Abstract
Tectonic family member 1 (TCTN1) encodes a member of the tectonic family which are evolutionarily conserved secreted and transmembrane proteins, involving in a diverse variety of developmental processes. It has been demonstrated that tectonics expressed in regions that participate in Hedgehog (Hh) signaling during mouse embryonic development and was imperative for Hh-mediated patterning of the ventral neural tube. However, the expression and regulation of tectonics in human tumor is still not clear. In this study, shRNA-expressing lentivirus was constructed to knockdown TCTN1 in medulloblastoma cell line Daoy. The results showed that knockdown of TCTN1 inhibited cell proliferation and colony formation in Daoy cell line, also caused cell cycle arrest at the G2/M boundary. Taken all together, our data suggest that TCTN1 might play an important role in the progression of medulloblastoma.
Keywords: Daoy, medulloblastoma, proliferation, RNA interference, tectonic family member 1
Introduction
Medulloblastoma (MB) is one of the most common intracranial malignant tumors in children, which is considered to be of neuroectodermal origin and has a propensity to disseminate throughout the central nervous system. Medulloblastoma is diagnosed in approximately 1 in 200,000 children less than 15 years old each year [1]. Although aggressive chemotherapy after craniospinal radiotherapy and stem-cell rescue have drastically improved survival rates in children with newly diagnosed MB, a considerable segment of patients are currently incurable [2]. In addition, the majority of survivors always suffer from long-term treatment-related side effects, including developmental, neurological, neuroendocrine and psychosocial deficits resulting from radiation therapy and high-dose of chemotherapy [3-5]. Considerable progress has been made in the treatment of MB over the past 90 years; unfortunately, the pathogenesis of MB is not entirely understood.
The current consensus support the existence of four main subgroups of medulloblastoma (Wnt, Shh, Group 3, and Group 4) [6-8]. These four subgroups vary extremely in cytogenetics, mutational spectra and gene expression signature [9,10], apart from disparate clinical phenotypes [11]. Up until very recently, a small fraction of genes are identified as recurrently mutated either in the germline or somatic cell in patients with medulloblastoma, including the tumor suppressor genes PTCH1, SUFU, TP53 and the oncogenes CTNNB1, MYC, and SMO [12]. The wingless (WNT) pathway coordinates a diverse array of developmental processes, including the proliferation and fate of neural progenitor cells. Mutations in proteins on the WNT pathway occur in about 15% of sporadic medulloblastomas [13-16]. The SHH pathway has also been implicated in the development of sporadic and heritable forms of medulloblastoma [17]. There is evidence that aberrant expression of ERBB2 in granule-neuron precursor cells of the cerebellum might have a role in medulloblastoma formation [18]. It has been lately demonstrated miRNA-31 suppresses medulloblastoma tumorigenesis by negatively regulating DNA replication via MCM2 [19]. Generally, mutations in these tumor suppressors as well as aberrant activation of oncogenes and the 17-92 clusters of microRNAs have been linked to the aetiology of medulloblastoma. Recent studies show that epigenetic alterations and aberrant expression of genes controlling epigenetic mechanisms have been identified in medulloblastoma [20-22]. In spite of these striking progress, the confirmed genes only account for a small portion of targets for all genetic damage that result in medulloblastomas [1]. The precise mechanisms underlying the pathogenesis of MB are still ambiguous, and the development of effective therapeutic strategies for MB is undoubtedly urgent. Therefore, identifying the crucial molecules and/or signal transduction pathways that regulate medulloblastoma carcinogenesis is extremely impending.
Tectonic family member 1 (TCTN1) is a member of the tectonic family which are evolutionarily conserved secreted and transmembrane proteins. A large amount of gene families of secreted and transmembrane proteins related by homology play pivotal roles in crucial biological processes including morphogenesis, cellular differentiation, angiogenesis, apoptosis, and modulation of the immune response, as well as disease processes such as non-Hodgkin’s lymphoma and breast carcinoma progression [23]. Recent studies find tectonic 1 is essential for ciliogenesis in some, but not all, tissues. TCTN1 forms a membrane-spanning transition zone complex with multiple ciliopathy proteins associated with Meckel and Joubert syndromes. This tectonic complex controls ciliogenesis and ciliary protein composition and a mutation in TCTN1 gives rise to Joubert syndrome [24]. In addition, it has been reported that during neural tube development, mouse tectonic is required for formation of the most ventral cell types and for full Hedgehog (Hh) pathway activation. It functions downstream of smoothened and rab23 to modulate hedgehog signal transduction [25]. Genomic database searches identify two other mammalian tectonic family members, Tect2 and Tect3, which are 49% and 58% similar to Tect1, respectively [25]. Tectonics participate in a variety of developmental processes, and are shown to regulate SHH signaling. TCTN1-/- and TCTN2-/- mouse exhibit neural tube defect and polydactyly [25,26]. No TCTN3 mouse model is reported as yet, but TCTN3 is indicated to form a complex with multiple ciliary proteins colocalizing at the transition zone, including TCTN1, TCTN2, and all known MKS proteins except RPGRIP1L [24]. Furthermore, studies have shown TCTN3 involves in the regulation of the key SHH signaling pathway and that its disruption brings about a severe form of ciliopathy, combining features of Meckel and Orofaciodigital IV syndromes [27].
