Abstract
The Interleukin-6 (IL-6) genetic polymorphism is associated with bronchial asthma, a number of studies have been conducted to investigate the association between IL-6 gene -174G/C polymorphism and bronchial asthma risk. However, the results are inconclusive. This meta-analysis aims to investigate whether -174G/C polymorphism is a potential risk factor for bronchial asthma. We searched Web of Science, PubMed, Google Scholar, China National Knowledge Infrastructure (CNKI) and Wanfang Database from inception through December 1st, 2014. Meta-analysis was performed using the STATA 12.0. Overall, a significantly reduced risk for asthma was found in IL-6 -174 CC genotype (CC vs. GG: OR = 0. 51, 95% CI = 0.27-0.96, P = 0.038). Furthermore, analysis by ethnicity indicated that there was a markedly reduced risk for asthma in IL-6 -174 CC genotype in Caucasian (CC vs. GG: OR = 0.51, 95% CI = 0.27-0.96, P = 0.038). Analysis by age indicated that there was a significantly reduced risk for asthma in IL-6 -174 CC genotype in adults (CC vs. GG: OR = 0.47, 95% CI = 0.23-0.97, P = 0.042). In conclusion, the current meta-analysis indicates that IL-6 -174 CC genotype may be a protective factor against asthma in Caucasian and adults.
Keywords: IL-6, polymorphism, asthma, risk, meta-analysis
Introduction
Bronchial asthma is one of the most common chronic inflammatory respiratory diseases, characterized by airway hyper responsiveness, airway inflammation and reversible airway obstruction. The development of asthma appears to be determined by genetic and environmental factors [1,2]. Numerous studies have reported that there is a significant correlation between the genetic polymorphism of cytokine genes and asthma [3-5]. Interleukin-6 (IL-6) functions as a pleiotropic pro-inflammatory and immunomodulatory cytokine, which regulates T cell differentiation and activation, induces Th2 cytokine production [6]. IL-6 is secreted by a variety of lymphoid and non-lymphoid cells including: T cells, B cells, monocytes, fibroblasts, hepatocytes, airway endothelial cells, alveolar macrophages, adipocytes, myocytes as well as other tissues and cells [7-9]. Accumulated evidences have shown that IL-6 is involved in the development of asthma [10-12].
A single-nucleotide polymorphism (SNP) at position -174 (also called rs1800795) in the promoter region of the IL-6 gene, which was firstly described by Fishman et al., is located in chromosome 7p21 [13]. This SNP presents guanosine (G) or cytosine (C) at position -174 of IL-6 gene and this variability could be functional, because it modifies transcriptional regulation and cytokine levels, and regulates inflammatory phenotype [13]. Several studies have reported the correlation between IL-6 -174G/C polymorphism and bronchial asthma risk [14-19]. However, the results were contradictory and all above studies were performed in an individual medical center with less sample size. Therefore, it is important to clarify the relationship between IL-6 -174G/C polymorphism and bronchial asthma susceptibility based on all published studies, and the aim of present study is to address this issue using meta-analysis.
Materials and methods
Identification of eligible studies
To identify all eligible studies that investigated the association between IL-6 -174G/C polymorphism and bronchial asthma risk, a comprehensive electronic search of Web of Science, PubMed, Google Scholar, China National Knowledge Infrastructure (CNKI) and Wanfang Database was performed until December 1st, 2014. We applied various combinations of the following medical subject headings and key words: (interleukin-6 or IL-6 or cytokine) and (polymorphism or variant or variation or SNP) and (asthma or asthmatic). Furthermore, the reference lists of reviews and retrieved articles were manually screened for additional studies. No restrictions were placed on language, and only published studies with full-text articles were included.
Inclusion and exclusion criteria
The studies identified from the above mentioned databases were screened by two independent authors (Fangwei Li and Xinming Xie) according to the following predesigned inclusion criteria: I) case-control design; II) evaluating the correlation of IL-6 -174G/C polymorphism with asthma risk; and III) providing sufficient data to calculate the odds ratio (OR) and its corresponding 95% confidence interval (CI). When several studies with overlapping data were eligible, those with smaller sample size or less reliability were excluded. In addition, studies without detailed information were excluded after the efforts to extract data from the original paper or contact the corresponding authors failed.
