Abstract
Studies examining the association between the cyclin D1 (CCND1) A870G (rs9344 G>A) polymorphism and esophageal squamous cell carcinoma (ESCC) have yielded inconsistent results. Here, we conducted a hospital-based case-control study in a Chinese Han population to assess the association between the CCND1 A870G polymorphism and ESCC. We then performed a meta-analysis to further investigate this association. We recruited 629 patients with ESCC and 686 cancer-free controls. Genotyping was performed with the polymerase chain reaction-ligase detection reactions (PCR-LDR) method. The meta-analysis was performed with the STATA 12.0 software. The case-control study showed no significant difference between the ESCC cases and controls in the allele frequencies or genotype distributions of the CCND1 A870G polymorphism. To obtain a more precise estimate of this relationship, we performed a meta-analysis of seven case-control studies involving a total of 2080 ESCC cases and 2833 controls. The meta-analysis suggested that the CCND1 A870G polymorphism is not associated with a risk of ESCC. A further subgroup analysis based on ethnicity also detected no association. This study suggests that the CCND1 A870G polymorphism is not associated with the risk of ESCC.
Keywords: cyclin D1, polymorphism, cancer susceptibility, meta-analysis, esophageal cancer
Introduction
It is estimated that 455,800 new cases of esophageal cancer (EC) and 400,200 deaths resulting from the disease occurred worldwide in 2012 [1]. In China, based on annual report on status of cancer estimates, 287,632 new cases of esophageal cancer (EC) and 208,473 deaths occurred in 2010 [2]. Each year, approximately half of the global cases of EC are diagnosed in China [3,4]. The mortality rate for EC patients is very high with a five-year survival rate of 12.3% [5]. In the highest-risk geographic areas, such as China and Iran, esophageal squamous cell carcinoma (ESCC) is the most frequent subtype of EC, with an estimated 90% of cases shown histologically to be ESCC [6]. However, the etiology of EC is mixed. Evidence is mounting that EC is a multifactorial disease resulting from interactions between genetic and environmental factors [7-12]. Contributing environmental risk factors to EC are not completely understood, but are deemed to include low intake of vegetables and fruits, poor nutritional status, smoking, alcohol consumption and eating at high temperatures [13-15]. Recently, a mass of molecular epidemiological studies demonstrated that susceptibility to EC is affected by both genetic and environmental factors [16]. Thus, it is quite likely that there are a number of genetic mutations contributing to the remaining indistinct EC susceptibility, which have not yet been found.
The cyclin D1 (CCND1) gene is located on chromosome 11q13. It encodes a subunit of the holoenzyme that phosphorylates the protein, subsequently affects the transcription of genes and promotes progression from G1 to S phase of the cell cycle during cell differentiation [17]. Thus, it is regarded as a key regulatory protein in cell proliferation and differentiation. The accumulating evidence highlights that over-expression of CCND1 were observed in many malignances, which might be correlated with metastases and the poor prognoses [18-20].
In the last decade, the cyclin D1 (CCND1) A870G polymorphism has been widely investigated, and several studies have tested the hypothesis that genetic variants of CCND1 A870G are associated with a risk of ESCC [21-25]. Two meta-analyses indicated that the CCND1 A870G polymorphism might be a risk factor for the development of EC, especially in Asian populations [26,27]. However, results of other previous studies suggested that the CCND1 G870A polymorphism were not associated with the risk of ESCC [28,29], so the issue remains unclear. Although a single study could be underpowered, an analysis of the accumulated data from all ESCC epidemiological studies could provide evidence of an association between CCND1 A870G polymorphism and the risk of ESCC. Therefore, to further investigate this potential relationship, we conducted a hospital-based case-control study followed by a comprehensive meta-analysis to establish a more detailed understanding of this relationship.
Materials and methods
Subjects
In total, 629 consecutive ESCC patients and 686 cancer-free controls were recruited from the Affiliated People’s Hospital of Jiangsu University and the Affiliated Hospital of Jiangsu University (Zhenjiang City, Jiangsu Province, China), between October 2008 and December 2010 as described previously [8,9,30]. All subjects were from the city of Zhenjiang and its surrounding regions. The controls were well matched with ESCC cases on average age (±5 years), gender and ethnicity described previously [8,9,31]. The control individuals were selected from the two hospitals for cure of fracture. The selection process was described in previous molecular epidemiological study [9,30,32]. This investigation was approved by the Ethics Committee of Jiangsu University (Zhenjiang, China). A structured questionnaire was used to extract detailed information on the demographics and risk factors of the study subjects and the results are given in Table 1. Each subject completed an in-person interview and gave his/her written informed consent.
Table 1.
Distribution of selected demographic variables and risk factors in the ESCC cases and controls
| Variable | Cases (n = 629) | Controls (n = 686) | P a | |||
|---|---|---|---|---|---|---|
|
|
|
|||||
| n | % | n | % | |||
| Age (years) mean ± SD | 62.85 (±8.13) | 62.58 (±7.89) | 0.541 | |||
| Age (years) | 0.155 | |||||
| < 63 | 310 | 49.28 | 365 | 53.21 | ||
| ≥ 63 | 319 | 50.72 | 321 | 46.79 | ||
| Sex | 0.185 | |||||
| Male | 444 | 70.59 | 461 | 67.20 | ||
| Female | 185 | 29.41 | 225 | 32.80 | ||
| Tobacco use | < 0.001 | |||||
| Never | 355 | 56.44 | 499 | 72.74 | ||
| Ever | 274 | 43.56 | 187 | 27.26 | ||
| Alcohol use | < 0.001 | |||||
| Never | 428 | 68.04 | 526 | 76.68 | ||
| Ever | 201 | 31.96 | 160 | 23.32 | ||
Two-sided x2 test and Student’s t test; bold values are statistically significant (P < 0.05).
