Abstract
Long non-coding RNAs (lncRNAs) UCA1 have been shown to paly an important regulatory roles in cancer biology, and UCA1 dysfunction is related to TNM stage, metastasis and postoperative survival in several cancers. However, the biological role and clinical significance of UCA1 in the carcinogenesis of prostate cancer (PC) remain largely unclear. Herein, we found that UCA1 was abnormally upregulated in tumor tissues from PC patients, and patients with high UCA1 levels had a significantly poorer prognosis. Intriguingly, the mRNA and protein levels of KLF4 were significantly increased in tumor tissues, which was highly correlated to UCA1 levels. Moreover, UCA1 depletion inhibited the growth and induced apoptosis in PC3 and LNCaP cell lines. In addition, UCA1 loss-of-function could decrease KLF4 expression, subsequently, the downregulation of KRT6 and KRT13. Taken together, our study indicated that UCA1 had a crucial role in the tumorigenesis of PC. Moreover, UCA1 loss-of-function inhibited cell proliferation and induced cell apoptosis, at least partially, through inactivation KLF4-KRT6/13 cascade.
Keywords: Prostate cancer, long non-coding RNA, UCA1, KLF4, KRT6/13
Introduction
Prostate cancer (PC) is the most commonly diagnosed malignancy and the third leading cause of cancer death among men in developed countries [1]. In the United States alone, an estimated 242,000 will be diagnosed with PC in 2012 and 28,000 will die from their disease [2]. Intriguingly, PC-related deaths have declined over the past decade, owing to improved methods for early detection and diagnosis and more-effective therapeutic strategies [3]. However, the more accurate predictive biomarkers are essential to be exploited in PC patients.
Eukaryotic genomes encode numerous long non-coding RNAs (lncRNAs), which are defined as endogenous cellular RNAs with length longer than 200 nucleotides, but lack open reading frames of significant length [4]. Estimates suggest the number of human lncRNAs ranging from 10,000 to 20,000 [5]. Despite these large numbers, only a handful of lncRNAs have been characterized. Notably, most characterized lncRNAs display deregulated expression in cancer cells, where they play oncogenic or tumor suppressive functions [4]. Urothelial carcinoma associated 1 (UCA1), the entire sequence consists of three exons with 1.4 kb in length, is an lncRNA originally identified in bladder transitional cell carcinoma [6] and found highly expressed in some carcinomas [7,8]. Moreover, upregulated UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway [8]. In bladder cancer cell, UCA1 can enhance cell proliferation and metastasis through PI3K, Wnt or Akt signaling pathway [9-11]. These results indicate that UCA1 plays an important role in the occurrence and development progress of malignant tumors. Nevertheless, there is no relevant report about the interaction between UCA1 and the progression of PC. Thus, the role of UCA1 in PC and its underlying mechanism remain to be determined.
In the present study, we performed a hierarchical cluster analysis of the differentially expressed lncRNAs in the tumor tissues of PC patients to identify the role of UCA1 in the developmental progress of PC. The results showed that UCA1 was overexpressed in PC tumor tissues, and it might play an oncogenic role in promoting cell proliferation and anti-apoptosis in vitro. Futhermore, the underlying mechanism that UCA1 promoted cell proliferation in vitro through regulation Kruppel-like factor 4 (KLF4) and keratin 6/13 (KRT6/13) was investigated.
Materials and methods
Patients and specimens
Forty human prostate cancer tumor tissues and matched adjacent non-tumor tissues were collected from the Department of Urology Surgery, Longsai hospital (Ningbo, China) between Jan 2008 and June 2014. All patients recruited in this study were not subjected to preoperative radiotherapy or chemotherapy and diagnosed with prostate cancer based on histopathological evaluation. All collected tissue samples were immediately stored at liquid nitrogen until use. Human samples were obtained with written informed consent from all patients. The study was approved by the Ethics Committee of the Longsai Hospital, China.
Cell culture
Three prostate cancer cell lines (22RV1, PC3 and LNCaP) and a human prostatic epithelial cells line (RWPE1) were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in DMEM (Dulbecco’s modified Eagle’s medium; Invitrogen), supplemented with 10% fetal bovine serum (FBS) (HyClone, Logan, UT), 100 units/ml penicillin, and 100 mg/ml streptomycin (Invitrogen) at 37°C in a humidified, 5% CO2, 95% air atmosphere.
