Abstract
Background: miRNAs have been participated in human carcinogenesis as tumor oncogenes or suppressors, and have a prognostic significant for patients with cancers. In recent years, miR-25 was found associated with clinical value of cancerous patients. Meanwhile the miR-25 showed a different expression in related studies. Therefore, we summarize the results from a variety of studies using a meta-analysis, in order to explain the prognostic value of miR-25 in many human cancers. Methods: The articles were retrieved from the on-line databases, PubMed, EMBASE and CNKI. We extracted and estimated the hazard ratios (HRs) for overall survival (OS), which compared the high and low expression levels of miR-25 in patients with a different of cancers. Pooled HRs and 95% confidence intervals (CIs) were calculated. Results: Eight studies of 868 patients were selected into the final meta-analysis after a strict filtering and qualifying process. Fixed model or random model method was chosen depending on the heterogeneity between the studies. For OS, higher miR-25 expression could significantly predict worse outcome with the pooled HR of 2.434 (95% CI 1.330-3.539, P=0.000). The subgroup analysis showed that high expressed miR-25 could worsen OS in Chinese patients with pooled HR of 1.895 (95% CI 1.096-2.693, P=0.007). The sensitive analysis showed that removing the document one by one, there no obvious change of HR of 0.823 (95% CI 0.385-1.260). No bias existed in the group. Conclusion: Our study indicates that high expression level of miR-25 corresponds with poor survival in cancerous patients, and the expression of miR-25 could be a promising prognostic biomarker in the future.
Keywords: miR-25, cancer, biomarker, prognostic factor, meta-analysis
Introduction
microRNAs (miRNAs) are small noncoding endogenous RNA molecules of 19-25 nucleotides. They could bind to complementary sequences in the 3’ untranslated region of target mRNAs, leading to mRNA degradation or translational repression. They suppress gene expression by binding to the complementary regions of mRNA, which either blocks translation or degrades mRNA through the RNA-induced silencing complex [1]. Recently, evidence from clinical studies has demonstrated that many miRNAs deregulated in variety of cancers, and the expression levels of some miRNAs correlated with clinicopathological features of diseases [2,3]. Thus, these miRNAs could be potential biomarkers for cancer.
miR-25 is a member of the miR-106b-25 cluster which consists of miR-106b, miR-25 and miR-93, and is located within intron 13 of the minichromosome maintenance complex component 7 (MCM7) gene on chromosome 7q22.1 [4,5]. The effects of miR-25 on cancer are complicated because it can be oncogenic or tumor suppressive in different types of cancer. Therefore, the prognostic value of circulating miR-25 in patients with cancer remains controversial. In terms of the limits of the single study, the aim of this meta-analysis was to evaluate the relationship between miR-25 and the survival outcome in patients with cancer.
Materials and methods
Search strategy
We performed a meta-analysis following the guidelines of the Meta-analysis of Observational Studies in Epidemiology group (MOOSE) [14]. We carefully searched online database PubMed, EMBASE and CNKI to identify relevant published studies from Jan 1st, 1993 to Dec 12, 2014. Two sets of key words were used among that process, namely “miR-25 and cancer” and “microRNA-25 and cancer”. Annual review of the references of relevant publications was also performed to obtain additional studies.
Inclusion and exclusion criteria
We used references manager software EndNote to check out duplications. And according the follow inclusive criteria to choose studies: (1) it studied miR-25 in a variety of human cancers; (2) it must be human samples; (3) it studied the association between miR-25 and survival outcome. The candidate articles were manually chosen by 2 authors (JJ Qu and W Zhong) independently. The studies were excluded if they were: (1) investigation of a set of miRNAs but not miR-25 alone; (2) review or letters; (3) non-dichotomous miR-25 expression levels; (4) or lack of important information such as hazard ratio (HR), 95% CI and P value. If we needed addition information and data which cannot been found in article, we will e-mailed the authors. M Li supervised the entire process.
Quality assessment
All of the quality of studies were assessed by the two authors according to a basic standard as follows: (1) clear report of study origin of country and population, (2) clear method of study design, (3) clear definition of outcome assessment, (4) clear definition of type of cancer. The study was removed if not including the basic standard in order to enhance the quality of the meta-analysis.
Data extraction
Two reviewers independently extracted the required information from all eligible studies to rule out any discrepancy. The following data were extracted: first author, year of publication, country of origin, sample size, tumor type, method of testing miR-25, and HR of miR-25 for overall survival (OS), as well as the corresponding 95% CI. If the HR and CI were not reported directly, the total observed death events and the numbers of patients in each group were extracted to calculate HR and its variance indirectly [15]. If only Kaplan-Meier curves are available, data was extracted from the graphical survival plots. Disagreements were resolved by discussion. All the data were resolved by consensus.