DNA extraction and genotyping
Peripheral blood (2 ml) was collected from each subject. DNA was extracted from the peripheral blood lymphocytes with the QIAamp DNA Blood Mini Kit (Qiagen, Berlin, Germany) following the manufacturer’s protocol. CCND1 A870G polymorphism was genotyped using the polymerase chain reaction-ligation detection reaction (PCR-LDR) method [30,33]. As a measure of quality control, 160 DNA assays were randomly selected and tested reciprocally, with 100% concordance.
Statistical analysis
The x2 test was used to assess the differences in the categorical variables including genotypes, demographic characteristics, tobacco use, and alcohol use between the ESCC cases and the controls. Deviation from Hardy-Weinberg equilibrium (HWE) in the controls was assessed with an internet-based calculator (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). The association between the CCND1 A870G polymorphism and the risk of ESCC was estimated with logistic regression for crude odds ratios (ORs) and adjusted ORs, where appropriate. All statistical analyses were conducted with the SAS 9.1.3 software (SAS Institute, Cary, NC). A P < 0.05 (two-sided probability) was considered as the criterion for statistical significance.
Meta-analysis
We searched the Embase and PubMed for publications that investigated the association between the CCND1 A870G polymorphism and EC risk. The last search update was on September 26, 2014. The combination terms were ‘esophageal’ or ‘esophagus’ or ‘cancer’ or ‘neoplasm’ or ‘carcinoma’ or ‘tumor’ and ‘cyclin D1’ or ‘CCND1’, annexed with ‘polymorphism’ or ‘SNP’ or ‘mutation’ or ‘variant’. In addition, the search was limited to English and Chinese language publications, and all studies carried out in human subjects were included. Additional publications were identified with a manual search of all references listed in these studies and reviews. Studies were eligible for the meta-analysis if they met the major inclusion criteria: (1) designed as a case-control study; (2) evaluated the CCND1 A870G polymorphism and ESCC risk; (3) provide genotype counts of the CCND1 A870G polymorphism for the ESCC cases and controls; and (4) the control genotype distributions were consistent with HWE. The exclusion criteria were: (1) not designed as a case-control study, (2) a review, and (3) overlapping data.
The information was carefully and independently extracted by three reviewers (W. Tang, P. Yu and Y. Wang), and any differences were resolved after further discussion among all the reviewers. The following data were extracted: (1) first author, year of publication; (2) country, ethnicity of study subjects; (3) number of cases and controls, genotype method, allele and genotype frequencies; and (4) HWE in the controls. For literatures that did not provide sufficient data, the reviewers attempted to obtain these information by correspondence with the authors. If these could not be obtained, the publications were excluded.
The quality of studies included in our study was assessed by three reviewers (W. Tang, P. Yu and Y. Wang) according to the “methodological quality assessment scale” described previously [34,35]. If the quality scores ≥ 6, studies were categorized as “high quality”, and others were defined as “low quality”.
Deviation from HWE in the controls was tested with an internet-based calculator (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). In this meta-analysis, the strength of the association between the CCND1 A870G polymorphism and ESCC risk was evaluated with a crude OR and the corresponding 95% confidence intervals (95% CI). The significance of the pooled ORs was measured with the Z-test and P-value, and P < 0.05 (two-tailed) was considered the criterion for statistical significance. The heterogeneity across studies was estimated with a Q statistic test. If P < 0.10, the ORs were pooled with a random-effects model (the DerSimonian-Laird method) [36]; otherwise, a fixed-effects model was used (the Mantel-Haenszel method) [37]. Subgroup analyses were conducted to assess the specific effect of different factors. A funnel plot and Egger’s test were used to assess potential publication bias. A one-way sensitivity analysis was conducted to evaluate the stability of the meta-analysis results. Further stratified analyses were used to detect the source of heterogeneity. In this meta-analysis, all statistical analyses were conducted with the STATA software (version 12.0) and all P values were two sided.
Results
Baseline characteristics
The demographics and risk factors of the subjects are given in Table 1. There was no significant difference between the ESCC cases and controls in term of their age or sex distribution, suggesting that the frequency matching was complete. However, there were significant differences between the ESCC cases and controls in their tobacco and alcohol consumption (P < 0.001). The primary data about the CCND1 A870G polymorphism are shown in Table 2. The genotyping success rate was 98.48% for all samples. The observed genotypic frequencies in the control subjects indicated that there are no significant departures from HWE (P = 0.388; Table 2).
Table 2.
Primary information for the CCND1 A870G (rs9344) polymorphism
| Genotyped SNPs | CCND1 A870G (rs9344) |
|---|---|
| Chromosome | 11 |
| Gene official symbol | CCND1 (cyclin D1) |
| Function | Cds-synon |
| Chr pos (Genome Build 36.3) | 69172091 |
| Regulome DB Scorea | 3a |
| MAFb for Chinese in database | 0.427 |
| MAF in our controls (n = 686) | 0.470 |
| P value for HWEc test in our controls | 0.388 |
| Genotyping methodd | LDR |
| % Genotyping value | 98.48% |
http://www.regulomedb.org/;
MAF: minor allele frequency;
HWE: Hardy-Weinberg equilibrium;
LDR: ligation detection reaction;
Bold values are statistically significant (P < 0.05).