Cell viability detection by CCK8
Prostate cancer cells (1.0 × 104/well) were plated and treated in 96-well plates (three wells per group). 10 μL of CCK8 (Dojindo, Kumamoto, Japan) was added to the cells, and the viability of the cells was measured at 450 nm using an ELISA reader (BioTek, Winooski, VT, USA) according to the manufacturer’s instructions.
Cell apoptosis assay
Apoptosis was assessed using annexin V, a protein that binds to phosphatidylserine (PS) residues which are exposed on the cell surface of apoptotic cells. Cells were washed twice with PBS (pH = 7.4), and re-suspended in staining buffer containing 1 μg/ml PI and 0.025 μg/ml annexin V-FITC. Double-labeling was performed at room temperature for 10 min in the dark before the flow cytometric analysis. Cells were immediately analyzed using FACScan and the Cellquest program (Becton Dickinson). Quantitative assessment of apoptotic cells was also assessed by the terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick endlabeling (TUNEL) method, which examines DNA-strand breaks during apoptosis by using BD ApoAlertTM DNA Fragmentation Assay Kit. The cells were trypsinized, fixed with 4% paraformaldehyde, and permeabilized with 0.1% Triton-X-100 in 0.1% sodiumcitrate. After being washed, the cells were incubated with the reaction mixture for 60 min at 37°C. The stained cells were then analyzed with flow cytometer.
RNA interference
Small interfering RNAs (siRNAs) were synthesized from Genepharma (China) and transfected into PC3 and LNCaP cells for suppressing the function of UCA1 using Lipofectamine 2000 (Invitrogen, USA). The small interfering RNA with the following primers: si-UCA1, GGACAACAGUACACGCAUA; si-KLF4: GGACCUGGACUUUAUUCUCUCCAAU; scramble, UAAUCGGCAGCUUGCAAGCUGCCGC.
Real-time and reverse transcription PCR
Total RNA was extracted by Trizol reagent (Invitrogen, Carlsbad, CA, USA). Reaction mixture (20 μl) containing 2 μg of total RNA was reversely transcribed to cDNA by using PrimeScript RT-polymerase (Takara, Dalian, China). Quantitative PCR was performed on the cDNA using specific primers (Sangon, Shanghai, China). The first strand cDNAs served as the template for the regular polymerase chain reaction (PCR) performed using a DNA Engine (ABI 9700). Glyceraldehyde-phosphate dehydrogenase (GAPDH) as an internal control was used to normalize the data to determine the relative expression of the target genes. The reaction conditions were set according to the kit instructions. After completion of the reaction, the amplification curve and melting curve were analyzed. Gene expression values are represented using the 2-ΔΔCt method. PCR with the following primers: UCA1, Forward 5’-TTTGCCAGCCTCAGCTTAAT-3’ and Reverse 5’-TTGTCCCCATTTTCCATCAT-3’; KLF4, Forward 5’-ATCTTTCTCCACGTTCGCGTCTG-3’ and Reverse 5’-AAGCACTGGGGGAAGTCGCTTC-3’; GAPDH, Forward 5’-GTCAACGGATTTGGTCTGTATT-3’ and Reverse 5’-AGTCTTCTGGGTGGCAGTGAT-3’.
Western blotting
The human prostate cancer tumor tissues and cells were homogenized and extracted in NP-40 buffer, followed by 5-10 min boiling and centrifugation to obtain the supernatant. Protein extracted from cells were separated by 10% SDS-PAGE and transferred to PVDF membranes (Millipore, Germany). Membranes were blocked and then incubated with primary antibodies specific for KLF4, KRT6, KRT13, JNK and p-JNK (Santa Cruz Biotechnoogy, CA, USA). β-actin (Bioworld Technology, USA) was used as protein loading control. The membranes were next incubated with the appropriate HRP (horseradish peroxidase)-conjugated antibody visualized with the and detected by chemiluminescence (Thermo, USA).
Statistical analysis
All statistical analyses were performed using SPSS version 18.0 software. Data were analyzed using independent two-tailed t test. Categorical data were analyzed using the two-side chi-square test. Overall survival was estimated by using Kaplan-Meier method, and univariate analysis was conducted by log-rank test. The Cox proportional hazards model was used in the multivariate analysis. Values of P < 0.05 were considered statistically significant.