Data extraction
Data of eligible studies were extracted by two reviewers (Fangwei Li and Shaojun Li) independently and in duplicate according to the predesigned data-collection form. The following information was extracted: first author, publication year, country of origin, ethnicity, age, number of both cases and control subjects and phenotypic distribution in both groups. Different ethnic descents were categorized as Caucasian, Asian. Different age groups were categorized as adult, children. Discrepancies occurring during the process of study inclusion and data extraction were resolved by discussion with a third reviewer (Rui Ke), and consensus on each item was achieved eventually.
Data analysis
The statistical analysis was conducted using STATA12.0 software (Stata Corporation, College Station, Texas, USA). Hardy-Weinberg equilibrium (HWE) in controls was tested by the chi-square test, and it was considered statistically significant when P-value was less than 0.05. Heterogeneity among studies was examined with the X2-based Q testing and I2 statistics [20]. If there was a significant heterogeneity (P < 0.10), a random-effects model (the DerSimonian and Laird method) was selected to pool the data [21]. Otherwise, a fixed-effects model (the Mantel-Haenszel method) was selected to pool the data [22]. The pooled odds ratio (OR) of asthma was performed for recessive model (CC vs. GC + GG), homozygote comparison (CC vs. GG), allele model (C allele vs. G allele) and dominant model (CC + GC vs. GG) respectively. The significance of the combined OR determined by a Z-test and two-sided P value < 0.05 was considered significant. To explore the source of the heterogeneity, Galbraith plot was used. Publication bias was examined with Begg’s test and Egger’s test [23,24] and the P value < 0.10 indicated significant publication bias.
Results
Characteristics of eligible studies
A total of 1165 papers were obtained with the initial search of databases. After screening, six case-control studies with a total of 1373 participants (479 cases and 894 controls) fulfilled the inclusion criteria, from which genotype data of IL-6 -174G/C polymorphism were obtained. As for ethnicity, 5 studies were of Caucasian, 1 study was of Asian. As for age, 4 studies were of adults, 1 study was of children, and 1 study was of mixed (1.5-33 years, Table 1). The qualities of these studies were considered accessible for the meta-analysis. The flow chart of selection of studies and reasons for exclusion are presented in Figure 1.
Table 1.
Main characteristics of all eligible studies
| First author | Year | Country | Ethnicity | Age group | No. of case | No. of control | Case | Control | P for HWE | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||
| GG | GC | CC | GG | GC | CC | ||||||||
| Kosugi | 2013 | Brazil | Caucasian | Adults | 90 | 144 | 69 | 20 | 1 | 70 | 67 | 7 | 0.070 |
| Daneshmandi | 2012 | Iran | Caucasian | Adults | 81 | 124 | 60 | 15 | 6 | 83 | 32 | 9 | 0.027 |
| Mahdaviani | 2009 | Iran | Caucasian | Mixed | 57 | 139 | 5 | 51 | 1 | 42 | 93 | 4 | 0.000 |
| Settin | 2008 | Egypt | Caucasian | Children | 69 | 98 | 11 | 52 | 6 | 5 | 87 | 6 | 0.000 |
| Trajkov | 2008 | Macedonia | Caucasian | Adults | 74 | 301 | 46 | 25 | 3 | 144 | 132 | 25 | 0.492 |
| Liu | 2008 | China | Asian | Adults | 108 | 88 | 72 | 36 | 0 | 71 | 17 | 0 | 0.316 |
HWE: Hardy-Weinberg equilibrium.
Figure 1.

Flowchart shows study selection procedure.
Meta-analysis of IL-6 -174G/C polymorphism and bronchial asthma
A summary of association between IL-6 -174 G/C polymorphism and bronchial asthma risk is shown in Table 2.
Table 2.