Statistical analysis
Firstly, HR with 95% CI was used to combine the pooled data. We used the chi-square based Q-test to test the statistical heterogeneity of studies, then identified the absence of heterogeneity across studies, after used fixed-effects model (the Mantel-Haenszel method), otherwise, we will performed the random effect model. Heterogeneity was defined as P<0.10 or I2>50%. Secondly, subgroup and sensitivity analysis was carried out by investigating the influence of a single study on the overall HR. Finally, we used the methods of Begg plots and Egger test to analysis publication bias (P<0.05 was considered representative of statistically significant publication bias). All of the analyses were carried out using STATA v11.0 (Stata Corp., College Station, TX).
Results
Study inclusion and characteristics
A total of 211 articles were identified by initial search. After screening titles, abstracts and key data, 119 articles were excluded for not meeting the inclusion criteria. After reading the full texts of the remaining, 84 reports were excluded because they were not records without survival data. Then, 8 potential candidate studies were evaluated between 2011 and 2014 (Figure 1).
Figure 1.
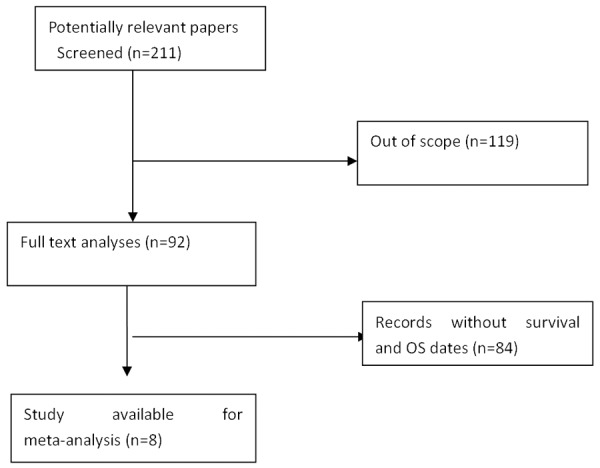
Flow diagram of the identification and selection of studies.
Table 1 shows the main results of this meta-analysis. A total of 868 participants were enrolled in the 8 studies. Seven studies evaluated patients from China, 1 evaluated from Korea, The types of cancers in these studies included ovarian cancer, hepatocellular cancer, non-small cell lung cancer, colorectal cancer, acute myelocytic leukemia, esophageal cancer, gastric cancer. The method of miR-25 detection was all quantitative real-time polymerase chain reaction (qRT-PCR). miR-25 expression levels were measured in plasma or tissue.
Table 1.
Characteristics of the studies included in this meta-analysis
| Study | Year | Country | Disease | Number | Sample | Assay | Survival | HR Reported |
|---|---|---|---|---|---|---|---|---|
| X Wang [11] | 2014 | China | Ovarian cancer | 86 | Tissue | qRT-PCR | OS | 2.119 (1.568-3.221) |
| Zhongxue Su [12] | 2014 | China | Hepatocellular | 131 | Tissue | qRT-PCR | OS | 2.179 (1.876-4.335) |
| Fangxiu Xu [9] | 2014 | China | Lung adenocarcinoma | 100 | Tissue+Plasma | qRT-PCR | OS | 2.34 (1.02-5.35) |
| Xiaojun Li [8] | 2014 | China | Colorectal | 186 | Tissue | qRT-PCR | OS | 1.90 (1.22-2.97) |
| Yungui Wang [10] | 2011 | China | AML | 53 | Bone marrow samples | qRT-PCR | OS | 0.648551 (0.36-1.45) |
| Caiyun Wu [7] | 2014 | China | Oesophageal Squamous cell carcinoma | 111 | Serum | qRT-PCR | OS | 3.84 (1.02-14.41) |
| Baek-Hui Kim [6] | 2012 | Korea | Gastric carcinoma | 91 | Tissue | qRT-PCR | OS | 4.52 (3.39-5.38) |
| Xiaohui Xu [13] | 2013 | China | Esophageal Squamous cell carcinoma | 86 | Tissue | qRT-PCR | OS | 3.87 (0.65-5.83) |
Meta-analysis
For evaluating the association between miR-25 expression and OS, there appeared obvious heterogeneity among those studies for miR-25 (P=0.000, I2=86.0%). Therefore, the random-effect model was used to calculate the pooled HR with the corresponding 95% CI. The result showed that a higher expression level of miR-25 significantly predicted worse OS in various carcinomas, with the pooled HR of 2.434 (95% CI 1.330-3.539, P=0.000) (Figure 2). Interestingly, subgroup analysis was performed in the ethnicity, it was found that the I2=66.1%, pooled HR of OS was 1.895 (95% CI 1.096-2.693, P=0.007) in China population with cancers, indicating racial maybe the source of heterogeneity (Figure 3). Meanwhile, the sensitive analysis was performed by omitting one study at each time to measure its effect on the pooled HR. As presented in Figure 4, no individual study influenced the overall HR dominantly.