Single-locus analysis
The allele frequencies and genotype distributions of the CCND1 A870G polymorphism are summarized in Table 3. In the controls, the frequencies of the AA, AG and GG genotype were 27.2%, 51.5% and 21.3%, respectively, which did not differ significantly from those of the ESCC cases (31.0%, 49.8% and 19.2%, respectively; P = 0.289; Table 3). Logistic regression analyses were conducted and showed that the CCND1 A870G single nucleotide polymorphism (SNP) was not associated with the risk of ESCC.
Table 3.
Logistic regression analysis of the association between the CCND1 A870G, polymorphism and risk of ESCC
| Genotype | Cases (n = 629) | Controls (n = 686) | Crude OR (95% CI) | p | Adjusted ORa (95% CI) | p | ||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| n | % | n | % | |||||
| AA | 192 | 31.0 | 184 | 27.2 | 1.00 | 1.00 | ||
| AG | 308 | 49.8 | 348 | 51.5 | 0.85 (0.66-1.09) | 0.204 | 0.85 (0.66-1.10) | 0.213 |
| GG | 119 | 19.2 | 144 | 21.3 | 0.79 (0.58-1.09) | 0.148 | 0.80 (0.58-1.10) | 0.164 |
| GG vs. AG vs. AA | 0.289 | |||||||
| AG+GG | 427 | 69.0 | 492 | 72.8 | 0.83 (0.65-1.06) | 0.133 | 0.83 (0.65-1.06) | 0.144 |
| AA+AG | 500 | 80.8 | 532 | 78.7 | 1.00 | 1.00 | ||
| GG | 119 | 19.2 | 144 | 21.3 | 0.88 (0.67-1.15) | 0.355 | 0.88 (0.67-1.17) | 0.380 |
| A allele | 692 | 55.9 | 716 | 53.0 | 1.00 | - | ||
| G allele | 546 | 44.1 | 636 | 47.0 | 0.89 (0.76-1.04) | 0.134 | - | - |
Adjusted for age, sex, smoking and drinking status; bold values are statistically significant (P < 0.05).
Genotyping for CCND1 A870G was successful in 619 (98.4%) ESCC cases, and 676 (98.5%) controls.
Article eligible for the meta-analysis
As shown in Figure 1, an extensive search yielded 136 potentially relevant publications. After additional filters were applied, six case-control studies and our study were eligible for the analysis.
Figure 1.
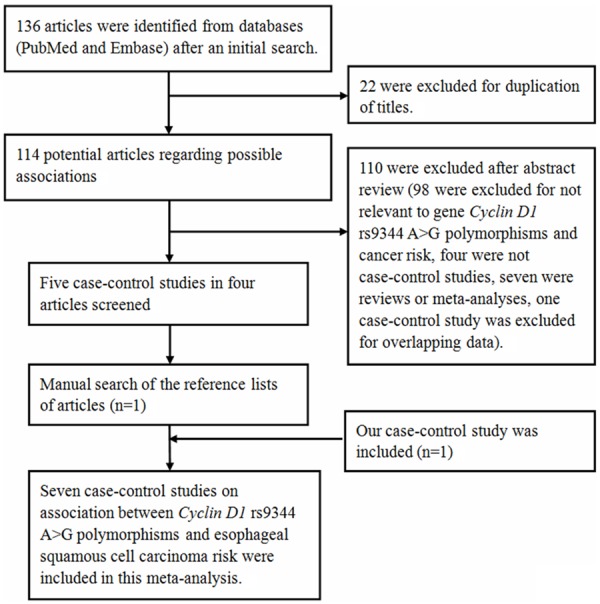
Flow diagram of articles selection process for CCND1 A870G polymorphism and esophageal cancer risk meta-analysis.
Study characteristics
There were two subgroups in a publication conducted by Akbari et al. [25], we considered them separately. In total, six separate case-control studies and our study involving 2080 ESCC cases and 2833 controls were recruited for this meta-analysis [21-25]. Among them, four were Asian [21-23] and three were Caucasian studies [24,25]. The relevant information for each study is listed in Table 4. The frequencies of the CCND1 A870G polymorphism and allele among the cases and controls are listed in Table 5. Results of the quality scores are summarized in Table 5.
Table 4.
Characteristics of populations and cancer types of the individual studies included in meta-analysis
| Study | Year | Country | Ethnicity | No. of cases/controls | Histological type of EC | Genotype method |
|---|---|---|---|---|---|---|
| Djansugurova et al. [24] | 2013 | Kazakhstan | Caucasians | 115/100 | ESCC | Direct sequencing |
| Hussain et al. [21] | 2011 | India | Asians | 151/151 | ESCC | PCR-RFLP |
| Akbari et al. [25] | 2009 | Iran | Caucasians | 195/250 | ESCC | MALDI-TOF MS |
| Akbari et al. [25] | 2009 | Iran | Caucasians | 549/1118 | ESCC | MALDI-TOF MS |
| Zhang et al. [22] | 2003 | China | Asians | 120/183 | ESCC | PCR-SSCP |
| Yu et al. [23] | 2003 | China | Asians | 321/345 | ESCC | PCR-RFLP |
| Our study | 2013 | China | Asians | 629/686 | ESCC | LDR-PCR |
PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism; MALDI-TOF MS: Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry; ESCC: esophageal squamous cell carcinoma; LDR-PCR: ligation detection reaction polymerase chain reaction; PCR-SSCP: polymerase chain reaction single-stranded conformation polymorphism.