The results of meta-analysis
| CC vs. GC + GG | CC vs. GG | C allele vs. G allele | CC + GC vs. GG | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| OR (95% CI) | P(Z) | I2 (%) | OR (95% CI) | P(Z) | I2 (%) | OR (95% CI) | P(Z) | I2 (%) | OR (95% CI) | P(Z) | I2 (%) | |
| Overall | 0.72 (0.40, 1.29) | 0.270 | 0.0 | 0.51 (0.27, 0.96) | 0.038 | 2.8 | 0.86 (0.54, 1.35) | 0.504 | 81.0 | 0.81 (0.38, 1.74) | 0.586 | 86.2 |
| Caucasian | 0.72 (0.40, 1.29) | 0.270 | 0.0 | 0.51 (0.27, 0.96) | 0.038 | 2.8 | 0.75 (0.47, 1.18) | 0.209 | 79.1 | 0.61 (0.31, 1.21) | 0.297 | 83.6 |
| Adults | 0.62 (0.29, 1.33) | 0.221 | 0.0 | 0.47 (0.23, 0.97) | 0.042 | 28.4 | 0.74 (0.40, 1.36) | 0.334 | 82.4 | 0.69 (0.32, 1.48) | 0.339 | 84.7 |
OR: odds ratio; CI: confidence interval; P(Z): P-value of pooled effect. Significant results (P < 0.05) for overall effect were marked in bold characters.
Overall, a significantly reduced risk was observed in CC genotype (CC vs. GG: OR = 0.51, 95% CI = 0.27-0.96, P = 0.038; Figure 2; Table 2) and no markedly increased or reduced risk for bronchial asthma under other genetic models was detected (CC vs. GC + GG: OR = 0.72, 95% CI = 0.40-1.29, P = 0.270; C allele vs. G allele: OR = 0.86, 95% CI = 0.54-1.35, P = 0.504; CC + GC vs. GG: OR = 0.81, 95% CI = 0.38-1.74, P = 0.586; Table 2).
Figure 2.
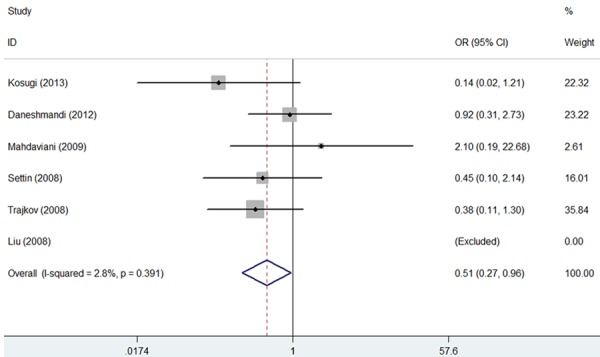
Forest plot for the association between IL-6-174G/C polymorphism and bronchial asthma risk in total analysis (CC vs. GG).
In the subgroup analysis by ethnicity, results indicated a significantly reduced risk for asthma in CC genotype in Caucasian (CC vs. GG: OR = 0.51, 95% CI = 0.27-0.96, P = 0.038; Figure 3; Table 2). There was only one study [19] in Asian, in which CC genotype was not found, so no subgroup analysis was performed. After subgroup analysis by age, results indicated a markedly reduced risk for asthma in CC genotype in adults (CC vs. GG: OR = 0.47, 95% CI = 0.23-0.97, P = 0.042; Figure 4; Table 2). There was only one study in children [17] or mixed (1.5-33 years) [16] respectively, so no subgroup analysis was performed.
Figure 3.
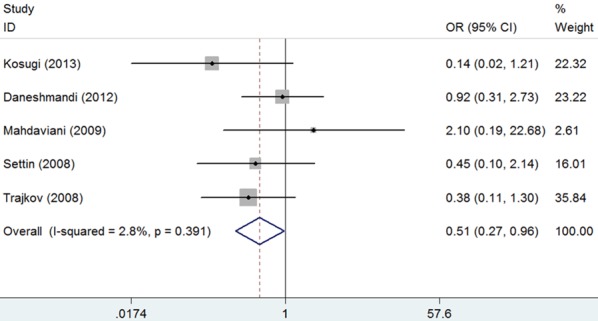
Forest plot for the association between IL-6 -174G/C polymorphism and bronchial asthma risk in Caucasian (CC vs. GG).
Figure 4.