Figure 2.
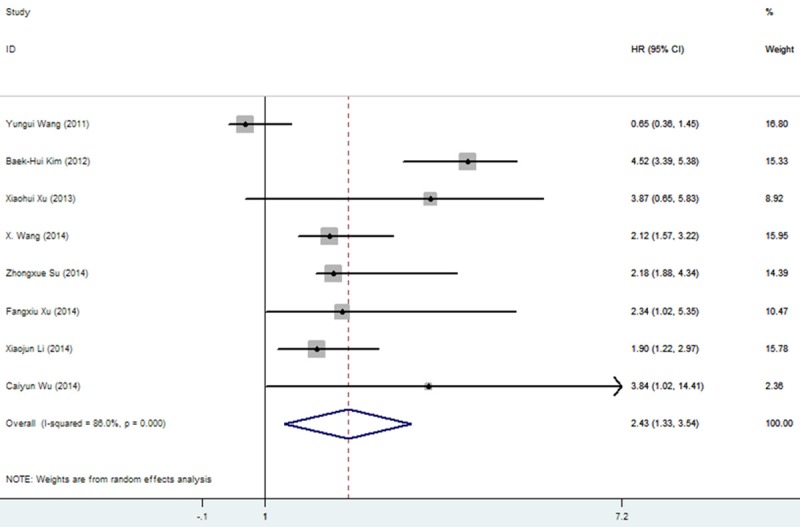
Forest plots of studies evaluating HR of overall survival comparing high- and low miR-25 expression.
Figure 3.
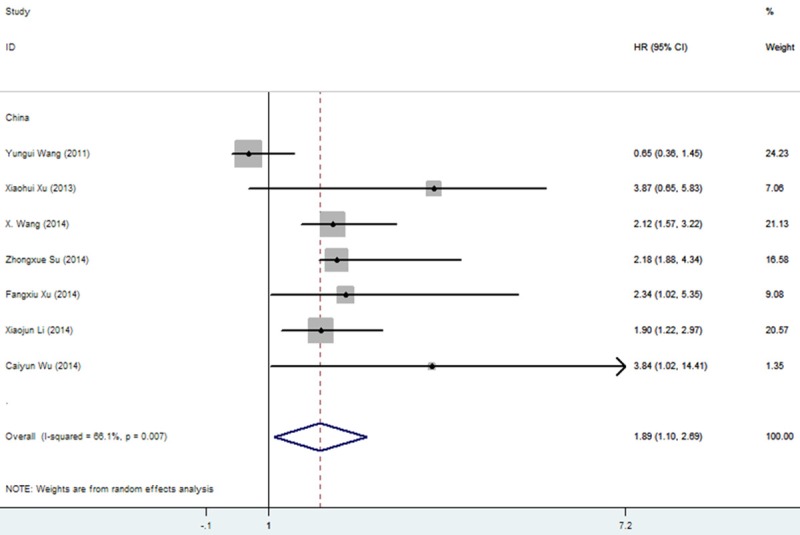
Subgroup analysis evaluating HR of overall survival comparing high- and low-miR-25 expression in Chinese.
Figure 4.
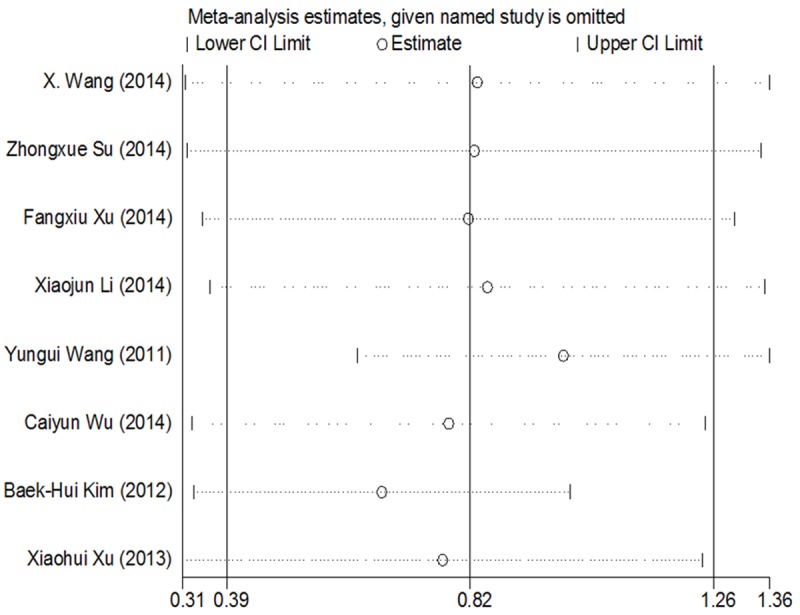
Sensitivity analysis. Removing the document one by one, there no obvious change.
