Abstract
Background: Circulating microRNAs are potential markers for disease detection. A novel class of small non-coding RNAs called Piwi-interacting RNAs (piRNAs) has been recently reported to participate in the epigenetic regulation of cancers and other diseases. This study aims to discover blood-based piRNAs which can be used as markers for disease detection and monitoring. Materials and methods: We selected five piRNAs for detection, namely, has-piR-651, has-piR-823, has-piR-36707, has-piR-36741 and has-piR-57125. Serum or plasma samples were used to isolate small RNAs, including the piRNAs. The extracted small RNAs were reverse-transcribed in the presence of a poly-A polymerase with an oligo-dT adaptor, and quantitative real-time PCR (qRT-PCR) was applied to measure the levels of piRNAs. Room-temperature incubation and repetitive freeze–thaw cycles were performed to measure the stability of the piRNAs. Results: Unlike the four other piRNAs, has-piR-57125 was present in both the serum and plasma samples. Regardless of the serum or plasma samples, qRT-PCR analysis indicated that the Ct values showed no remarkable variation with prolonged incubation time (P > 0.05). We also detected the Ct values of the samples with repetitive freeze-thaw cycles and observed a similar trend (P > 0.05) among the samples with diverse freeze-thaw cycles. Conclusion: This study is the first to report that piRNAs are stably expressed in human serum or plasma samples. Therefore, piRNAs can serve as valuable blood-based biomarkers for disease detection and monitoring.
Keywords: piRNA, serum, plasma, circulating, blood, biomarker
Introduction
Detecting diseases before they spread and become incurable can significantly reduce the health burden of a wide spectrum of diseases worldwide [1]. Several early detection strategies (e.g. proteomics [2], genomics [3] and metabonomics) have shown promise, but clinically valid disease markers remain to be discovered.
Non-coding RNAs (ncRNAs) play important roles in epigenetics by contributing to the genesis and progression of human disorders [4]. The most widely studied class of ncRNAs are microRNAs (miRNAs), which are involved in regulating several cellular processes [5]. Other ncRNAs, such as PIWI-interacting RNAs (piRNAs), small nucleolar RNAs, transcribed ultra-conserved regions and large intergenic non-coding RNAs, are also emerging as key elements of cellular homeostasis [5].
The piRNAs are small non-coding RNAs which are 26-32 nucleotides long and are produced via a Dicer-independent mechanism [6]. These non-coding small RNAs can bind to PIWI proteins to form complexes and induce a silencing effect via DNA methylation, which can maintain the genomic integrity of germline stem cells [6,7]. Recent studies have implicated piRNA in tumour formation, migration and invasion [8,9]. Various expression profiles of cancer type-specific piRNAs have emerged in the past several years; thus, piRNAs are potential markers for the diagnosis of cancer and other diseases [10].
Recent studies have proposed that circulating miRNAs are potential disease biomarkers that may be used for risk assessment, diagnosis, prognosis and monitoring of treatment response. piRNAs are short small RNAs similar to miRNAs; different from long RNAs, piRNAs are not easily degraded and can easily pass through the cell membrane [10]. These characteristics imply that piRNAs can be detected in easily obtained samples, such as serum, plasma, saliva and urine. Therefore, this study aims to discover circulating piRNAs which can serve as markers for the early detection of human diseases.
Materials and methods
Ethics statement
This study was approved by the Institutional Review Board of the First Affiliated Hospital of Nanjing Medical University, Nanjing, China. Written informed consent was obtained from all participants involved in this study.
Sample collection and RNA isolation
Human plasma and serum samples from three healthy donors were obtained with informed consent in accordance with institutional review board-approved protocols at the First Affiliated Hospital of Nanjing Medical University, China. The serum samples were collected in tubes containing a separating gel and clot activator. The plasma samples were collected in EDTA-anticoagulant tubes before they were immediately centrifuged at 2000 rpm for 15 min at 4°C. The total RNA of three healthy donors was isolated from 200 μL of the serum or plasma samples by using an miRNeasy Serum/Plasma Kit (Qiagen) in accordance with the manufacturer’s protocol.