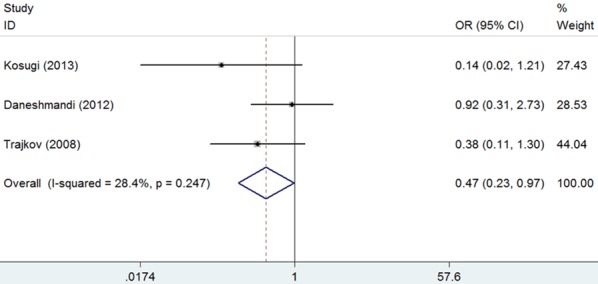
Forest plot for the association between IL-6 -174G/C polymorphism and bronchial asthma risk in adults (CC vs. GG).
Sensitivity analysis
Sensitivity analysis was also performed to explore the potential influence of each individual study on the overall results by deleting one single study each time from the pooled analysis. No substantial change was demonstrated in the overall studies, indicating that no individual study could affect the pooled OR significantly (figure not shown).
Heterogeneity and publication bias
Among the four genetic models, significant heterogeneity was observed in allele model (C allele vs. G allele) and dominant model (CC + GC vs. GG) with P-value < 0.10. After stratifying for ethnicity and age respectively, significant heterogeneity was observed in Caucasian and adults (Table 2). So the Galbraith plot was used to find the source of the heterogeneity. As shown in Figure 5, three studies were the outliers [14,16,19]. For publication bias, results were observed with all P-value > 0.1 of Begg’s test and Egger’s test (Table 3).
Figure 5.
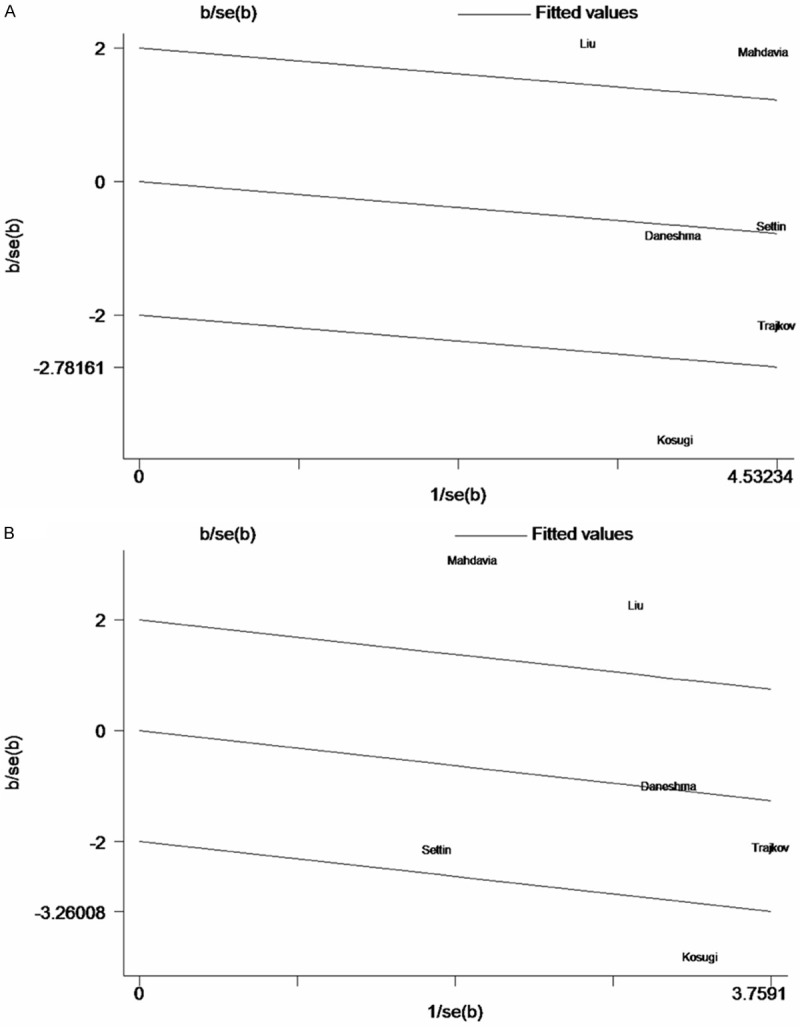
Galbraith plots for the association between IL-6 -174G/C polymorphism and bronchial asthma. A. C allele vs. G allele; B. CC + GC vs. GG.