Table 5.
Distribution of CCND1 A870G polymorphisms genotype and allele among esophageal squamous cell carcinoma patients and controls
| Case | Control | Case | Control | HWE, P value | Quality scores | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||||||
| Study | Year | AA | AG | GG | AA | AG | GG | A (%) | G (%) | A (%) | G (%) | ||
| Djansugurova et al. | 2013 | 44 | 49 | 22 | 18 | 54 | 28 | 137 (59.57) | 93 (40.43) | 90 (45.00) | 110 (55.00) | 0.363 | 6.5 |
| Hussain et al. | 2011 | 32 | 99 | 20 | 23 | 72 | 56 | 163 (53.97) | 139 (46.03) | 118 (39.07) | 184 (60.93) | 0.986 | 7.5 |
| Akbari et al. | 2009 | 53 | 83 | 59 | 79 | 116 | 55 | 189 (48.46) | 201 (51.54) | 274 (54.80) | 226 (45.20) | 0.316 | 6.5 |
| Akbari et al. | 2009 | 158 | 276 | 115 | 355 | 550 | 213 | 592 (53.92) | 506 (46.08) | 1260 (56.35) | 976 (43.65) | 0.999 | 7.5 |
| Zhang et al. | 2003 | 35 | 74 | 11 | 43 | 102 | 38 | 144 (60.00) | 96 (40.00) | 188 (51.37) | 178 (48.63) | 0.118 | 7.0 |
| Yu et al. | 2003 | 96 | 157 | 68 | 110 | 177 | 58 | 349 (54.36) | 293 (45.64) | 397 (57.54) | 293 (42.46) | 0.354 | 6.0 |
| Our study | 2013 | 192 | 308 | 119 | 184 | 348 | 144 | 692 (55.90) | 546 (44.10) | 716 (52.96) | 636 (47.04) | 0.388 | 8.0 |
HWE: Hardy-Weinberg equilibrium.
Meta-analysis results
When all eligible studies were combined, there was no significant difference in the allele frequencies or genotype distributions of the CCND1 A870G polymorphism in the ESCC cases and controls, even when different ethnic groups were involved (Table 6; Figure 2).
Table 6.
Summary of results of the meta-analysis from different comparative genetic models in the subgroup analysis
| No. (cases/controls) | G vs. A | GG vs. AA | GG+AG vs. AA | GG vs. AG+AA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||||||
| OR (95% CI) | P | P (Q-test) | OR (95% CI) | P | P (Q-test) | OR (95% CI) | P | P (Q-test) | OR (95% CI) | P | P (Q-test) | ||
| Total | 2080/2833 | 0.87 (0.71-1.08) | 0.205 | 0.000 | 0.73 (0.47-1.14) | 0.173 | 0.000 | 0.88 (0.69-1.12) | 0.296 | 0.007 | 0.79 (0.54-1.16) | 0.232 | 0.000 |
| Ethnicity | |||||||||||||
| Asians | 1221/1365 | 0.81 (0.62-1.07) | 0.133 | 0.001 | 0.60 (0.31-1.15) | 0.123 | 0.000 | 0.87 (0.73-1.04) | 0.119 | 0.370 | 0.61 (0.32-1.18) | 0.143 | 0.000 |
| Caucasians | 859/1468 | 0.96 (0.66-1.39) | 0.817 | 0.001 | 0.93 (0.46-1.88) | 0.832 | 0.003 | 0.85 (0.48-1.52) | 0.592 | 0.002 | 1.08 (0.72-1.63) | 0.694 | 0.060 |
| Sample sizes | |||||||||||||
| < 600 | 581/684 | 0.71 (0.47-1.06) | 0.097 | 0.000 | 0.45 (0.18-1.13) | 0.089 | 0.000 | 0.71 (0.42-1.18) | 0.183 | 0.003 | 0.53 (0.24-1.13) | 0.100 | 0.000 |
| ≥ 600 | 1499/2149 | 1.07 (0.85-1.34) | 0.548 | 0.030 | 1.16 (0.74-1.81) | 0.528 | 0.032 | 0.97 (0.81-1.15) | 0.711 | 0.178 | 1.18 (0.83-1.68) | 0.361 | 0.053 |
| Publication year | |||||||||||||
| > 2009 | 895/937 | 0.66 (0.46-0.96) | 0.030 | 0.006 | 0.43 (0.19-0.95) | 0.037 | 0.005 | 0.62 (0.38-1.01) | 0.055 | 0.045 | 0.53 (0.25-1.13) | 0.100 | 0.001 |
| ≤ 2009 | 1185/1896 | 1.06 (0.88-1.29) | 0.543 | 0.037 | 1.09 (0.70-1.69) | 0.696 | 0.017 | 1.10 (0.94-1.30) | 0.223 | 0.457 | 1.07 (0.72-1.59) | 0.754 | 0.010 |
Figure 2.
