4.
Sensitivity analysis of the summary odds ratio coefficients on the relationships between GST polymorphisms and breast cancer risk. Results were computed by omitting each study in turn. The two ends of the dotted lines represent the 95% CI. A. GSTT1 polymorphism. B. GSTM1 polymorphism. C. GSTP1 polymorphism under dominant model (GG+AG vs AA), D GSTP1 polymorphism under allelic model (G vs A).
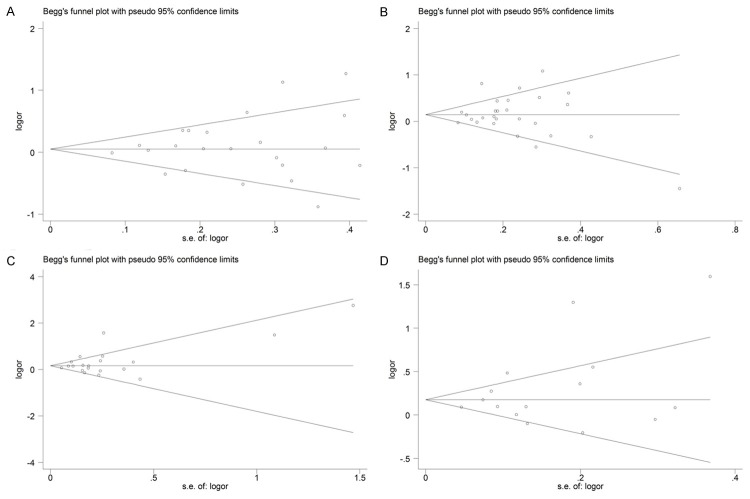
Publication bias
Begg’s funnel plot and Egger’s test were performed to access the publication bias of literatures. As shown in Figure 5, the shapes of the funnel plots did not show obvious asymmetry. In addition, the results of Egger’s test also revealed the absence of publication bias in the GSTT1 (P = 0.493 for Null vs Present model), GSTM1 (P = 0.836 for Null vs Present model) and GSTP1 (P = 0.204 for dominant model GG+AG vs AA and P = 0.170 for allelic model G vs A) polymorphisms.
Figure 5.
Begg’s funnel plot for publication bias in selection of studies on GST polymorphisms. A. GSTT1 polymorphism. B. GSTM1 polymorphism. C. GSTP1 polymorphism under dominant model (GG+AG vs AA). D. GSTP1 polymorphism under allelic model (G vs A).
Discussion
The glutathione S-transferase (GST) family is an important phase II isoenzyme which can implicate in the inactivation of procarcinogens and detoxify environmental carcinogens and toxins [7,49]. Given the important roles of GST in breast cancer etiology makes it possible that genetic variations of the GST genes may affect the susceptibility to the development of breast cancer. At present, some studies found that some mutant sites of the GSTT1, GSTM1 and GSTP1 might play roles in the multifunctional physiological processes in breast cancer. However, results on the associations of these polymorphisms with breast cancer risk have been controversial since the first investigation was reported. In our study, we evaluated whether the GSTT1 and GSTM1 null/present and GSTP1 A/G polymorphisms could become valuable indicators to predict the risk of breast cancer, and tried to derive a more stable conclusion using meta-analysis method.
To the best of our knowledge, the present study is the first meta-analysis of the literature performed to explore the association between GST polymorphisms and breast cancer risk in Asian population. This analysis of pooled individual data revealed no noteworthy associations between GSTT1 null genotype and breast cancer risk in Asian population, while significantly increased risks for GSTM1 null and GSTP1 GG/AG genotypes were observed in breast cancer.