Table 3.
Results of publication bias test
| Polymorphism | Compared genotype | Begg’s test | Egger’s test | ||
|---|---|---|---|---|---|
|
|
|
||||
| z value | P value | t value | P value | ||
| -174G/C | CC vs. GC + GG | 0.73 | 0.462 | -1.38 | 0.261 |
| CC vs. GG | -0.24 | 1.000 | -0.26 | 0.815 | |
| C allele vs. G allele | 0.38 | 0.707 | 0.33 | 0.755 | |
| CC + GC vs. GG | 0.75 | 0.452 | 0.70 | 0.525 | |
Discussion
Human genetic study is an indispensable approach in understanding the molecular basis of diseases including bronchial asthma. Many candidate genes, including IL-6 [14-16], IL-4 [25], IL-13 [26,27], tumor necrosis factor [27], beta 2-adrenoceptor [28,29] and so on, have been reported to be involved in bronchial asthma susceptibility. IL-6 is a potential mediator of inflammation in asthma, which has been shown to be increased in serum and bronchoalveolar lavage fluid (BALF) during stable condition and exacerbation, predicting a faster decline in FEV1 [9,11]. IL-6 -174G > C has been shown to be associated with increased risk of cardiovascular events and factors such as C-reactive protein and hypertension [30-32]. The results of the relationship between IL-6 -174G > C polymorphism and bronchial asthma risk are inconsistent. Therefore, it is critical to systematically evaluate all studies from different groups and to quantify the overall association between IL-6 -174G > C polymorphism and bronchial asthma.
In this study, our results indicated that IL-6 -174 CC genotype carriers had a reduced risk for bronchial asthma. Furthermore, analysis by ethnicity indicated that IL-6 -174 CC genotype carriers had a significantly reduced risk for asthma in Caucasian. There was only one study [19] in Asian, in which the CC genotype was not found, so no subgroup analysis was performed. In addition, analysis by age indicated that IL-6 -174 CC genotype carriers had a markedly reduced risk for asthma in adults. We could not perform analysis in children due to only one study [17] included.
To our knowledge, this is the first meta-analysis about IL-6 gene polymorphism and bronchial asthma susceptibility. In the present meta-analysis, we have combined six eligible studies with 479 cases and 894 controls to yield summary results, indicating that IL-6 -174 G/C polymorphism might be related to asthma risk in the overall population. Furthermore, we considered that CC genotype was a protective factor against asthma in Caucasian according to ethnicity and a protective factor against asthma in adults according to age. Our results are supported by several studies, which found that IL-6 -174G/C polymorphism modified transcriptional regulation and IL-6 levels [13], meanwhile IL-6 might be involved in the pathophysiology of bronchial asthma [10,33]. However, some other studies did not find evidence for association between IL-6 -174G/C polymorphism and circulating IL-6 Level and considered IL-6 a byproduct of ongoing inflammation in the airways [34-37]. It is suggested that studies based on larger well-designed populations are still needed to clarify the relationships between IL-6 gene polymorphism and bronchial asthma susceptibility.
The heterogeneity needs to be mentioned when interprets the meta-analysis results. For the stable results of -174G/C polymorphism in recessive model (CC vs. GC + GG) and homozygote comparison (CC vs. GG), there was no or low heterogeneity (indicated by I2-statistics) among studies. However, obvious heterogeneity was observed in allele model (C allele vs. G allele) and dominant model (CC + GC vs. GG). After stratifying by ethnicity and age respectively, relatively significant heterogeneity was still observed in Caucasian and adults, which may have been responsible for the negative results. Furthermore, Galbraith plot was performed to explore the source of heterogeneity. As shown in Figure 5, it is possible that three studies [14,16,19] may have been the main source of heterogeneity.