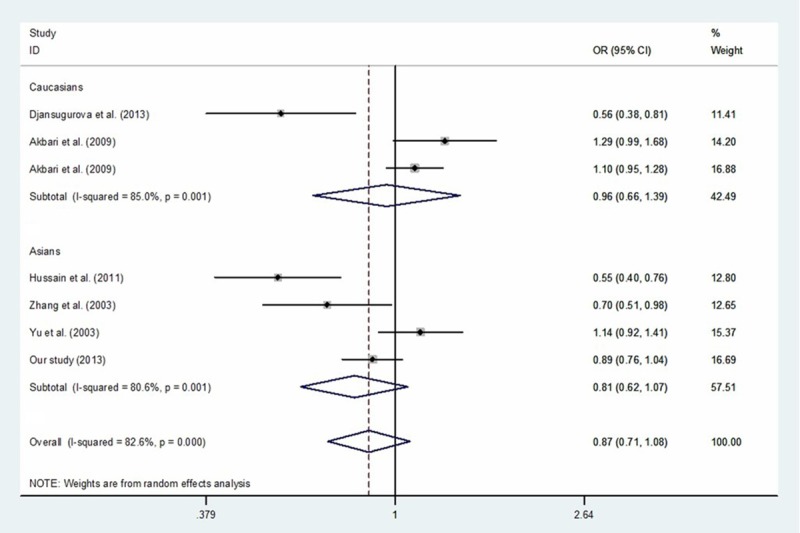
Meta-analysis with a random-effects model for the association between the risk of esophageal cancer and the CCND1 A870G polymorphism (G vs. A).
Tests for publication bias, sensitivity analyses, and heterogeneity
Begg’s funnel plots and Egger’s tests were used to assess the publication bias, and the shape of the funnel was symmetrical in all genetic models (Figure 3). This suggested that our findings were robust (G vs. A: Begg’s test P = 0.133, Egger’s test P = 0.165; GG vs. AA: Begg’s test P = 0.368, Egger’s test P = 0.109; GG+AG vs. AA: Begg’s test P = 0.133, Egger’s test P = 0.157; GG vs. AG+AA: Begg’s test P = 0.368, Egger’s test P = 0.166).
Figure 3.
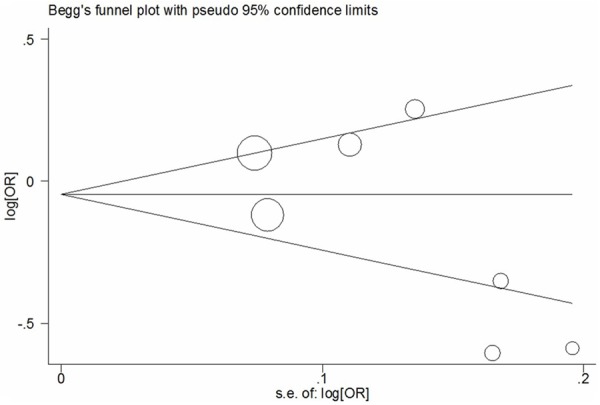
Begg’s funnel plot of meta-analysis of between the CCND1 A870G polymorphism and the risk of cancer (G vs. A).
We used a one-way sensitivity analysis to assess the influence of each study on the pooled ORs and CIs. The results demonstrated that our findings were stable (Figure 4; data not shown).
Figure 4.
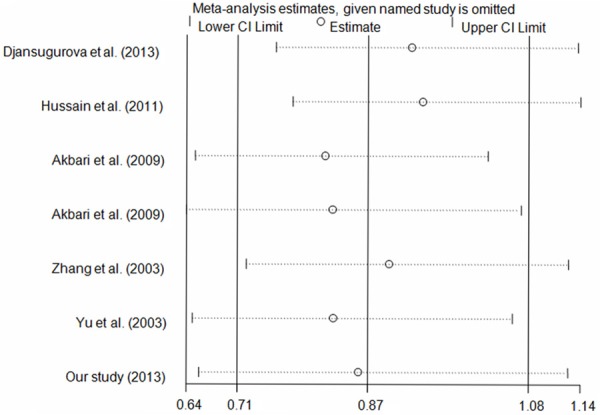
Sensitivity analysis of the influence of G vs. A genetic model (random-effects estimates).
As shown in Table 6, significant heterogeneity was observed among the studies in all the genetic comparison models. Because ethnicity, sample size and publication year can influence the results of a meta-analysis, we conducted subgroup analyses and the results are shown in Table 6. The results confirmed association between significant heterogeneity and Asian populations, studies published after 2009, and small samples (< 600 subjects) (Table 6).
Discussion
Studies investigating the association between the CCND1 A870G polymorphism and the risk of ESCC have yielded inconclusive results. We conducted a case-control study in a Chinese Han population, together with a meta-analysis of several studies to determine the association between the CCND1 A870G polymorphism and ESCC. To the best of our knowledge, this is so far the most comprehensive meta-analysis to investigate the association. The results of the case-control study showed no significant differences in the allele frequencies or genotype distributions of the CCND1 A870G polymorphism between the ESCC cases and controls. The results of the meta-analysis suggested that the CCND1 A870G polymorphism is not associated with a risk of ESCC, even in different populations.