Results
Identification of tumor tissues-enriched lncRNA implicated in PC patients
To identify that lncRNAs were potentially involved in the progression of NSCLC, we searched for lncRNAs, which were enriched in tumor tissues. Firstly, the Agilent G3 Human GE Microarray (8 × 60 K) was used to analyze lncRNAs expression profiles in 3 PC tumor tissues and paired corresponding conjunctive non-tumor tissues. Fold change greater than 2 and P value less than 0.05 between tumor tissues and adjacent non-tumor tissues were set as the criteria in filtering differently expressed lncRNAs. After the removal of redundant and unannotated sequences, 17 lncRNAs were found to be significantly down-regulated and 17 lncRNAs to be significantly up-regulated in the PC tumor tissues, and we finally focused on UCA1 in our study (Figure 1A). To evaluate whether UCA1 was differentially expressed in PC tumor tissues, a total of 5 paired tumor tissues and adjacent normal tissues were analyzed for UCA1 expression using RT-PCR. The agarose electrophoretogram results showed that UCA1 expression in tumor tissues from PC patients was significantly increased as compared to adjacent non-tumor tissues (Figure 1B). In addition, to understand the prognostic significance of UCA1 upregulation in PC, we analyzed the the relationship between the expression of UCA1 and patients survival time and found that patients with high UCA1 levels had a significantly poorer prognosis than those with low expression patients (Figure 1C, P < 0.001).
Figure 1.
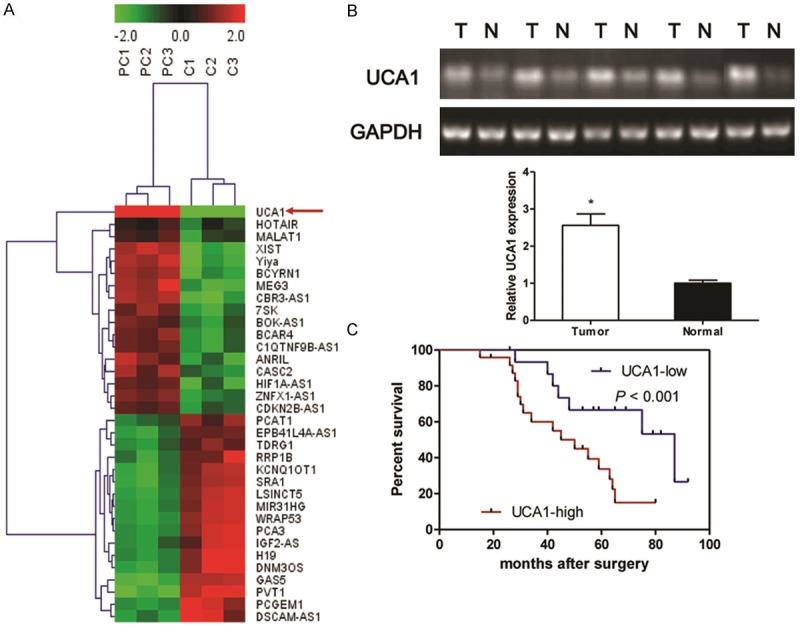
Identification of tumor tissues-enriched lncRNA implicated in PC patients. Microarray and hierarchical cluster analysis of the differentially expressed long non-coding RNAs (lncRNA) in tumor tissues from PC patients and corresponding nontumourous tissues. The figure is drawn by MeV software (version 4.2.6). Differentially expressed lncRNAs chosen from lncRNA and disease database. Correlation similarity matrix and average linkage algorithms are used in the cluster analysis. Each row represents an individual lncRNA, and each column represents a sample. The dendrogram at the left side and the top displays similarity of expression among lncRNAs and samples individually. The color legend at the top represents the level of RNA expression, with red indicating high expression levels and green indicating low expression levels (A). Semi-quantitative reverse transcription polymerase chain reaction (RT-PCR) is used to detect the expression of UCA1 in the tumor tissues and corresponding non-tumourous specimens from PC patients (B). Kaplan-Meier survival curve and log-rank test are used to evaluate whether UCA1 expression level is associated with 5-years overall survival rate. Patients were segregated into UCA1-high group and UCA1-low according to the median of UCA1 expression in tumor tissues (C). Values are expressed as mean ± SD, *P < 0.05 versus non-tumourous group.