Some limitations should be addressed when interpreting the results: i) the limited number of bronchial asthma cases and control subjects may lead to a relatively small power; ii) in some cases, heterogeneity was not resolved after subgroup analyses, suggesting that other factors such as the differences in gender, lifestyle factors or clinical characteristics might have caused heterogeneity; not all the control subjects were age or sex matched to cases, which was likely one of the causes for heterogeneity; iii) it is well acknowledged that many other factors, such as gene-gene or gene-environment interactions, may affect the risk for bronchial asthma; and iv) only published studies with sufficient data were included and publication bias may have occurred even though the result of Begg’s test or Egger’s test did not detect it.
In conclusion, the current meta-analysis indicates that IL-6 -174 CC genotype may be a protective factor against asthma in Caucasian and adults. Larger and well-designed studies based on different ethnic groups are needed to confirm our result.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant No. 81070045) and the Key Clinical Project for Affiliated Hospital of Ministry of Public Health of China (Grant No. 111). The authors would like to acknowledge the reviewers for their helpful comments on this article.
Disclosure of conflict of interest
None.
References
- 1.Sandford A, Weir T, Pare P. The Genetics of Asthma. Am J Respir Crit Care Med. 1996;153:1749–65. doi: 10.1164/ajrccm.153.6.8665031. [DOI] [PubMed] [Google Scholar]
- 2.McLeish S, Turner SW. Gene-environment interactions in asthma. Arch Dis Child. 2007;92:1032–1035. doi: 10.1136/adc.2006.112185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freidin MB, Puzyrev VP, Ogorodova LM, Kobiakova OS, Kulmanakova IM. Polymorphism of interleukins and interleukin receptor genes: population distribution and association with atopic bronchial asthma. Genetika. 2002;38:1710–1718. [PubMed] [Google Scholar]
- 4.Tam EK, Jourdan-LeSaux C, Stauder S, Bollt O, Reber B, Yamamoto F, Haymer D. Polymorphisms in the interleukin-4 receptor alpha chain: association with traits of allergy and asthma in an admixed population in Hawaii. Cell Mol Biol (Noisy-le-grand) 2003;49:1345–1349. [PubMed] [Google Scholar]
- 5.Heinzmann A, Bauer E, Ganter K, Kurz T, Deichmann KA. Polymorphisms of the TGF-beta1 gene are not associated with bronchial asthma in Caucasian children. Pediatr Allergy Immunol. 2005;16:310–314. doi: 10.1111/j.1399-3038.2005.00287.x. [DOI] [PubMed] [Google Scholar]
- 6.Neurath MF, Finotto S. IL-6 signaling in autoimmunity, chronic inflammation and inflammation-associated cancer. Cytokine Growth Factor Rev. 2011;22:83–89. doi: 10.1016/j.cytogfr.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Fredj S, Bescond J, Louault C, Delwail A, Lecron JC, Potreau D. Role of interleukin-6 in cardiomyocyte/cardiac fibroblast interactions during myocyte hypertrophy and fibroblast proliferation. J Cell Physiol. 2005;204:428–436. doi: 10.1002/jcp.20307. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi R, Yamashita N, Matsui S, Fujita T, Araya J, Sassa K, Arai N, Yoshida Y, Kashii T, Maruyama M, Sugiyama E, Kobayashi M. Bradykinin stimulates IL-6 and IL-8 production by human lung fibroblasts through ERK- and p38 MAPK-dependent mechanisms. Eur Respir J. 2000;16:452–458. doi: 10.1034/j.1399-3003.2000.016003452.x. [DOI] [PubMed] [Google Scholar]