CCND1 plays a crucial role in the transition from the G1 to S phase of the cell cycle during mitosis, and regulates cell proliferation and differentiation [38,39]. The stabilized over-expression of CCND1 can accelerate cell proliferation, disrupt the cell cycle, alter the capacity of cells to undergo DNA repair, which all promote tumorigenesis [40,41]. With a growing interest in the association between the CCND1 A870G and ESCC risk, several recent studies have tested the hypothesis that the CCND1 A870G polymorphism is associated with a risk of ESCC [21-25], but the findings have been inconsistent. Because common SNPs are usually low-penetrance cancer susceptibility variants, our meta-analysis examined seven studies to comprehensively assess the association between the CCND1 A870G genetic variant and ESCC risk. Several investigations have reported a negative correlation between the CCND1 A870G polymorphism and ESCC [21,22,24]; whereas others have reported no link between the polymorphism and ESCC [23,25]. Based on our meta-analysis of 4913 subjects (2080 ESCC cases and 2833 controls), the CCND1 A870G polymorphism may not be associated with a risk of ESCC. However, it is possible that other genetic or environmental factors may have diluted or obscured the effects of the CCND1 A870G polymorphism, and these important factors should not be ignored. Because only seven case-control studies, involving 4913 subjects were included in this meta-analysis with most studies designed for small samples, further investigations with larger samples and detailed individual data must be performed to validate or refute our findings.
Heterogeneity, publication bias and sensitivity analysis were assessed in this study. Significant heterogeneity was observed between the publications regarding the CCND1 A870G polymorphism (Table 6). Potential sources of heterogeneity include ethnicity, publication year, sample size, and so on. When stratified analyses were conducted according to these potential biasing factors, heterogeneity was reduced or removed in some subgroups, confirming the effects of ethnicity, publication year, and sample size, even for the same polymorphism (Table 6). Begg’s funnel plots and Egger’s tests were used to detect publication bias, and indicated that there was no significant publication bias in the meta-analysis, suggesting that our results are robust. One-way sensitivity analyses indicated that our findings are stable (Figure 4).
The specific merits of the present study should be noted. First, the sample size in this case-control study is the largest used to investigate the association between the CCND1 A870G polymorphism and ESCC risk, and the meta-analysis is the most comprehensive exploration of the association between CCND1 A870G polymorphism and ESCC risk to date. Second, although our case-control study was hospital-based, the control genotype distributions were consistent with HWE.
Some limitations in interpreting the results should be acknowledged. In this study, the control subjects were recruited from two local hospitals, which might have led to an unavoidable selection bias. Moreover, all case-control studies included in the meta-analysis were of Asian or Caucasian subjects; no data for other ethnicities (e.g., African) were available. Only published studies and our investigation were included, which might have increased the chance of publication bias. Furthermore, because individual data were limited in this review, we did not analyze other factors (e.g., age, sex, smoking, alcohol consumption, other lifestyle factors, or environmental factors). Finally, significant heterogeneity was observed among the publications.
In conclusion, this meta-analysis of seven case-control studies suggests that the CCND1 A870G polymorphism is not involved in the pathogenesis of ESCC. Further investigations with larger samples and detailed individual information should be undertaken in future to validate or refute our results.
Acknowledgements
We appreciate all patients who participated in this study. We wish to thank Dr. Yiqun Chen (Biowing Applied Biotechnology Company, Shanghai, China) for technical support. This study was supported in part by National Natural Science Foundation of China (81472332, 81370001, 81300037, 81101889, 81000028, 81341006), Jiangsu Province Natural Science Foundation (BK2010333, BK2011481), Fujian Province Natural Science Foundation (2013J01126, 2013J05116), Fujian Medical University professor fund (JS12008) and Fujian Province science and technology programmed fund (2012Y0030), Social Development Foundation of Zhenjiang (SH2010017) and Changzhou Young Talents and Science-Technology Foundation of Health Bureau (QN201102).
Disclosure of conflict of interest
None.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Zheng L, Tang W, Shi Y, Chen S, Wang X, Wang L, Shao A, Ding G, Liu C, Liu R, Yin J, Gu H. p21 rs3176352 G>C and p73 rs1801173 C>T polymorphisms are associated with an increased risk of esophageal cancer in a Chinese population. PLoS One. 2014;9:e96958. doi: 10.1371/journal.pone.0096958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, He Y, Zheng R, Zhang S, Zeng H, Zou X, He J. Esophageal cancer incidence and mortality in China, 2009. J Thorac Dis. 2013;5:19–26. doi: 10.3978/j.issn.2072-1439.2013.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang L, Parkin DM, Li L, Chen Y. Time trends in cancer mortality in China: 1987-1999. Int J Cancer. 2003;106:771–783. doi: 10.1002/ijc.11300. [DOI] [PubMed] [Google Scholar]