In this study, we had investigated the function of TCTN1 in medulloblastoma growth and progression. Our findings manifested that decreased expression of TCTN1 restrained the proliferation of medulloblastoma cell. Moreover, our study indicated that TCTN1 was implicated in the regulation of cell cycle. In conclusion, TCTN1 may play a significant part in medulloblastoma tumorgenesis.
Materials and methods
Cell culture medium MEME and DMEM were purchased from Sigma (St. Louis, MO) and Hyclone, respectively. Fetal bovine serum was from Biowest (France). Lipofectamine 2000 was from Invitrogen. For Western blotting, we used primary antibodies to: TCTN1 from Sigma (St. Louis, MO); GAPDH from Proteintech Group, Inc. HRP-linked secondary antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell culture
Human medulloblastoma cell line Daoy and human embryonic kidney cell line 293T (HEK293T) were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). Daoy cells were cultured in M5650 medium (Sigma, USA) with 1% L-glutamine. HEK293T cells were cultured with Dulbecco’s modified Eagle’s medium (DMEM, Hyclone, USA). All the culture fluid was supplemented with 10% fetal bovine serum (FBS) and all cells were cultured with 5% CO2 at 37°C in a humidified chamber.
Construction of recombinant lentivirus and gene silencing
shRNA for TCTN1 (5’-GCTCAGATGCATCAGTTCCTTCTCGAGAAGGAACTGATGCATCTGAGCTTTTTT-3’, NM_001082537.2) was inserted into the lentiviral expression vector pFH-L (Shanghai Hollybio, China) which contains a green fluorescent protein (GFP) gene as a reporter gene. Non-silencing shRNA (5’-GCGGAGGGTTTGAAAGAATATCTCGAGATATTCTTTCAAACCCTCCGTTTTTT-3’) was used as control. We generated shRNAs corresponding to shRNA sequences for TCTN1 and non-silencing. Recombinant lentivirus was generated by triple transfection of 80% confluent HEK293T cells with modified pFH-L vector and packing plasmids pVSVG-I and pCMVΔR8.92 (Shanghai Hollybio, China) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendation. For lentivirus infection, Daoy cells (5×104 cells per well) were seeded into 6-well plates and transfected with TCTN1 shRNA (Lv-shTCTN1) or control shRNA (shCon) expressing lentivirus at a multiplicity of infection (MOI) of 25. Lentiviruses were harvested in serum-free medium after 4 days, filtered, and concentrated in primed Centricon Plus-20 filter devices (Millipore).
RNA extraction and real-time quantitative PCR
Total RNA was prepared from cultured Daoy cells after 4 days infection using Trizol (Gibco, USA). 5 μg of total RNA was reverse transcribed to cDNA with the use of SuperScript II RT 200 U/µl (Invitrogen). The SYBR green-based real time reverse transcription-polymerase chain reaction (RT-PCR) assay was used to determine the mRNA level of TCTN1 in infected Daoy cells (reaction system: 2× SYBR premix ex taq 10 µL, forward and reverse primers (2.5 µM) 0.8 µL, cDNA 5 µL, ddH2O 4.2 µL). After initial denaturation at 95°C for 1 min, a total of 40 cycles (denaturation 95°C, 5 s; annealing extension of 60°C, 20 s) was carried out. Receiver operating characteristic (ROC) analysis was used to select the optimal cut-off point for statistical analysis. ACTIN was applied as the input reference. The PCR primers were as following: TCTN1: CCTTTGCGTGAATGTTGTTC as forward and AGAGGGACTGGCTGGGTATT as reverse; ACTIN: GTGGACATCCGCAAAGAC as forward and AAAGGGTGTAACGCAACTA as reverse. Results are presented as CT values, defined as the threshold PCR cycle number at which an amplified product is first detected. The average CT was calculated for both TCTN1 and ACTIN, and ΔCT was determined as the mean of the triplicate CT values for CTCN1 minus the mean of the triplicate CT values for ACTIN.