The expression of KLF4 in tumor tissues from PC patients
As shown in Figure 2A-C, the mRNA (Figure 2A and 2C) and protein (Figure 2B and 2C) expression of KLF4 in tumor tissues were significantly higher than that of the corresponding non-tumor adjacent tissues (P < 0.05). To demonstrate the interaction between UCA1 and KLF4 in PC, the linear correlation between UCA1 and KLF4 was measured in PC tumor tissues. The results showed that there was a high positive correlation comparing the RNA expression between UCA1 and KLF4 in tumor tissues (Figure 2D, r = 0.781).
Figure 2.
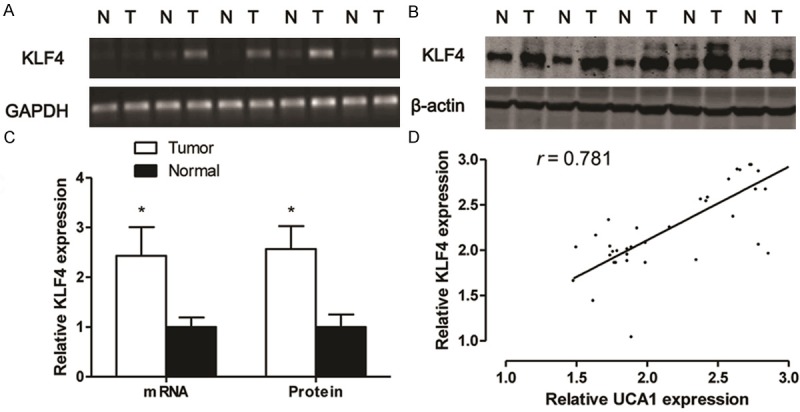
The expression of KLF4 in tumor tissues from PC patients. RT-PCR (A) and western blotting (B) are used to detect the expression of KLF4 in the tumor tissues and corresponding nontumourous specimens from RCC patients, and densitometric quantification for RT-PCR and western blotting (C). Linear correlation plot of RNA expression between UCA1 and KLF4 (D). Values are expressed as mean ± SD, *P < 0.05 versus non-tumourous group. N = 40 in each group.
UCA1 loss-of-function induces cell apoptosis
Based on above observations, the expression of UCA1 was measured among 3 different PC cell lines (22RV1, PC3 and LNCaP) and a human prostatic epithelial cells line (RWPE1). We found that UCA1 was obviously overexpressed in 3 PC cell lines than that of RWPE1 cells, especially in PC3 and LNCaP cell lines (Figure 3A). Thus, PC3 and LNCaP cell lines were selected as research represents of PC cells in the following studies. To assess the effects of si-UCA1 on the in vitro growth of PC cell lines, we examined the cell viability of PC3 and LNCaP transfected with si-UCA1. The results showed that the cell viability was significantly inhibited in PC3 and LNCaP cells at 48 or 72 h after treatment with si-UCA1, however, no significant difference was observed in NC and siRNA-NC transfected cells at each time point (Figure 3B). Consistent with the CCK8 assay, the Annexin V-PI double labeling results showed that inhibition the function of UCA1 with siRNA could increase the proportion of the apoptosis cells (Figure 3C, P < 0.05).
Figure 3.
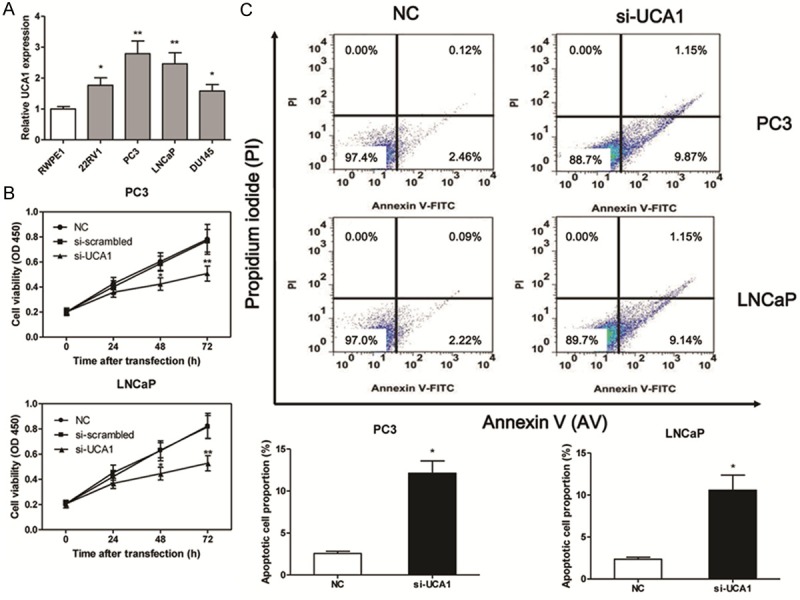
UCA1 loss-of-function induces cell apoptosis. The expression of UCA1 in prostate cancer cell lines (A). The cell viability is measured by CCK8 in PC3 and LNCaP cell lines after si-UCA1 transfection (B). The apoptotic cell proportion is measured by flow cytometric analysis of annexin V/PI double staining after the si-UCA1 transfection for 48 h, upper right and lower right in the cell distribution pictures represent the late and early apoptotic cells proportion respectively (C). Values are expressed as mean ± SD, *P < 0.05, **P < 0.01 versus control group. N = 3 in each group.