- 9.He JQ, Foreman MG, Shumansky K, Zhang X, Akhabir L, Sin DD, Man SF, DeMeo DL, Litonjua AA, Silverman EK, Connett JE, Anthonisen NR, Wise RA, Pare PD, Sandford AJ. Associations of IL6 polymorphisms with lung function decline and COPD. Thorax. 2009;64:698–704. doi: 10.1136/thx.2008.111278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokoyama A, Kohno N, Fujino S, Hamada H, Inoue Y, Fujioka S, Ishida S, Hiwada K. Circulating interleukin-6 levels in patients with bronchial asthma. Am J Respir Crit Care Med. 1995;151:1354–1358. doi: 10.1164/ajrccm.151.5.7735584. [DOI] [PubMed] [Google Scholar]
- 11.Tillie-Leblond I, Pugin J, Marquette CH, Lamblin C, Saulnier F, Brichet A, Wallaert B, Tonnel AB, Gosset P. Balance between proinflammatory cytokines and their inhibitors in bronchial lavage from patients with status asthmaticus. Am J Respir Crit Care Med. 1999;159:487–494. doi: 10.1164/ajrccm.159.2.9805115. [DOI] [PubMed] [Google Scholar]
- 12.Virchow JC Jr, Kroegel C, Walker C, Matthys H. Inflammatory determinants of asthma severity: mediator and cellular changes in bronchoalveolar lavage fluid of patients with severe asthma. J Allergy Clin Immunol. 1996;98:S27–33. discussion S33-40. [PubMed] [Google Scholar]
- 13.Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S, Woo P. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102:1369–1376. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kosugi EM, de Camargo-Kosugi CM, Hirai ER, Mendes-Neto JA, Gregorio LC, Guerreiro-da-Silva ID, Weckx LL. Interleukin-6 -174 G/C promoter gene polymorphism in nasal polyposis and asthma. Rhinology. 2013;51:70–76. doi: 10.4193/Rhino12.166. [DOI] [PubMed] [Google Scholar]
- 15.Daneshmandi S, Pourfathollah AA, Pourpak Z, Heidarnazhad H, Kalvanagh PA. Cytokine gene polymorphism and asthma susceptibility, progress and control level. Mol Biol Rep. 2012;39:1845–1853. doi: 10.1007/s11033-011-0927-7. [DOI] [PubMed] [Google Scholar]
- 16.Mahdaviani SA, Rezaei N, Moradi B, Dorkhosh S, Amirzargar AA, Movahedi M. Proinflammatory cytokine gene polymorphisms among Iranian patients with asthma. J Clin Immunol. 2009;29:57–62. doi: 10.1007/s10875-008-9232-1. [DOI] [PubMed] [Google Scholar]
- 17.Settin A, Zedan M, Farag M, Ezz El Regal M, Osman E. Gene polymorphisms of IL-6(-174) G/C and IL-1Ra VNTR in asthmatic children. Indian J Pediatr. 2008;75:1019–1023. doi: 10.1007/s12098-008-0161-z. [DOI] [PubMed] [Google Scholar]
- 18.Trajkov D, Mirkovska-Stojkovikj J, Arsov T, Petlichkovski A, Strezova A, Efinska-Mladenovska O, Sandevska E, Gogusev J, Spiroski M. Association of Cytokine Gene Polymorphisms with Bronchial Asthma in Macedonians. Iran J Allergy Asthma Immunol. 2008;7:143–156. [PubMed] [Google Scholar]
- 19.Xiaomin L, Fenglin C, Jianmin H, Yuzhi S, Binsheng G, Yingmei Z. Correlation between genetic polymorphism of cytokine genes, plasma protein levels and bronchial asthma in the Han people in northern China. J Asthma. 2008;45:583–589. doi: 10.1080/02770900802032925. [DOI] [PubMed] [Google Scholar]
- 20.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 21.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 22.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 23.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 24.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Lin LH, Wang J, Peng X, Dai HR, Xiao H, Li F, Wang YP, Yang ZJ, Li L. Interleukin-4 and interleukin-13 pathway genetics affect disease susceptibility, serum immunoglobulin E levels, and gene expression in asthma. Ann Allergy Asthma Immunol. 2014;113:173–179.e1. doi: 10.1016/j.anai.