With regard to the subregion, we concluded that GSTT1, GSTM1 and GSTP1 polymorphisms conferred significant increase in the risk of breast cancer in East Asian, and we also detected a 44% increase in the risk of breast cancer under allelic model in South Asian for GSTP1. In contrast to these findings, however, there was a suggestion that the carriers of GSTT1 null genotype had a 27% lowered risk of breast cancer in Southeast Asian. In addition, our results indicated the lack of association between the all three polymorphisms and breast cancer risk in West Asian. These results could be due to the fact that almost half of the studies were about East Asian people (weighted more than 40% in all comparisons for the all three polymorphisms), therefore the analyses on Southeast Asian and West Asian might be insufficient. And there was another explanation that the geographically diverse populations might contribute to the possible presence of heterogeneity between the studies and affect the results of genetic association studies. By analyzing the subgroup by menopausal status, our results indicated that GSTT1 and GSTM1 polymorphisms were obviously associated with premenopausal breast cancer, while no evidence of positive estimates was observed in both premenopausal and postmenopausal groups for GSTP1. Possible explanation to these different results may be that the GST genes are, almost in part, under the control of sex hormones which may have association with the risk of breast cancer and the premenopausal women have a higher level of sex hormones than the postmenopausal women, which may cause a high susceptibility to breast cancer in premenopausal women [50]. There are also other explanations. For example, our case patients were slightly younger and, therefore, the proportion of premenopausal women in cases may be higher than that in controls. In addition, there might be more premenopausal women in case patients of our studies exposed to cigarette smoking and alcohol which contain a wide variety of potentially carcinogenic compounds. These two factors would cause a bias toward a false positive finding. Unluckily, no adequate data were available for stratified analyses by smoking status, drinking status, age and hormone levels. Data from future indepth research regarding the gene-environment interactions and the role of hormone levels in the development of premenopausal breast cancer among Asian women may further interpret this issue. When summarizing the results of stratification analysis by study design, the HCC group was more strongly associated with the risk of breast cancer in GSTT1, GSTM1 and GSTP1 polymorphisms compared with PCC group. This reason may be that the hospital-based studies have some biases because such controls may be just the representative of a sample of ill-defined reference population, and may not represent the general population very well.
Heterogeneity is one of the potential problems when elucidating the results of the present meta-analysis. Although we minimized the likelihood by performing a careful search for published studies, using the explicit criteria for study inclusion, performing data extraction and data analysis strictly, the significant between-study heterogeneity still existed not only in null/present model for GSTT1 and GSTM1, but also in both dominant and allelic models for GSTP1. After subgroup analyses by subregion, menopausal status and study design, the heterogeneity was effectively removed in Southeast Asian group or decreased in East Asian and PCC groups for all 3 polymorphisms. The presence of heterogeneity can result from genetic heterogeneity between the samples that were drawn from geographically diverse populations. Another important factor contributing to heterogeneity was that homogeneity in either the case or control groups was uncertain. Although most of the controls were selected from healthy populations, some studies had selected controls among friends or family of breast cancer patients or patients with other diseases. In addition, we attempted to determine if the heterogeneity might also be explained by other variables such as stages of breast cancer, smoking status, older age at first birth, and environmental factors included in the different studies, but are unable to provide a reliable answer to this question because of insufficient information for these variables.
Several limitations of this meta-analysis should be acknowledged when explaining our results. Firstly, in our meta-analysis, as only certain published studies written in English or Chinese were included, which indicates that some potential published studies in other languages or unpublished studies could be missed, publication bias is very likely to occur in GSTT1, GSTM1 and GSTP1 polymorphisms, although it was not shown in the statistical test. Secondly, the overall outcomes were based on individual unadjusted ORs, while a more precise estimation should be conducted adjusted by confounding factors such as smoking status, age and environmental factors if individual data were available. Thirdly, the results should be cautiously interpreted because participants of some studies draw from different populations were not uniformly defined, which could cause some biases and might distort the results. And the last, in the subgroup analyses, the number of Southeast and West Asian population were relatively small, not having enough statistical power to explore the real association. Therefore, more subjects of different subregions would be required to accurately clarify whether subregion has a biological influence on the susceptibility of breast cancer.