UCA1 regulates the expression of KLF4 and KRT6/13
The mRNA and protein expression of KLF4 were examined by RT-PCR and western blotting respectively. As expected, UCA1 loss-of-function in PC3 and LNCaP cells triggered a markedly decreasing in KLF4 mRNA and protein expression (Figure 4A). To further explore the potential mechanism that UCA1 involved in the progression of PC, we examined the levels of KRT6, KRT13, JNK and p-JNK in PC3 and LNCaP cell lines. The results indicated that the protein expression levels of KRT6 and KRT13 were markedly reduced in the si-UCA1 transfected PC3 and LNCaP cells as compared to the NC or siRNA-NC group. However, the protein levels of JNK and p-JNK had no significant changes in three experimental groups (Figure 4B). KLF4 is a member of the KLF family of transcription factors and is involved in both cellular proliferation and differentiation. KLF4 promotes esophageal squamous cell carcinoma (ESCC) differentiation by upregulating KRT13 expression [12]. In this study, we demonstrated that KLF4 loss-of-function could significantly inhibit the cell viability in PC3 and LNCaP cells (Figure 5A). Moreover, the results indicated that the protein expression levels of KRT6 and KRT13 were decreased in the si-KLF4 transfected PC3 and LNCaP cells as compared to the NC. However, the protein levels of JNK and p-JNK had no significant changes after si-KLF4 transfection in PC3 and LNCaP cells (Figure 5B).
Figure 4.
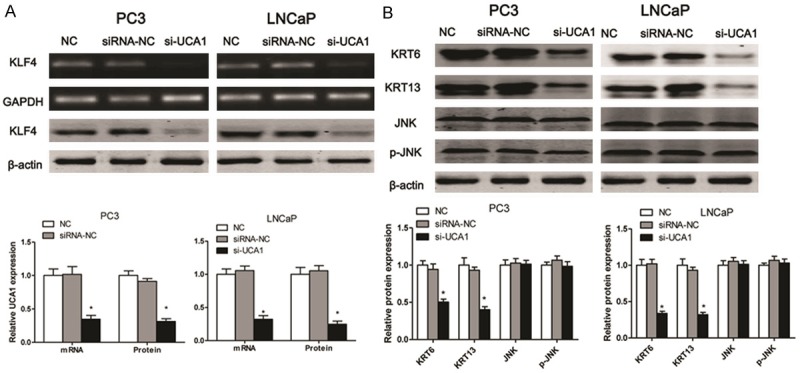
UCA1 regulates the expression of KLF4 and KRT6/13. After the cells transfected with si-UCA1 for 48 h, RT-PCR and western blotting are used to detect the expression of KLF4 in PC3 and LNCaP cell lines (A), and the protein exprossion of KRT6, KRT13, JNK and p-JNK is measured in PC3 and LNCaP cell lines (B). Values are expressed as mean ± SD, *P < 0.05 versus control group. N = 3 in each group.
Figure 5.