Molecular cloning of piRNA and qRT-PCR
In brief, 1 μL of extracted RNA was reverse-transcribed to cDNA in the presence of poly-A polymerase with an oligo-dT adaptor (Takara Bio, Dalian, China). PCR amplification was performed using Taq PCR Mastermix (TianGen Biotech, Beijing, China) with a forward primer for the piRNA sequence and a universal-adaptor reverse primer. The reaction was performed under the following conditions: 95°C for 3 min; 40 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 40 s; and a final extension at 72°C for 5 min. The sequences of the upstream PCR primers for the piRNAs were as follows: 5’-AGAGAGGGGCCCGTGCCTTG-3’ (piR-651); 5’-AGCGTTGGTGGTATAGTGGT-3’ (piRNA-823); 5’-GTTAAGATGGCAGAGCCCGG-3’ (piR-36707); 5’-GTTTAGACGGGCTCACATCAC-3’ (piR-36741); and 5’-TGGTCGTGGTTGTAGTCCGTG-3’ (piR-57125). The amplified PCR products were analysed on ethidium bromide-stained 3% agarose gels. The universal TA cloning strategy was used to identify the sequence of the PCR products. The SYBR green qRT-PCR assay (Takara Bio, Dalian, China) was conducted to quantify piRNA with an Applied Biosystems Step One Plus System.
Statistical analysis
All analyses were conducted with SPSS 13.0 (SPSS Inc., Chicago, IL, USA). The stability of piRNAs in peripheral blood was analysed using one-way ANOVA. Two-sided P values less than 0.05 were considered to indicate statistical significance.
Results
Detection of piRNAs in the peripheral blood
The levels of piR-651, piR-823, piR-36707, piR-36741 and piR-57125 in the serum and plasma from individual 001 were amplified. In accordance with the principle of reverse transcription, the poly-A tail was added to the small RNA template, and cDNA was synthesised using a poly-T primer. Thus, the expected length of the PCR product was approximately 90 bp. Agarose gel electrophoresis showed the length of the PCR products, and only three piRNAs (piR-651, piR-36707 and piR-57125) had a product length of approximately 90 bp (Figure 1A). Universal TA cloning and sequencing showed that the product sequence of piR-57125 was in accordance with that in the PubMed database (http://www.ncbi.nlm.nih.gov/nuccore/108081267; Figure 1B). Furthermore, qRT-PCR suggested that piR-57125 was efficiently amplified (Figure 1C). Melting curve analysis showed that only one peak appeared at the highest T m. This result indicates that no primer-dimers or nonspecific products were formed (Figure 1D).
Figure 1.
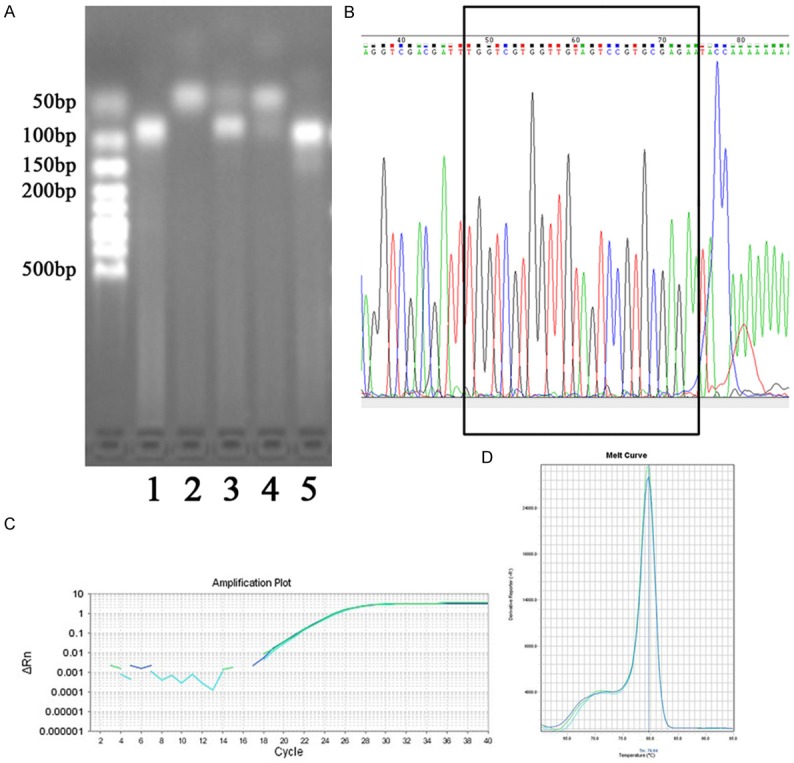
Detection of circulating piRNAs from individual 001. A. Agarose electrophoresis of PCR products. Lane 1: piR-651; Lane 2: piR-823; Lane 3: piR-36707; Lane 4: piR-36741; Lane 5: piR-57125. B. TA cloning sequencing showed the successful amplification of piR-57125. C. Representative amplification plots of piR-57125. D. Dissociation curve analyses of piR-57125.