Conclusions
The present meta-analysis revealed that the GSTM1 and GSTP1 polymorphisms can obviously increase the risk of breast cancer in Asian population, especially in East Asian and HCC groups, while the association between GSTT1 null genotype and breast cancer risk is not significant. Thus, our results may have important practical significance for further medical research concerning breast cancer and personalized therapy for breast cancer patients. To further assess gene-to-gene and gene-to-environment combined effects on GST polymorphisms and breast cancer, future large-scale studies in Asian population with different environmental backgrounds are urgently needed.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 4.Kim Y, Yoo KY, Goodman MT. Differences in incidence, mortality and survival of breast cancer by regions and countries in Asia and contributing factors. Asian Pac J Cancer Prev. 2015;16:2857–70. doi: 10.7314/apjcp.2015.16.7.2857. [DOI] [PubMed] [Google Scholar]
- 5.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer-analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 6.Zhong SL, Zhang J, Hu Q, Chen WX, Ma TF, Zhao JH. C1420T Polymorphism of Cytosolic Serine Hydroxymethyltransferase and Risk of Cancer: a Meta-analysis. Asian Pac J Cancer Prev. 2014;15:2257–62. doi: 10.7314/apjcp.2014.15.5.2257. [DOI] [PubMed] [Google Scholar]
- 7.Hayes JD, Pulford DJ. The Glut athione S-Transferase Supergene Family: Regulation of GST and the Contribution of the lsoenzymes to Cancer Chemoprotection and Drug Resistance Part II. Crit Rev Biochem Mol Biol. 1995;30:521–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 8.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 9.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 10.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 11.Zhang H, Xu Y, Zhang Z, Liu R, Ma B. Association between COX-2 rs2745557 polymorphism and prostate cancer risk: a systematic review and meta-analysis. BMC Immunol. 2012;13:14. doi: 10.1186/1471-2172-13-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu KD, Fan L, Di GH, Yuan WT, Zheng Y, Huang W, Chen AX, Yang C, Wu J, Shen ZZ. Genetic variants in GSTM3 gene within GSTM4-GSTM2-GSTM1-GSTM5-GSTM3 cluster influence breast cancer susceptibility depending on GSTM1. Breast Cancer Res Treat. 2010;121:485–496. doi: 10.1007/s10549-009-0585-9. [DOI] [PubMed] [Google Scholar]
- 14.Saxena A, Dhillon VS, Shahid M, Khalil HS, Rani M, Das TP, Hedau S, Hussain A, Naqvi RA, Deo S. GSTP1 methylation and polymorphism increase the risk of breast cancer and the effects of diet and lifestyle in breast cancer patients. Exp Ther Med. 2012;4:1097–1103. doi: 10.3892/etm.2012.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajkumar T, Samson M, Rama R, Sridevi V, Mahji U, Swaminathan R, Nancy NK. TGFβ1 (Leu10Pro), p53 (Arg72Pro) can predict for increased risk for breast cancer in south Indian women and TGFβ1 Pro (Leu10Pro) allele predicts response to neo-adjuvant chemo-radiotherapy. Breast Cancer Res Treat. 2008;112:81–87. doi: 10.1007/s10549-007-9821-3. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Jihu Q, Wang H. Relationship Between GSTM1 Gene Deletion and Susceptibility to Breast Cancer. Acta Academ e Medicinae Nantong. 2002;22:11–12. [Google Scholar]