Publication bias
Finally, Begg’s funnel plot and Egger’s test were used to evaluate the publication bias of the included studies. The shape of the funnel plot did not reveal any evidence of obvious asymmetry. The P value of Egger’s regression intercept was 0.242, indicating that there was no significant publication bias in the meta-analysis.
Discussion
miRNAs regulate gene expression at post-transcriptional level and play a pivotal role in the regulation of cell development, metabolism, proliferation, differentiation and apoptosis. Thus, they may serve as potential biomarkers to indicate different disease courses and outcomes [16,17]. Previous studies have shown that the expression of miR-25 was up-regulated significantly in several types of cancers, such as small cell lung cancer, gastric cancer, liver cancer [18-20]. Zhao and his colleagues [18] find that miR-25 was overexpressed in both SCLC cells and human SCLC tumor tissues. Down-regulation of miR-25 significantly reduced cancer cell growth, invasive capability and resistance to cisplatin. Also, it induced G1 cell cycle arrest and downregulated cell cycle related proteins cyclin E2 and CDK2. Li and his colleagues [19] find that miR-25 was overexpressed in plasma and primary tumor tissues of gastric cancer (GC) patients with tumor node metastasis stage (III or IV) or lymph node metastasis. miR-25 inhibition significantly decreased the metastasis, invasion and proliferation of GC cells in vitro, and reduced their capacity to develop distal pulmonary metastases and peritoneal dissemination in vivo. In the study by Li [20], more than 50% of the hepatocellular carcinoma samples showed a greater than two fold increase in the expression of the miR-106b-25 cluster when compared with the corresponding paired (or counterpart) non-tumor samples. Knock-down studies for the miR-106b-25 cluster, which includes miR-106b, miR-93 and miR-25, showed that the expression of the cluster was necessary for cell proliferation and for inhibited independent growth. However, some investigators found that the expression of miR-25 had a reverse expression in other cancers. For example, Zhou [21] found that high expression of miR-25 resulted in inhibit colony formation, metasomatic in both cell lines. It is reported the overexpression of miR-25 led to a decreased ANGPTL2 mRNA and protein expression, whereas there was an adverse efficiency when down regulated miR-25. Although miR-25 was found to be expressed in most human cancers and widely studied in tissue or plasma, indicating it can be used as a potential biomarker for cancer. However the prognostic value of miR-25 in cancers is still a puzzle. Therefore, we conduct this meta-analysis to answer this question, which is the first evaluation we know concerning the relationship between miR-25 expression and the survival in patients with various cancers.
The present meta-analysis showed that miR-25 indeed predict poor survival in carcinomas patients. Furthermore, the subgroup analysis showed that the association was more prominent in Chinese. Because there was some limitations exist in present meta-analysis. Since the number of included studies was relatively small with only about 868 cases and no random clinical trials had been found. Finally, the heterogeneity existed in this meta-analysis and was probably because of the differences in types of cancer, demographic characters of population, disease stages etc. It would be more valuable if data are from different sample origins. However, it is difficult to perform a meta-analysis in subgroups currently due to the limited sample size.
In summary, the present meta-analysis shows that the highly expressed miR-25 is significantly associated with poor survival in patients with various types of carcinomas. However, the role that miR-25 acts in the process of oncogenes is still unclear to us. Our study could aid in the delineation of this issue by demonstrating miR-25 performance in clinic and provide clues for future investigations.
Acknowledgements
This study was supported in part by grants from Central South University Innovation Foundation For Postgraduates (2015zzts114); National Key Scientific & Technology Support Program: Collaborative innovation of Clinical Research for chronic obstructive pulmonary disease and lung cancer (NO. 2013BAI09B09).