Western blot analysis
Dayo cells were harvested and lysed with 2X SDS sample buffer (100 mM Tris-HCl (pH 6.8), 10 mM EDTA, 4% SDS, 10% Glycine) on ice for 30 min. The lysates were clarified by centrifugation at 13,000 g for 15min at 4°C. Proteins were quantified using the BCA protein assay kit. After protein quantification and normalization, equivalent amounts of proteins (60 μg) were electrophoresed on 10% SDS-PAGE gels and transferred to PVDF membranes (Millipore, Bedford, MA). After blocking with 5% nonfat milk in TBST for 1 h at room temperature, membranes were immunoblotted with primary antibodies at 4°C overnight, followed by HRP-linked secondary antibodies. The signals were detected using the ECL system (Millipore, Billerica, WI, USA). Protein levels were normalized to GAPDH. The following antibodies were used: TCTN1 (1:2000 dilution; sigma SAB3500518), GAPDH (1:40000 dilution; Proteintech Group, Inc. 10494-1-AP), anti-rabbit HRP (1:5000 dilution; Santa Cruz SC-2054).
Cell survival analysis
Daoy cells from different groups were seeded at a density of 2500 cells/well in 96-well plates. At predetermined time, MTT was added into each well and then acidic isopropanol (10% SDS, 5% isopropanol and 0.01 mol/L HCl) was added to stop the reaction. Absorbance was measured with an ELISA reader at a wavelength of 595 nm. Viability of cells was expressed relative to theoretical absorbance.
Colony formation assay
To assay monolayer colony formation, Daoy cells were plated at 500 cells per well in a 6-well plate, allowed to adhere overnight. After 9 days of incubation, the cells were stained with 0.2% crystal violet after methanol fixation, and the numbers of colonies containing more than 50 cells were counted.
Cell cycle analysis
After 4 days of infection, Daoy cells were seeded on 6 cm-diameter plates at density of 2×105 cells per dish and then cultured for 40 h. Cells were covered at a cell confluence of 80%. After washing with ice-cold PBS, cells were suspended in about 0.5 ml of 70% alcohol and kept at 4°C for 30 min. The suspension was filtered through a 50-mm nylon mesh, and the DNA content of stained nuclei with PI was analyzed by a flow cytometer. Each experiment was performed in triplicate.
Statistical analysis
Each experiment was repeated independently three times. The results were given as a mean ± standard deviation (SD) and compared using the unpaired t-test or ANOVA followed by the Bonferroni post-test using Graph Pad InStat v3.02 software. Results with P<0.05 were considered statistically significant.
Results
Inhibition of TCTN1 expression mediated by lentivirus
Daoy, a main representative cell lines, is widely used in studies of medulloblastoma [28]. To investigate the role of TCTN1 in the progression of medulloblastoma, we constructed a recombinant lentivirus to silence the expression of the target gene in Daoy cell lines. The rate of lentivirus infection was highly effective as visualized by GFP expression 4 days after infection (Figure 1A). TCTN1 mRNA expression was then measured with real-time PCR. As shown in Figure 1B, the expression of endogenous TCTN1 mRNA was remarkably reduced in Lv-shTCTN1 group. Further confirmation was demonstrated by Western blot analysis, verifying that TCTN1 protein reduced notably following TCTN1 knockdown treatment (Figure 1C). The results demonstrated that expression of TCTN1 in Daoy cell was evidently inhibited by recombinant lentivirus mediated RNA interference.
Figure 1.