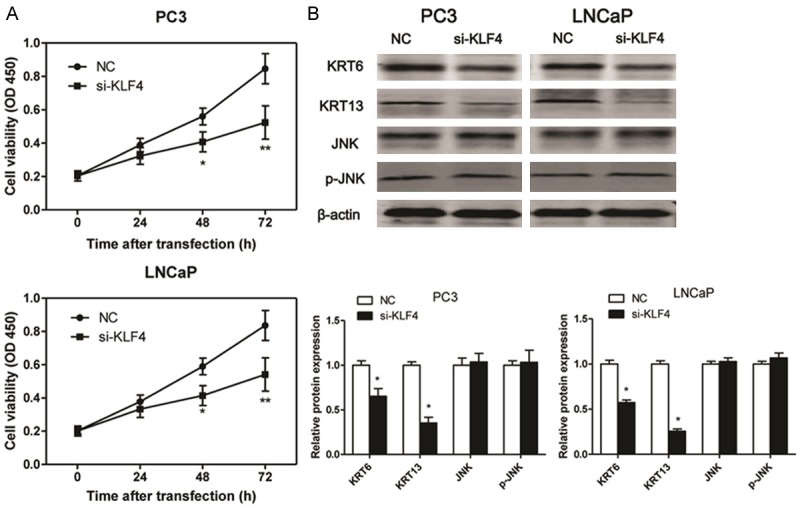
KLF4 loss-of-function decreases the protein expression of KRT6/13. The cell viability is measured by CCK8 in PC3 and LNCaP cell lines after si-KLF4 transfection (A). The protein exprossion of KRT6, KRT13, JNK and p-JNK is measured in PC3 and LNCaP cell lines after UCA1 transfection for 48 h. Values are expressed as mean ± SD, *P < 0.05, **P < 0.01 versus control group. N = 3 in each group.
Discussion
Recent genome-wide studies have indicated that the mammalian genome is abundantly transcribed, and that at least 80% of this transcription is exclusively associated with lncRNAs. Emerging data strongly implicate lncRNAs in the basal regulation of protein-coding genes, which are central to normal development and oncogenesis, at both the transcriptional and the post-transcriptional levels [13]. Mounting evidences have showed that UCA1 play a central role in the regulation of cell development, differentiation, proliferation and apoptosis [9-11]. Moreover, UCA1 acts as an oncogene through activating miR-216b-FGFR1-ERK signaling pathway promating the proliferation of hepatoma cell [8]. Here we revealed that UCA1 was upregulated in PC tissues compared to the adjacent non-tumor tissues, and patients with high UCA1 levels had a significantly poorer prognosis than those with low expression patients. Next, the results demonstrated that the mRNA and protein expression of KLF4 in tumor tissues were significantly higher than that of the corresponding non-tumor adjacent tissues, and there was a high positive correlation comparing the RNA expression between UCA1 and KLF4 in tumor tissues. Furthermore, UCA1 loss-of-function inhibited PC3 and LNCaP cells proliferation and induced cell apoptosis through suppression KLF4/KRT signaling pathways. In general, these results suggested that UCA1 might be involved in development, progression and prognosis of the majority of human PC.
To investigate the underlying molecular mechanisms of UCA1-induced cell apoptosis in PC3 and LNCaP cells, we particularly focused on KLF4 for further studies. KLF4 is a zinc-finger transcription factor, which plays a key role in cancer cellular differentiation and proliferation [14]. In prostate cancer, upregulated KLF4 mediates lysophosphatidic acid (LPA)-stimulated migration and proliferation of PC3M cells, furthermore, silencing of KLF4 expression results in the abolishment of LPA-stimulated migration and proliferation prostatic cancer cells [15]. In contrast to that KLF4 is considered to a potential tumor-suppressing factor, and overexpression of KLF4 enhances the DNA synthesis suppressive and proapoptotic effects of selenium in prostate cancer cells [16]. In this study, we showed that the expression of KLF4 increased in tumor tissues from PC patients. Moreover, we observed that there was a positive correlation between the levels of KLF4 mRNA expression and the levels of UCA1 expression in PC tumor tissues. In PC3 and LNCaP cells, UCA1 loss-of-function triggered a markedly silencing effect on KLF4 mRNA and protein expression. In general, these results suggested that the downregulation of KLF4 could inhibit cell proliferation and induce cell apoptosis in PC3 and LNCaP cells. In order to better understand the underlying molecular mechanisms of KLF4-induced growth inhibition in prostate cancer cells, we particularly focused on KRT6 and KRT13 for further studies. The molecular investigation demonstrates that KLF4 transcriptionally regulates KRT13 and the expression of the two molecules appreciably correlates in ESCC tissues and cell lines [12]. In the present study, we found that KLF4 loss-of-function resulted in downregulating expression of KRT6 and KRT13 in PC3 and LNCaP cells. Therefore, the findings provide a new clue for understanding the pathogenesis of PC and provide an intriguing approach for KLF4 regulating the expression of KRT6 and KRT13.