- 17.Wu SH, Tsai SM, Hou MF, Lin HS, Hou LA, Ma H, Lin JT, Yeh FL, Tsai LY. Interaction of genetic polymorphisms in cytochrome P450 2E1 and glutathione S-transferase M1 to breast cancer in Taiwanese woman without smoking and drinking habits. Breast Cancer Res Treat. 2006;100:93–98. doi: 10.1007/s10549-006-9226-8. [DOI] [PubMed] [Google Scholar]
- 18.Yu KD, Di GH, Fan L, Wu J, Hu Z, Shen ZZ, Huang W, Shao ZM. A functional polymorphism in the promoter region of GSTM1 implies a complex role for GSTM1 in breast cancer. FASEB J. 2009;23:2274–2287. doi: 10.1096/fj.08-124073. [DOI] [PubMed] [Google Scholar]
- 19.Ge J, Tian AX, Wang QS, Kong PZ, Yu Y, Li XQ, Cao XC, Feng YM. The GSTP1 105Val allele increases breast cancer risk and aggressiveness but enhances response to cyclophosphamide chemotherapy in North China. PLoS One. 2013;8:e67589. doi: 10.1371/journal.pone.0067589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geng Y, Chen Y, Zhao W, Wang L, Wang L, Yang J. Mutations and Polymorphisms of GSTP1 Gene in Benign and Malignant Breast Lesions. Chinese Journal of Clinical Oncology. 2010;37:1028–1031. [Google Scholar]
- 21.Khabaz MN. Polymorphism of the glutathione S-transferase P1 gene (GST-pi) in breast carcinoma. Pol J Pathol. 2014;65:141–146. doi: 10.5114/pjp.2014.43964. [DOI] [PubMed] [Google Scholar]
- 22.Khabaz MN, Gari MA, Al-Maghrabi JA, Nedjadi T, Bakarman M. Association between GSTP1 genotypes and hormone receptor phenotype in invasive ductal carcinomas of breast. Asian Pac J Cancer Prev. 2015;16:1707–13. doi: 10.7314/apjcp.2015.16.5.1707. [DOI] [PubMed] [Google Scholar]
- 23.Kim SU, Lee KM, Park SK, Yoo KY, Noh DY, Choe KJ, Ahn SH, Hirvonen A, Kang D. Genetic polymorphism of glutathione S-transferase P1 and breast cancer risk. J Biochem Mol Biol. 2004;37:582–585. doi: 10.5483/bmbrep.2004.37.5.582. [DOI] [PubMed] [Google Scholar]
- 24.Lee SA, Fowke JH, Lu W, Ye C, Zheng Y, Cai Q, Gu K, Gao YT, Shu XO, Zheng W. Cruciferous vegetables, the GSTP1 Ile105Val genetic polymorphism, and breast cancer risk. Am J Clin Nutr. 2008;87:753–760. doi: 10.1093/ajcn/87.3.753. [DOI] [PubMed] [Google Scholar]
- 25.Chacko P, Joseph T, Mathew BS, Rajan B, Pillai MR. Role of xenobiotic metabolizing gene polymorphisms in breast cancer susceptibility and treatment outcome. Mutat Res. 2005;581:153–163. doi: 10.1016/j.mrgentox.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 26.Cheng TC, Chen ST, Huang CS, Fu YP, Yu JC, Cheng CW, Wu PE, Shen CY. Breast cancer risk associated with genotype polymorphism of the catechol estrogen-metabolizing genes: A multigenic study on cancer susceptibility. Int J Cancer. 2005;113:345–353. doi: 10.1002/ijc.20630. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Long Q, Tao P, Hu R, Li H, Lei F, Zhou W, Li S. Using MSR model to analyze the impact of gene-gene interaction with related to the genetic polymorphism of metabolism enzymes on the risk of breast cancer. Sichuan Da Xue Xue Bao Yi Xue Ban. 2008;39:780–783. [PubMed] [Google Scholar]