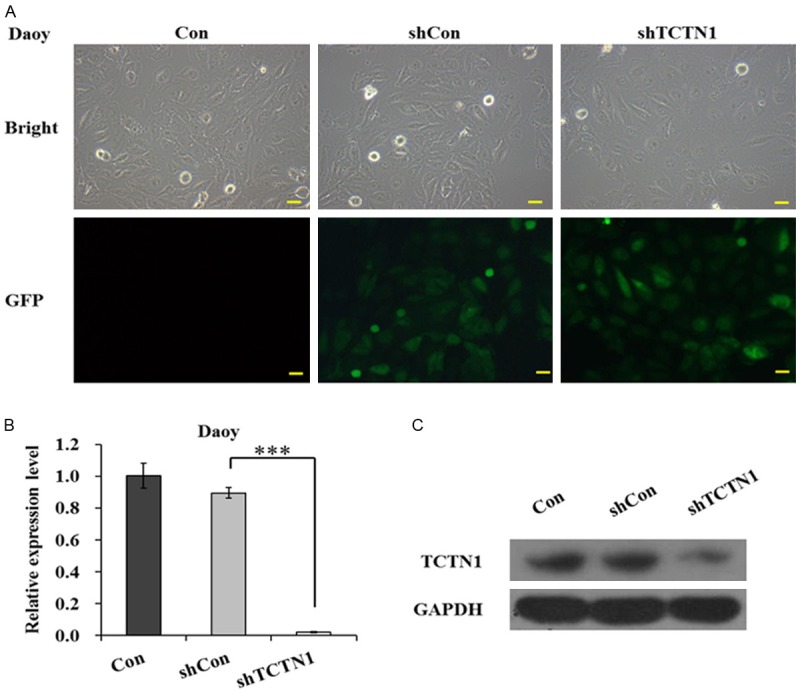
Depletion of TCTN1 expression mediated by lentivirus. (A) Representative images of Daoy cells infected with indicated lentivirus. Efficiency of lentivirus infection was evidenced by reporter gene GFP expression. Scale bar represented 100 μm. (B) Total RNA was extracted and TCTN1 mRNA level was measured with real-time PCR in different groups of Daoy cells. Histogram represented the average value of three independent experiments (***P<0.001), and (C) protein levels were detected by Western blot analysis, GAPDH was used as loading control.
Suppression of TCTN1 gene inhibits proliferation of Daoy cells
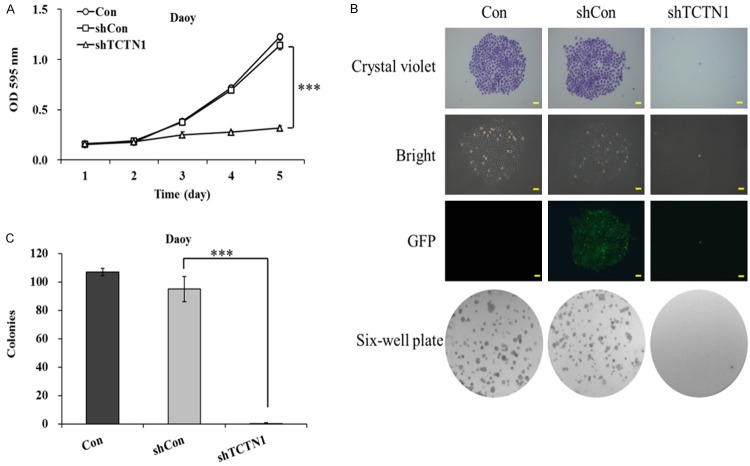
To illuminate whether TCTN1 play a crucial part in medulloblastoma proliferation and tumorigenesis, the growth of lentivirus infected Daoy cells was first examined by MTT assay. As shown in Figure 2A, the growth curves for TCTN1 knockdown cells were dramatically lower than those for control cells (P<0.001), indicating that TCTN1 was potentially involved in Daoy cell proliferation. In addition, images recorded under a fluorescence microscope after crystal violet staining and photographs of plates vividly declared the suppression of colony formation in Lv-shTCTN1 group (Figure 2B). Furthermore, monolayer colony formation assay revealed the number of surviving colonies in TCTN1 knockdown group were significantly decreased compared to those in negative control group (95 ± 9 vs 0 ± 1, P<0.01) (Figure 2C). In conclusion, our data strongly suggest that TCTN1 is prerequisite for Daoy cell growth and proliferation.
Figure 2.
Knockdown of TCTN1 inhibits Daoy cells proliferation and colony formation. A. Different groups of Daoy cells were treated for 4 days and proliferation was determined by MTT assay. Statistical plots represent three independent cell proliferation assays (***P<0.0001). B. Photographs of plates and representative colonies stained with crystal violet were shown. Scale bar represented 250 μm. C. Suppression of TCTN1 inhibited colony forming ability. Histogram was the average number of colonies in each plate (***P<0.0001).
TCTN1 depletion impaired cell cycle distribution of Daoy cells
To further explore the mechanism by which TCTN1 downregulation suppressed Daoy cell proliferation, the cell cycle progression during Daoy cell proliferation was subsequently detected by flow cytometry (Figure 3A). As illustrated in Figure 3B, a significantly reduced ratio of Daoy cells treatment with Lv-shTCTN1 was observed in the S phase in comparison with those in Con or shCon group (P<0.001). More intriguingly, we found that percentage of cells from Lv-shTCTN1 group in G2/M phase dramatically augmented, indicating that cell cycle was blocked in G2/M phase when TCTN1 was silenced in Daoy cells (P<0.01). Additionally, we examined the percentage of sub-G1 phase cells which signified the occurrence of apoptosis and found that downregulation of TCTN1 in Daoy cells caused a remarkable increase in the number of cells in sub-G1 phase. Taken all together, our data implies that TCTN1 may regulate Daoy cell proliferation by controlling cell cycle progression and inducing apoptosis.