Disclosure of conflict of interest
None.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Ferracin M, Veronese A, Negrini M. Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev Mol Diagn. 2010;10:297–308. doi: 10.1586/erm.10.11. [DOI] [PubMed] [Google Scholar]
- 3.Nana-Sinkam P, Croce CM. MicroRNAs in diagnosis and prognosis in cancer: what does the future hold? Pharmacogenomics. 2010;11:667–669. doi: 10.2217/pgs.10.57. [DOI] [PubMed] [Google Scholar]
- 4.Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68:8191–8194. doi: 10.1158/0008-5472.CAN-08-1768. [DOI] [PubMed] [Google Scholar]
- 5.Savita U, Karunagaran D. MicroRNA-106b-25 cluster targets beta-TRCP2, increases the expression of Snail and enhances cell migration and invasion in H1299 (non small cell lung cancer) cells. Biochem Biophys Res Commun. 2013;434:841–847. doi: 10.1016/j.bbrc.2013.04.025. [DOI] [PubMed] [Google Scholar]
- 6.Kim BH, Hong SW, Kim A, Choi SH, Yoon SO. Prognostic implications for high expression of oncogenic microRNAs in advanced gastric carcinoma. J Surg Oncol. 2013;107:505–510. doi: 10.1002/jso.23271. [DOI] [PubMed] [Google Scholar]
- 7.Wu C, Li M, Hu C, Duan H. Clinical significance of serum miR-223, miR-25 and miR-375 in patients with esophageal squamous cell carcinoma. Mol Biol Rep. 2014;41:1257–1266. doi: 10.1007/s11033-013-2970-z. [DOI] [PubMed] [Google Scholar]
- 8.Li X, Yang C, Wang X, Zhang J, Zhang R, Liu R. The expression of miR-25 is increased in colorectal cancer and is associated with patient prognosis. Med Oncol. 2014;31:781. doi: 10.1007/s12032-013-0781-7. [DOI] [PubMed] [Google Scholar]
- 9.Xu FX, Su YL, Zhang H, Kong JY, Yu H, Qian BY. Prognostic implications for high expression of MiR-25 in lung adenocarcinomas of female non-smokers. Asian Pac J Cancer Prev. 2014;15:1197–1203. doi: 10.7314/apjcp.2014.15.3.1197. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Li Z, He C, Wang D, Yuan X, Chen J, Jin J. MicroRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol Dis. 2010;44:191–197. doi: 10.1016/j.bcmd.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Meng X, Li H, Liu W, Shen S, Gao Z. MicroRNA-25 expression level is an independent prognostic factor in epithelial ovarian cancer. Clin Transl Oncol. 2014;16:954–958. doi: 10.1007/s12094-014-1178-6. [DOI] [PubMed] [Google Scholar]
- 12.Su ZX, Zhao J, Rong ZH, Geng WM, Wu YG, Qin CK. Upregulation of microRNA-25 associates with prognosis in hepatocellular carcinoma. Diagn Pathol. 2014;9:47. doi: 10.1186/1746-1596-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu XH, Liu JP, Zhang L. Esophageal squamous cell carcinoma patients with up-regulation of microRNA-25 have poor prognosis. Modern Oncology. 2013;21:1047–1049. [Google Scholar]
- 14.Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, Moher D, Becker BJ, Sipe TA, Thacker SB. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283:2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 15.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17:2815–2834. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 16.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 17.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Z, Liu J, Wang C, Wang Y, Jiang Y, Guo M. MicroRNA-25 regulates small cell lung cancer cell development and cell cycle through cyclin E2. Int J Clin Exp Pathol. 2014;7:7726–7734. [PMC free article] [PubMed] [Google Scholar]
- 19.Li BS, Zuo QF, Zhao YL, Xiao B, Zhuang Y, Mao XH, Wu C, Yang SM, Zeng H, Zou QM, Guo G. MicroRNA-25 promotes gastric cancer migration, invasion and proliferation by directly targeting transducer of ERBB2, 1 and correlates with poor survival. Oncogene. 2015;34:2556–2565. doi: 10.1038/onc.2014.214. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Tan W, Neo TW, Aung MO, Wasser S, Lim SG, Tan TM. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci. 2009;100:1234–1242. doi: 10.1111/j.1349-7006.2009.01164.x. [DOI] [PubMed] [Google Scholar]
- 21.Zhou J, Wang J, Wu S, Zhu S, Wang S, Zhou H, Tian X, Tang N, Nie S. Angiopoietin-like protein 2 negatively regulated by microRNA-25 contributes to the malignant progression of colorectal cancer. Int J Mol Med. 2014;34:1286–1292. doi: 10.3892/ijmm.2014.1909. [DOI] [PubMed] [Google Scholar]