- 28.Luo J, Gao YT, Chow WH, Shu XO, Li H, Yang G, Cai Q, Li G, Rothman N, Cai H. Urinary polyphenols, glutathione S-transferases copy number variation, and breast cancer risk: Results from the Shanghai women’s health study. Mol Carcinog. 2012;51:379–388. doi: 10.1002/mc.20799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma J, Cui Z, Sun Z. A Case Contro-study on the Associations between Polymorphism of GSTTI, GSTM1 and Susceptibility for Breast Cancer. Chinese Journal of Prevention and Control of Chronic Non-Communicable Diseases. 2007;15:123–126. [Google Scholar]
- 30.Masoudi M, Saadat I, Omidvari S, Saadat M. Additive effects of genetic variations of xenobiotic detoxification enzymes and DNA repair gene XRCC1 on the susceptibility to breast cancer. Breast Cancer Res Treat. 2010;120:263–265. doi: 10.1007/s10549-009-0521-z. [DOI] [PubMed] [Google Scholar]
- 31.Nosheen M, Malik F, Kayani M. Lack of influence of glutathione S-transferase gene deletions in sporadic breast cancer in Pakistan. Asian Pac J Cancer Prev. 2011;12:1749–52. [PubMed] [Google Scholar]
- 32.Park SK, Yim DS, Yoon KS, Choi IM, Choi JY, Yoo KY, Noh DY, Choe KJ, Ahn SH, Hirvonen A. Combined effect of GSTM1, GSTT1, and COMT genotypes in individual. Breast Cancer Res Treat. 2004;88:55–62. doi: 10.1007/s10549-004-0745-x. [DOI] [PubMed] [Google Scholar]
- 33.Park SK, Yoo KY, Lee SJ, Kim SU, Ahn SH, Noh DY, Choe KJ, Strickland PT, Hirvonen A, Kang D. Alcohol consumption, glutathione S-transferase M1 and T1 genetic polymorphisms and breast cancer risk. Pharmacogenetics. 2000;10:301–309. doi: 10.1097/00008571-200006000-00004. [DOI] [PubMed] [Google Scholar]
- 34.Wu FY, Lee YJ, Chen DR, Kuo HW. Association of DNA-protein crosslinks and breast cancer. Mutat Res. 2002;501:69–78. doi: 10.1016/s0027-5107(02)00006-4. [DOI] [PubMed] [Google Scholar]
- 35.Sakoda LC, Blackston CR, Xue K, Doherty JA, Ray RM, Lin MG, Stalsberg H, Gao DL, Feng Z, Thomas DB. Glutathione S-transferase M1 and P1 polymorphisms and risk of breast cancer and fibrocystic breast conditions in Chinese women. Breast Cancer Res Treat. 2008;109:143–155. doi: 10.1007/s10549-007-9633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samson M, Swaminathan R, Rama R, Sridevi V, Nancy KN, Rajkumar T. Role of GSTM1 (null/present), GSTP1 (Ile105Val) and P53 (Arg72Pro) genetic polymorphisms and the risk of breast cancer-a case control study from south India. Asian Pac J Cancer Prev. 2007;8:253–7. [PubMed] [Google Scholar]
- 37.Ceschi M, Sun CL, Van Den Berg D, Koh WP, Mimi CY, Probst-Hensch N. The effect of cyclin D1 (CCND1) G870A-polymorphism on breast cancer risk is modified by oxidative stress among Chinese women in Singapore. Carcinogenesis. 2005;26:1457–1464. doi: 10.1093/carcin/bgi093. [DOI] [PubMed] [Google Scholar]
- 38.Chang TW, Wang SM, Guo YL, Tsai PC, Huang CJ, Huang W. Glutathione S-transferase polymorphisms associated with risk of breast cancer in southern Taiwan. Breast. 2006;15:754–761. doi: 10.1016/j.breast.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Egan KM, Cai Q, Shu XO, Jin F, Zhu TL, Dai Q, Gao YT, Zheng W. Genetic Polymorphisms in GSTM1, GSTP1, and GSTT1 and the Risk for Breast Cancer Results from the Shanghai Breast Cancer Study and Meta-Analysis. Cancer Epidemiol Biomarkers Prev. 2004;13:197–204. doi: 10.1158/1055-9965.epi-03-0294. [DOI] [PubMed] [Google Scholar]