Figure 3.
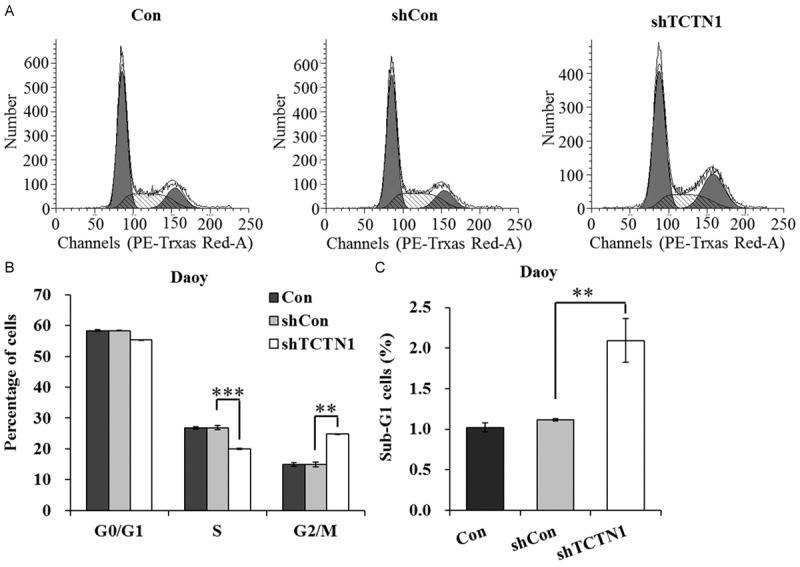
Downregulation of TCTN1 impaired cell cycle progression and induced apoptosis in Daoy cells. A. Different groups of Daoy cells were harvested after treatment and subjected to fluorescence activated cell sorter (FACS) analysis. B. Statistical representation of data of three independent experiments from flow cytometry analysis. The percentages of G0/G1, S, G2/M phase cells were indicated. C. The percentage of Daoy cells in sub-G1 phase was significantly higher in Lv-TCTN1 group than that in control groups (**P<0.01, ***P<0.001).
Discussion
Tectonic is a novel gene which is identified through a screen for genes encoding secreted and transmembrane proteins [25]. Secreted and transmembrane proteins play a fundamental role in intercellular communication during the development of multicellular organisms. A large number of genes encoding these proteins involved in cell-cell contact and signaling in the development of organisms [29]. A mountain of work has been done in identifying these genes and elucidating their possible function [29,30]. Up to now, tectonic family members contain Tect1, Tect2 and Tect3. TCTN2 absence would cause neural tube defect and polydactyly and TCTN3 is involved in the regulation of the key SHH signaling pathway in vivo. Researches on TCTN1 are rare. TCTN1 is involved in the development of neural tube and it affects downstream of smoothened and rab23 to regulate hedgehog signal transduction. However, its function in human tumorigenesis has not been reported.
In the present study, we found that TCTN1 was implicated in medulloblastoma progression and it was essential for medulloblastoma cells proliferation. We demonstrated for the first time that knockdown of endogenous TCTN1 expression could inhibit growth and proliferation of Daoy cells by inducing cell cycle arrest and apoptosis. A series of evidence have demonstrated that constitutively activated STAT3 serves as a crucial role in medulloblastoma tumorigenesis by controlling the expression of target genes, which can protect apoptosis and enhance cell proliferation [31]. Accordingly, whether TCTN1 involves in STAT3 signaling network in medulloblastoma tumorigenesis is a question worth exploring. In addition, Studies have shown the IGF-1R/PI3K pathway impairs cell proliferation and survival in subsets of medulloblastoma [32]. The receptor tyrosine kinase (RTK)/phosphoinositide 3-kinase (PI3K) pathway is fundamental for cancer cell proliferation and is known to be frequently altered and activated in neoplasia. Therefore, it is intriguing to think that there is a link between the activated RTK/PI3K/mTOR signaling and the mechanism underlying TCTN1 induction of medulloblastoma cells proliferation. The apoptosis is a normal and programmed cell death and is a clearing means of abnormal cells or senescent cells. Inducing the apoptosis of tumor cells is one of the most important strategy to discover anti-tumor drugs based on apoptosis mechanism [33]. It has been reported that BMP-2 mediates retinoid-induced apoptosis in medulloblastoma cells through a paracrine effect [34]. TCTN1 can inhibit apoptosis and promote proliferation of medulloblastoma cells obviously, and it may be a potential anticancer molecular target. Furthermore, the SHH pathway which could be regulated by TCTN3 has also been implicated in the development of medulloblastoma, suggesting that TCTN1 might modulate medulloblastoma cell growth through SHH signaling pathway.