In conclusion, our present work indicated that UCA1, as an oncogene, promoted malignant progression of PC. Moreover, the mechanistic analysis revealed that UCA1 loss-of-function could inhibit cell proliferation and induce cell apoptosis in PC3 and LNCaP cell lines, at least partially, through inactivation KLF4-KRT6/KRT13 cascade.
Disclosure of conflict of interest
None.
References
- 1.Sutcliffe S, Colditz GA. Prostate cancer: is it time to expand the research focus to early-life exposures? Nat Rev Cancer. 2013;13:208–518. doi: 10.1038/nrc3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Nacusi LP, Tindall DJ. Targeting 5alpha-reductase for prostate cancer prevention and treatment. Nat Rev Urol. 2011;8:378–384. doi: 10.1038/nrurol.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crea F, Watahiki A, Quagliata L, Xue H, Pikor L, Parolia A, Wang Y, Lin D, Lam WL, Farrar WL, Isogai T, Morant R, Castori-Eppenberger S, Chi KN, Wang Y, Helgason CD. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget. 2014;5:764–774. doi: 10.18632/oncotarget.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang F, Li X, Xie X, Zhao L, Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008;582:1919–1927. doi: 10.1016/j.febslet.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 7.Han Y, Yang YN, Yuan HH, Zhang TT, Sui H, Wei XL, Liu L, Huang P, Zhang WJ, Bai YX. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology. 2014;46:396–401. doi: 10.1097/PAT.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 8.Wang F, Ying HQ, He BS, Pan YQ, Deng QW, Sun HL, Chen J, Liu X, Wang SK. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6:7899–7917. doi: 10.18632/oncotarget.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang C, Li X, Wang Y, Zhao L, Chen W. Long non-coding RNA UCA1 regulated cell cycle distribution via CREB through PI3-K dependent pathway in bladder carcinoma cells. Gene. 2012;496:8–16. doi: 10.1016/j.gene.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Wu W, Zhang S, Li X, Xue M, Cao S, Chen W. Ets-2 regulates cell apoptosis via the Akt pathway, through the regulation of urothelial cancer associated 1, a long non-coding RNA, in bladder cancer cells. PLoS One. 2013;8:e73920. doi: 10.1371/journal.pone.0073920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J. 2014;281:1750–1758. doi: 10.1111/febs.12737. [DOI] [PubMed] [Google Scholar]
- 12.He H, Li S, Hong Y, Zou H, Chen H, Ding F, Wan Y, Liu Z. Kruppel-like Factor 4 Promotes Esophageal Squamous Cell Carcinoma Differentiation by Up-regulating Keratin 13 Expression. J Biol Chem. 2015;290:13567–13577. doi: 10.1074/jbc.M114.629717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Chen W, Yang C, Wu W, Wu S, Qin X, Li X. Long non-coding RNA UCA1a (CUDR) promotes proliferation and tumorigenesis of bladder cancer. Int J Oncol. 2012;41:276–284. doi: 10.3892/ijo.2012.1443. [DOI] [PubMed] [Google Scholar]
- 14.Lee HY, Ahn JB, Rha SY, Chung HC, Park KH, Kim TS, Kim NK, Shin SJ. High KLF4 level in normal tissue predicts poor survival in colorectal cancer patients. World J Surg Oncol. 2014;12:232. doi: 10.1186/1477-7819-12-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin SH, Kwon YW, Heo SC, Jeong GO, Kim BR, Seo EJ, Kim JH. Kruppel-like factor 4 mediates lysophosphatidic acid-stimulated migration and proliferation of PC3M prostate cancer cells. Exp Mol Med. 2014;46:e104. doi: 10.1038/emm.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S, Zhang H, Zhu L, Zhao L, Dong Y. Kruppel-like factor 4 is a novel mediator of selenium in growth inhibition. Mol Cancer Res. 2008;6:306–313. doi: 10.1158/1541-7786.MCR-07-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]