- 40.Gago-Dominguez M, Castelao JE, Sun CL, Van Den Berg D, Koh WP, Lee HP, Mimi CY. Marine n-3 fatty acid intake, glutathione S-transferase polymorphisms and breast cancer risk in post-menopausal Chinese women in Singapore. Carcinogenesis. 2004;25:2143–2147. doi: 10.1093/carcin/bgh230. [DOI] [PubMed] [Google Scholar]
- 41.Hashemi M, Eskandari-Nasab E, Fazaeli A, Taheri M, Rezaei H, Mashhadi M, Arbabi F, Kaykhaei M-A, Jahantigh M, Bahari G. Association between polymorphisms of glutathione S-transferase genes (GSTM1, GSTP1 and GSTT1) and breast cancer risk in a sample Iranian population. Biomark Med. 2012;6:797–803. doi: 10.2217/bmm.12.61. [DOI] [PubMed] [Google Scholar]
- 42.Kadouri L, Kote-Jarai Z, Hubert A, Baras M, Abeliovich D, Hamburger T, Peretz T, Eeles R. Glutathione-S-transferase M1, T1 and P1 polymorphisms, and breast cancer risk, in BRCA1/2 mutation carriers. Bri J Cancer. 2008;98:2006–2010. doi: 10.1038/sj.bjc.6604394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaushal M, Mishra AK, Raju B, Ihsan R, Chakraborty A, Sharma J, Zomawia E, Verma Y, Kataki A, Kapur S. Betel quid chewing as an environmental risk factor for breast cancer. Mutat Res. 2010;703:143–148. doi: 10.1016/j.mrgentox.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 44.Pongtheerat T, Treetrisool M, Purisa W. Glutathione s-transferase polymorphisms in breast cancers of Thai patients. Asian Pac J Cancer Prev. 2009;10:127–132. [PubMed] [Google Scholar]
- 45.Saxena A, Dhillon VS, Raish M, Asim M, Rehman S, Shukla N, Deo S, Ara A, Husain SA. Detection and relevance of germline genetic polymorphisms in glutathione S-transferases (GSTs) in breast cancer patients from northern Indian population. Breast Cancer Res Treat. 2009;115:537–543. doi: 10.1007/s10549-008-0098-y. [DOI] [PubMed] [Google Scholar]
- 46.Sohail A, Kanwal N, Ali M, Sadia S, Masood AI, Ali F, Iqbal F, Crickmore N, Shaikh RS, Sayyed AH. Effects of glutathione-S-transferase polymorphisms on the risk of breast cancer: A population-based case-control study in Pakistan. Environ Toxicol Pharmacol. 2013;35:143–153. doi: 10.1016/j.etap.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 47.Syamala VS, Sreeja L, Syamala V, Raveendran PB, Balakrishnan R, Kuttan R, Ankathil R. Influence of germline polymorphisms of GSTT1, GSTM1, and GSTP1 in familial versus sporadic breast cancer susceptibility and survival. Fam Cancer. 2008;7:213–220. doi: 10.1007/s10689-007-9177-1. [DOI] [PubMed] [Google Scholar]
- 48.Zgheib NK, Shamseddine AA, Geryess E, Tfayli A, Bazarbachi A, Salem Z, Shamseddine A, Taher A, El-Saghir NS. Genetic polymorphisms of CYP2E1, GST, and NAT2 enzymes are not associated with risk of breast cancer in a sample of Lebanese women. Mutat Res. 2013;747:40–47. doi: 10.1016/j.mrfmmm.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 49.Udomsinprasert R, Pongjaroenkit S, Wongsantichon J, Oakley AJ, Prapanthadara LA, Wilce MC, Ketterman AJ. Identification, characterization and structure of a new Delta class glutathione transferase isoenzyme. Biochem J. 2005;388:763–771. doi: 10.1042/BJ20042015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bellanti F, Matteo M, Rollo T, De Rosario F, Greco P, Vendemiale G, Serviddio G. Sex hormones modulate circulating antioxidant enzymes: impact of estrogen therapy. Redox Biol. 2013;1:340–346. doi: 10.1016/j.redox.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]