In conclusion, we demonstrated that knockdown of TCTN1 could inhibit the proliferation of Daoy cells by inducing cell cycle arrest and apoptosis. Since TCTN1 is a secreted protein, developing a neutralizing antibody that suppresses its protease activity should be highly feasible. There is a great possibility that TCTN1 is involved in other signaling pathways to regulate medulloblastoma cells progression in vitro or vivo. Thus, insight into the anticancer role of TCTN1 may open a new avenue for the development of medulloblastoma therapies.
Acknowledgements
We would like to thank all the people who made contributions to this study.
Disclosure of conflict of interest
None.
References
- 1.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, Gallia GL, Jallo GI, Binder ZA, Nikolsky Y, Hartigan J, Smith DR, Gerhard DS, Fults DW, VandenBerg S, Berger MS, Marie SK, Shinjo SM, Clara C, Phillips PC, Minturn JE, Biegel JA, Judkins AR, Resnick AC, Storm PB, Curran T, He Y, Rasheed BA, Friedman HS, Keir ST, McLendon R, Northcott PA, Taylor MD, Burger PC, Riggins GJ, Karchin R, Parmigiani G, Bigner DD, Yan H, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Packer RJ. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma. Curr Neurol Neurosci Rep. 2007;7:130, 132. [PubMed] [Google Scholar]
- 3.Mabbott DJ, Penkman L, Witol A, Strother D, Bouffet E. Core neurocognitive functions in children treated for posterior fossa tumors. Neuropsychology. 2008;22:159–168. doi: 10.1037/0894-4105.22.2.159. [DOI] [PubMed] [Google Scholar]
- 4.Spiegler BJ, Bouffet E, Greenberg ML, Rutka JT, Mabbott DJ. Change in neurocognitive functioning after treatment with cranial radiation in childhood. J. Clin. Oncol. 2004;22:706–713. doi: 10.1200/JCO.2004.05.186. [DOI] [PubMed] [Google Scholar]
- 5.Mabbott DJ, Spiegler BJ, Greenberg ML, Rutka JT, Hyder DJ, Bouffet E. Serial evaluation of academic and behavioral outcome after treatment with cranial radiation in childhood. J. Clin. Oncol. 2005;23:2256–2263. doi: 10.1200/JCO.2005.01.158. [DOI] [PubMed] [Google Scholar]
- 6.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S, Gajjar A, Ellison DW, Lichter P, Gilbertson RJ, Pomeroy SL, Kool M, Pfister SM. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, Chintagumpala M, Adesina A, Ashley DM, Kellie SJ, Taylor MD, Curran T, Gajjar A, Gilbertson RJ. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J. Clin. Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 8.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins CE, French P, Rutka JT, Pfister S, Taylor MD. Medulloblastoma comprises four distinct molecular variants. J. Clin. Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Northcott PA, Dubuc AM, Pfister S, Taylor MD. Molecular subgroups of medulloblastoma. Expert Rev Neurother. 2012;12:871–884. doi: 10.1586/ern.12.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA, Cho YJ, Koster J, Schouten-van Meeteren A, van Vuurden D, Clifford SC, Pietsch T, von Bueren AO, Rutkowski S, McCabe M, Collins VP, Backlund ML, Haberler C, Bourdeaut F, Delattre O, Doz F, Ellison DW, Gilbertson RJ, Pomeroy SL, Taylor MD, Lichter P, Pfister SM. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123:473–484. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Northcott PA, Korshunov A, Pfister SM, Taylor MD. The clinical implications of medulloblastoma subgroups. Nat Rev Neurol. 2012;8:340–351. doi: 10.1038/nrneurol.2012.78. [DOI] [PubMed] [Google Scholar]