- 5.Berrino F, De Angelis R, Sant M, Rosso S, Bielska-Lasota M, Coebergh JW, Santaquilani M EUROCARE Working group. Survival for eight major cancers and all cancers combined for European adults diagnosed in 1995-99: results of the EUROCARE-4 study. Lancet Oncol. 2007;8:773–783. doi: 10.1016/S1470-2045(07)70245-0. [DOI] [PubMed] [Google Scholar]
- 6.Gholipour C, Shalchi RA, Abbasi M. A histopathological study of esophageal cancer on the western side of the Caspian littoral from 1994 to 2003. Dis Esophagus. 2008;21:322–327. doi: 10.1111/j.1442-2050.2007.00776.x. [DOI] [PubMed] [Google Scholar]
- 7.Guzman VB, Yambartsev A, Goncalves-Primo A, Silva ID, Carvalho CR, Ribalta JC, Goulart LR, Shulzhenko N, Gerbase-Delima M, Morgun A. c New approach reveals CD28 and IFNG gene interaction in the susceptibility to cervical cancer. Hum Mol Genet. 2008;17:1838–1844. doi: 10.1093/hmg/ddn077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang W, Zhang S, Qiu H, Wang L, Sun B, Yin J, Gu H. Genetic variations in MTHFR and esophageal squamous cell carcinoma susceptibility in Chinese Han population. Med Oncol. 2014;31:915. doi: 10.1007/s12032-014-0915-6. [DOI] [PubMed] [Google Scholar]
- 9.Tang W, Qiu H, Jiang H, Sun B, Wang L, Yin J, Gu H. Lack of association between cytotoxic T-lymphocyte antigen 4 (CTLA-4)-1722T/C (rs733618) polymorphism and cancer risk: from a case-control study to a meta-analysis. PLoS One. 2014;9:e94039. doi: 10.1371/journal.pone.0094039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Du C, Guo X, Yuan L, Niu W, Yu W, Er L, Wang S. Interleukin-8-251A/T polymorphism and Helicobacter pylori infection influence risk for the development of gastric cardiac adenocarcinoma in a high-incidence area of China. Mol Biol Rep. 2010;37:3983–3989. doi: 10.1007/s11033-010-0057-7. [DOI] [PubMed] [Google Scholar]
- 11.Yin J, Liu C, Wang X, Wang L, Shi Y, Tang W, Ding G, Liu R, Chen S, Gu H, Zheng L. Interleukin 15 receptor alpha rs2228059 A>C polymorphism decreased risk of gastric cardiac adenocarcinoma in a Chinese population. Tumour Biol. 2014;35:6593–6600. doi: 10.1007/s13277-014-1872-6. [DOI] [PubMed] [Google Scholar]
- 12.Yin J, Wang X, Wei J, Wang L, Shi Y, Zheng L, Tang W, Ding G, Liu C, Liu R, Chen S, Xu Z, Gu H. Interleukin 12B rs3212227 T>G polymorphism was associated with an increased risk of gastric cardiac adenocarcinoma in a Chinese population. Dis Esophagus. 2015;28:291–8. doi: 10.1111/dote.12189. [DOI] [PubMed] [Google Scholar]
- 13.Islami F, Boffetta P, Ren JS, Pedoeim L, Khatib D, Kamangar F. High-temperature beverages and foods and esophageal cancer risk--a systematic review. Int J Cancer. 2009;125:491–524. doi: 10.1002/ijc.24445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu M, Liu AM, Kampman E, Zhang ZF, Van’t Veer P, Wu DL, Wang PH, Yang J, Qin Y, Mu LN, Kok FJ, Zhao JK. Green tea drinking, high tea temperature and esophageal cancer in high- and low-risk areas of Jiangsu Province, China: a population-based case-control study. Int J Cancer. 2009;124:1907–1913. doi: 10.1002/ijc.24142. [DOI] [PubMed] [Google Scholar]
- 15.Rasool S, A Ganai B, Syed Sameer A, Masood A. Esophageal cancer: associated factors with special reference to the Kashmir Valley. Tumori. 2012;98:191–203. doi: 10.1177/030089161209800203. [DOI] [PubMed] [Google Scholar]
- 16.Wang AH, Liu Y, Wang B, He YX, Fang YX, Yan YP. Epidemiological studies of esophageal cancer in the era of genome-wide association studies. World J Gastrointest Pathophysiol. 2014;5:335–343. doi: 10.4291/wjgp.v5.i3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: normal and abnormal functions. Endocrinology. 2004;145:5439–5447. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- 18.Elsheikh S, Green AR, Aleskandarany MA, Grainge M, Paish CE, Lambros MB, Reis-Filho JS, Ellis IO. CCND1 amplification and cyclin D1 expression in breast cancer and their relation with proteomic subgroups and patient outcome. Breast Cancer Res Treat. 2008;109:325–335. doi: 10.1007/s10549-007-9659-8. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura Y, Felizola SJ, Kurotaki Y, Fujishima F, McNamara KM, Suzuki T, Arai Y, Sasano H. Cyclin D1 (CCND1) expression is involved in estrogen receptor beta (ERbeta) in human prostate cancer. Prostate. 2013;73:590–595. doi: 10.1002/pros.22599. [DOI] [PubMed] [Google Scholar]
- 20.Balcerczak E, Pasz-Walczak G, Kumor P, Panczyk M, Kordek R, Wierzbicki R, Mirowski M. Cyclin D1 protein and CCND1 gene expression in colorectal cancer. Eur J Surg Oncol. 2005;31:721–726. doi: 10.1016/j.ejso.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Hussain S, M Y, Thakur N, Salam I, Singh N, Mir MM, Bhat MA, Siddiqi MA, Das BC, Bharadwaj M. Association of cyclin D1 gene polymorphisms with risk of esophageal squamous cell carcinoma in Kashmir Valley: a high risk area. Mol Carcinog. 2011;50:487–498. doi: 10.1002/mc.20732. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Li Y, Wang R, Wen D, Sarbia M, Kuang G, Wu M, Wei L, He M, Zhang L, Wang S. Association of cyclin D1 (G870A) polymorphism with susceptibility to esophageal and gastric cardiac carcinoma in a northern Chinese population. Int J Cancer. 2003;105:281–284. doi: 10.1002/ijc.11067. [DOI] [PubMed] [Google Scholar]