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Kabesch M, Schedel M, Carr D, Woitsch B, Fritzsch C, Weiland SK, von Mutius E. IL-4/IL-13 pathway genetics strongly influence serum IgE levels and childhood asthma. J Allergy Clin Immunol. 2006;117:269–274. doi: 10.1016/j.jaci.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 27.Undarmaa S, Mashimo Y, Hattori S, Shimojo N, Fujita K, Miyatake A, Doi S, Kohno Y, Okamoto Y, Hirota T, Tamari M, Hata A, Suzuki Y. Replication of genetic association studies in asthma and related phenotypes. J Hum Genet. 2010;55:342–349. doi: 10.1038/jhg.2010.32. [DOI] [PubMed] [Google Scholar]
- 28.Chiang CH, Lin MW, Chung MY, Yang UC. The association between the IL-4, ADRbeta2 and ADAM 33 gene polymorphisms and asthma in the Taiwanese population. J Chin Med Assoc. 2012;75:635–643. doi: 10.1016/j.jcma.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 29.Kohyama K, Abe S, Kodaira K, Yukawa T, Hozawa S, Morioka J, Inamura H, Ota M, Sagara H, Schwartz LB, Kurosawa M. Arg16Gly beta2-adrenergic receptor gene polymorphism in Japanese patients with aspirin-exacerbated respiratory disease. Int Arch Allergy Immunol. 2011;156:405–411. doi: 10.1159/000324463. [DOI] [PubMed] [Google Scholar]
- 30.Jenny NS, Tracy RP, Ogg MS, Luong le A, Kuller LH, Arnold AM, Sharrett AR, Humphries SE. In the elderly, interleukin-6 plasma levels and the -174G > C polymorphism are associated with the development of cardiovascular disease. Arterioscler Thromb Vasc Biol. 2002;22:2066–2071. doi: 10.1161/01.atv.0000040224.49362.60. [DOI] [PubMed] [Google Scholar]
- 31.Vickers MA, Green FR, Terry C, Mayosi BM, Julier C, Lathrop M, Ratcliffe PJ, Watkins HC, Keavney B. Genotype at a promoter polymorphism of the interleukin-6 gene is associated with baseline levels of plasma C-reactive protein. Cardiovasc Res. 2002;53:1029–1034. doi: 10.1016/s0008-6363(01)00534-x. [DOI] [PubMed] [Google Scholar]
- 32.Mayosi BM, Avery PJ, Baker M, Gaukrodger N, Imrie H, Green FR, Farrall M, Watkins H, Keavney B. Genotype at the -174G/C polymorphism of the interleukin-6 gene is associated with common carotid artery intimal-medial thickness: family study and meta-analysis. Stroke. 2005;36:2215–2219. doi: 10.1161/01.STR.0000182254.47941.96. [DOI] [PubMed] [Google Scholar]
- 33.Deetz DC, Jagielo PJ, Quinn TJ, Thorne PS, Bleuer SA, Schwartz DA. The kinetics of grain dust-induced inflammation of the lower respiratory tract. Am J Respir Crit Care Med. 1997;155:254–259. doi: 10.1164/ajrccm.155.1.9001321. [DOI] [PubMed] [Google Scholar]
- 34.Huang M, Wang L, Ma H, Wang J, Xiang M. Lack of an association between interleukin-6 -174G/C polymorphism and circulating interleukin-6 levels in normal population: a meta-analysis. DNA Cell Biol. 2013;32:654–664. doi: 10.1089/dna.2013.2148. [DOI] [PubMed] [Google Scholar]
- 35.Gao SP, Liang S, Pan M, Sun RL, Chen C, Luan H, Jiang MH. Interleukin-6 genotypes and serum levels in Chinese Hui population. Int J Clin Exp Med. 2014;7:2851–2857. [PMC free article] [PubMed] [Google Scholar]
- 36.Lin H, Joehanes R, Pilling LC, Dupuis J, Lunetta KL, Ying SX, Benjamin EJ, Hernandez D, Singleton A, Melzer D, Munson PJ, Levy D, Ferrucci L, Murabito JM. Whole blood gene expression and interleukin-6 levels. Genomics. 2014;104:490–495. doi: 10.1016/j.ygeno.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alizadeh-Navaei R, Rafiei A, Hedayatizadeh-Omran A, Mohammadzadeh I, Arabi M. Gene susceptibility in Iranian asthmatic patients: a narrative review. Ann Med Health Sci Res. 2014;4:837–840. doi: 10.4103/2141-9248.144871. [DOI] [PMC free article] [PubMed] [Google Scholar]