- 12.Northcott PA, Rutka JT, Taylor MD. Genomics of medulloblastoma: from Giemsa-banding to next-generation sequencing in 20 years. Neurosurg Focus. 2010;28:E6. doi: 10.3171/2009.10.FOCUS09218. [DOI] [PubMed] [Google Scholar]
- 13.Koch A, Hrychyk A, Hartmann W, Waha A, Mikeska T, Schuller U, Sorensen N, Berthold F, Goodyer CG, Wiestler OD, Birchmeier W, Behrens J, Pietsch T. Mutations of the Wnt antagonist AXIN2 (Conductin) result in TCF-dependent transcription in medulloblastomas. Int J Cancer. 2007;121:284–291. doi: 10.1002/ijc.22675. [DOI] [PubMed] [Google Scholar]
- 14.Yokota N, Nishizawa S, Ohta S, Date H, Sugimura H, Namba H, Maekawa M. Role of Wnt pathway in medulloblastoma oncogenesis. Int J Cancer. 2002;101:198–201. doi: 10.1002/ijc.10559. [DOI] [PubMed] [Google Scholar]
- 15.Zurawel RH, Chiappa SA, Allen C, Raffel C. Sporadic medulloblastomas contain oncogenic beta-catenin mutations. Cancer Res. 1998;58:896–899. [PubMed] [Google Scholar]
- 16.Eberhart CG, Tihan T, Burger PC. Nuclear Localization and Mutation of -Catenin in Medulloblastomas. J Neuropathol Exp Neurol. 2000;59:333–337. doi: 10.1093/jnen/59.4.333. [DOI] [PubMed] [Google Scholar]
- 17.Ruiz i Altaba A, Sánchez P, Dahmane N. Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat Rev Cancer. 2002;2:361–372. doi: 10.1038/nrc796. [DOI] [PubMed] [Google Scholar]
- 18.Gilbertson RJ. Medulloblastoma: signalling a change in treatment. Lancet Oncol. 2004;5:209–218. doi: 10.1016/S1470-2045(04)01424-X. [DOI] [PubMed] [Google Scholar]
- 19.Jin Y, Xiong A, Zhang Z, Li S, Huang H, Yu TT, Cao X, Cheng SY. MicroRNA-31 suppresses medulloblastoma cell growth by inhibiting DNA replication through minichromosome maintenance 2. Oncotarget. 2014;5:4821–4833. doi: 10.18632/oncotarget.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawlor ER, Thiele CJ. Epigenetic changes in pediatric solid tumors: promising new targets. Clin Cancer Res. 2012;18:2768–2779. doi: 10.1158/1078-0432.CCR-11-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hovestadt V, Jones DT, Picelli S, Wang W, Kool M, Northcott PA, Sultan M, Stachurski K, Ryzhova M, Warnatz HJ, Ralser M, Brun S, Bunt J, Jager N, Kleinheinz K, Erkek S, Weber UD, Bartholomae CC, von Kalle C, Lawerenz C, Eils J, Koster J, Versteeg R, Milde T, Witt O, Schmidt S, Wolf S, Pietsch T, Rutkowski S, Scheurlen W, Taylor MD, Brors B, Felsberg J, Reifenberger G, Borkhardt A, Lehrach H, Wechsler-Reya RJ, Eils R, Yaspo ML, Landgraf P, Korshunov A, Zapatka M, Radlwimmer B, Pfister SM, Lichter P. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature. 2014;510:537–541. doi: 10.1038/nature13268. [DOI] [PubMed] [Google Scholar]
- 22.Marino AM, Frijhoff J, Calero R, Baryawno N, Ostman A, Johnsen JI. Effects of epigenetic modificators in combination with small molecule inhibitors of receptor tyrosine kinases on medulloblastoma growth. Biochem Biophys Res Commun. 2014;450:1600–1605. doi: 10.1016/j.bbrc.2014.07.042. [DOI] [PubMed] [Google Scholar]
- 23.Clark HF, Gurney AL, Abaya E, Baker K, Baldwin D, Brush J, Chen J, Chow B, Chui C, Crowley C, Currell B, Deuel B, Dowd P, Eaton D, Foster J, Grimaldi C, Gu Q, Hass PE, Heldens S, Huang A, Kim HS, Klimowski L, Jin Y, Johnson S, Lee J, Lewis L, Liao D, Mark M, Robbie E, Sanchez C, Schoenfeld J, Seshagiri S, Simmons L, Singh J, Smith V, Stinson J, Vagts A, Vandlen R, Watanabe C, Wieand D, Woods K, Xie MH, Yansura D, Yi S, Yu G, Yuan J, Zhang M, Zhang Z, Goddard A, Wood WI, Godowski P, Gray A. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genome Res. 2003;13:2265–2270. doi: 10.1101/gr.1293003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Gonzalo FR, Corbit KC, Sirerol-Piquer MS, Ramaswami G, Otto EA, Noriega TR, Seol AD, Robinson JF, Bennett CL, Josifova DJ, Garcia-Verdugo JM, Katsanis N, Hildebrandt F, Reiter JF. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet. 2011;43:776–784. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reiter JF, Skarnes WC. Tectonic, a novel regulator of the Hedgehog pathway required for both activation and inhibition. Genes Dev. 2006;20:22–27. doi: 10.1101/gad.1363606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sang L, Miller JJ, Corbit KC, Giles RH, Brauer MJ, Otto EA, Baye LM, Wen X, Scales SJ, Kwong M, Huntzicker EG, Sfakianos MK, Sandoval W, Bazan JF, Kulkarni P, Garcia-Gonzalo FR, Seol AD, O’Toole JF, Held S, Reutter HM, Lane WS, Rafiq MA, Noor A, Ansar M, Devi AR, Sheffield VC, Slusarski DC, Vincent JB, Doherty DA, Hildebrandt F, Reiter JF, Jackson PK. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145:513–528. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas S, Legendre M, Saunier S, Bessieres B, Alby C, Bonniere M, Toutain A, Loeuillet L, Szymanska K, Jossic F, Gaillard D, Yacoubi MT, Mougou-Zerelli S, David A, Barthez MA, Ville Y, Bole-Feysot C, Nitschke P, Lyonnet S, Munnich A, Johnson CA, Encha-Razavi F, Cormier-Daire V, Thauvin-Robinet C, Vekemans M, Attie-Bitach T. TCTN3 mutations cause Mohr-Majewski syndrome. Am J Hum Genet. 2012;91:372–378. doi: 10.1016/j.ajhg.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peyrl A, Krapfenbauer K, Slavc I, Yang JW, Strobel T, Lubec G. Protein profiles of medulloblastoma cell lines DAOY and D283: identification of tumor-related proteins and principles. Proteomics. 2003;3:1781–1800. doi: 10.1002/pmic.200300460. [DOI] [PubMed] [Google Scholar]
- 29.Kopczynski CC, Noordermeer JN, Serano TL, Chen WY, Pendleton JD, Lewis S, Goodman CS, Rubin GM. A high throughput screen to identify secreted and transmembrane proteins involved in Drosophila embryogenesis. Proc Natl Acad Sci U S A. 1998;95:9973–9978. doi: 10.1073/pnas.95.17.9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang T, Li L, Tang J, Li Y, Lin WY, Martin F, Grant D, Solloway M, Parker L, Ye W, Forrest W, Ghilardi N, Oravecz T, Platt KA, Rice DS, Hansen GM, Abuin A, Eberhart DE, Godowski P, Holt KH, Peterson A, Zambrowicz BP, de Sauvage FJ. A mouse knockout library for secreted and transmembrane proteins. Nat Biotechnol. 2010;28:749–755. doi: 10.1038/nbt.1644. [DOI] [PubMed] [Google Scholar]
- 31.Kao CL, Huang PI, Tsai PH, Tsai ML, Lo JF, Lee YY, Chen YJ, Chen YW, Chiou SH. Resveratrol-induced apoptosis and increased radiosensitivity in CD133-positive cells derived from atypical teratoid/rhabdoid tumor. Int J Radiat Oncol Biol Phys. 2009;74:219–228. doi: 10.1016/j.ijrobp.2008.12.035. [DOI] [PubMed] [Google Scholar]
- 32.Wojtalla A, Salm F, Christiansen DG, Cremona T, Cwiek P, Shalaby T, Gross N, Grotzer MA, Arcaro A. Novel Agents Targeting the IGF-1R/PI3K Pathway Impair Cell Proliferation and Survival in Subsets of Medulloblastoma and Neuroblastoma. PLoS One. 2012;7:e47109. doi: 10.1371/journal.pone.0047109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou K, Ji H, Mao T, Bai Z. Effects of matrine on the proliferation and apoptosis of human medulloblastoma cell line D341. Int J Clin Exp Med. 2014;7:911–918. [PMC free article] [PubMed] [Google Scholar]
- 34.Hallahan AR, Pritchard JI, Chandraratna RA, Ellenbogen RG, Geyer JR, Overland RP, Strand AD, Tapscott SJ, Olson JM. BMP-2 mediates retinoid-induced apoptosis in medulloblastoma cells through a paracrine effect. Nat Med. 2003;9:1033–1038. doi: 10.1038/nm904. [DOI] [PubMed] [Google Scholar]