- 23.Yu C, Lu W, Tan W, Xing D, Liang G, Miao X, Lin D. Lack of association between CCND1 G870A polymorphism and risk of esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2003;12:176. [PubMed] [Google Scholar]
- 24.Djansugurova LB, Perfilyeva AV, Zhunusova GS, Djantaeva KB, Iksan OA, Khussainova EM. The determination of genetic markers of age-related cancer pathologies in populations from Kazakhstan. Front Genet. 2013;4:70. doi: 10.3389/fgene.2013.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akbari MR, Malekzadeh R, Shakeri R, Nasrollahzadeh D, Foumani M, Sun Y, Pourshams A, Sadjadi A, Jafari E, Sotoudeh M, Kamangar F, Boffetta P, Dawsey SM, Ghadirian P, Narod SA. Candidate gene association study of esophageal squamous cell carcinoma in a high-risk region in Iran. Cancer Res. 2009;69:7994–8000. doi: 10.1158/0008-5472.CAN-09-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhuo W, Zhang L, Wang Y, Zhu B, Chen Z. Cyclin D1 G870A polymorphism is a risk factor for esophageal cancer among Asians. Cancer Invest. 2012;30:630–636. doi: 10.3109/07357907.2012.726385. [DOI] [PubMed] [Google Scholar]
- 27.Wen L, Hu YY, Yang GL, Liu DX. CCND1 G870A polymorphism contributes to the risk of esophageal cancer: An updated systematic review and cumulative meta-analysis. Biomed Rep. 2014;2:549–554. doi: 10.3892/br.2014.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai W, Wang ZT, Zhong J, Zhang Y. Lack of association between Cyclin D1 gene G870A polymorphism and esophageal cancer: evidence from a meta-analysis. Genet Mol Res. 2013;12:6636–6645. doi: 10.4238/2013.April.26.1. [DOI] [PubMed] [Google Scholar]
- 29.He W, Zeng Y, Long J, Zhou Q, Hu Y, Chen M. Genetic polymorphism of G870A and esophageal cancer susceptibility: A meta-analysis. Biomed Rep. 2013;1:303–307. doi: 10.3892/br.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu H, Zheng L, Tang W, Yin P, Cheng F, Wang L. Programmed death-1 (PD-1) polymorphisms in Chinese patients with esophageal cancer. Clin Biochem. 2014;47:612–617. doi: 10.1016/j.clinbiochem.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 31.Yin J, Wang L, Zheng L, Wang X, Shi Y, Shao A, Ding G, Liu C, Chen S, Tang W, Gu H. TERT-CLPTM1L Rs401681 C>T polymorphism was associated with a decreased risk of esophageal cancer in a Chinese population. PLoS One. 2014;9:e100667. doi: 10.1371/journal.pone.0100667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin J, Wang X, Zheng L, Shi Y, Wang L, Shao A, Tang W, Ding G, Liu C, Liu R, Chen S, Gu H. Hsa-miR-34b/c rs4938723 T>C and hsa-miR-423 rs6505162 C>A Polymorphisms Are Associated with the Risk of Esophageal Cancer in a Chinese Population. PLoS One. 2013;8:e80570. doi: 10.1371/journal.pone.0080570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi P, Chen Z, Zhao Y, Guo J, Fu H, Zhou Y, Yu L, Li L. PCR/LDR/capillary electrophoresis for detection of single-nucleotide differences between fetal and maternal DNA in maternal plasma. Prenat Diagn. 2009;29:217–222. doi: 10.1002/pd.2072. [DOI] [PubMed] [Google Scholar]
- 34.Guo J, Jin M, Zhang M, Chen K. A genetic variant in miR-196a2 increased digestive system cancer risks: a meta-analysis of 15 case-control studies. PLoS One. 2012;7:e30585. doi: 10.1371/journal.pone.0030585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu MT, Hu JW, Ding XX, Yang X, Zhang Z, Yin R, Xu L. Hsa-miR-499 rs3746444 polymorphism contributes to cancer risk: a meta-analysis of 12 studies. PLoS One. 2012;7:e50887. doi: 10.1371/journal.pone.0050887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hua Z, Li D, Xiang G, Xu F, Jie G, Fu Z, Jie Z, Da P, Li D. PD-1 polymorphisms are associated with sporadic breast cancer in Chinese Han population of Northeast China. Breast Cancer Res Treat. 2011;129:195–201. doi: 10.1007/s10549-011-1440-3. [DOI] [PubMed] [Google Scholar]
- 37.Bayram S, Akkiz H, Ulger Y, Bekar A, Akgollu E, Yıldırım S. Lack of an association of programmed cell death-1 PD1.3 polymorphism with risk of hepatocellular carcinoma susceptibility in Turkish population: a case-control study. Gene. 2012;511:308–313. doi: 10.1016/j.gene.2012.09.119. [DOI] [PubMed] [Google Scholar]
- 38.Murray AW. Recycling the cell cycle: cyclins revisited. Cell. 2004;116:221–234. doi: 10.1016/s0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- 39.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 40.Antonaci A, Consorti F, Mardente S, Natalizi S, Giovannone G, Della Rocca C. Survivin and cyclin D1 are jointly expressed in thyroid papillary carcinoma and microcarcinoma. Oncol Rep. 2008;20:63–67. [PubMed] [Google Scholar]
- 41.Salimi M, Mozdarani H, Majidzadeh K. Expression pattern of ATM and cyclin D1 in ductal carcinoma, normal adjacent and normal breast tissues of Iranian breast cancer patients. Med Oncol. 2012;29:1502–1509. doi: 10.1007/s12032-011-0043-5. [DOI] [PubMed] [Google Scholar]