Stability of piRNAs in peripheral blood
To test the stability of piRNAs in the serum and plasma samples, we measured piR-57125 level in the serum and plasma samples collected from individuals 002 and 003. The Ct value was used to evaluate the level of piR-57125 in a 200 µL sample. As shown in Figure 2A, no significant difference (P > 0.05) in Ct values was observed among the samples incubated with diverse times. The Ct values also showed no obvious change with increasing freeze-thaw cycles (P > 0.05; Figure 2B).
Figure 2.
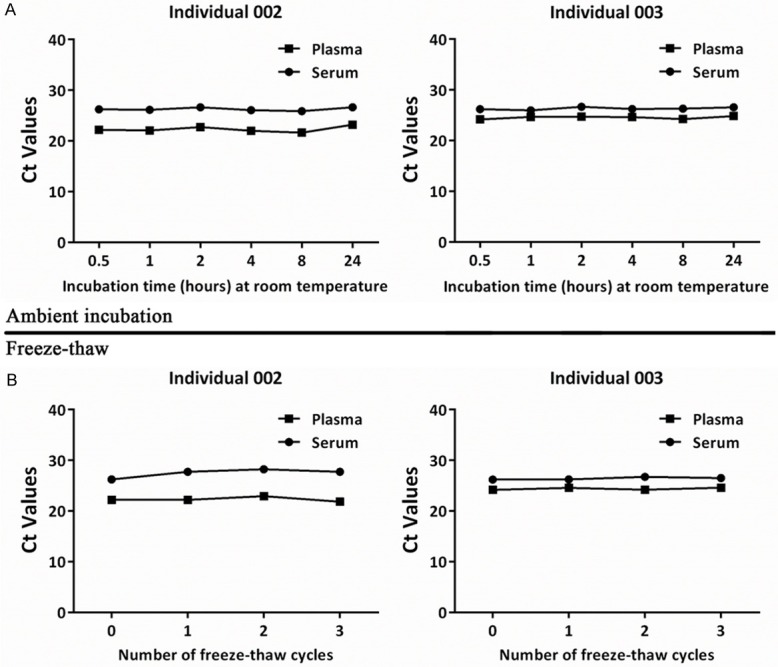
Characterisation of piRNA stability in human blood samples. piRNA levels remained stable when serum and plasma samples were subjected to prolonged room-temperature incubation (A) or freeze–thawed for multiple times (B). No significant differences were found among the samples (P > 0.05).
Comparison of piRNA levels between plasma and serum
We further determined whether or not piRNA levels are substantially different between the serum and plasma samples. Results showed that the piR-57125 levels detected from the plasma or serum samples strongly correlated (R 2 = 0.734, P < 0.001). These results indicate that both serum and plasma samples are suitable for investigating piRNAs as blood-based biomarkers (Figure 3).
Figure 3.
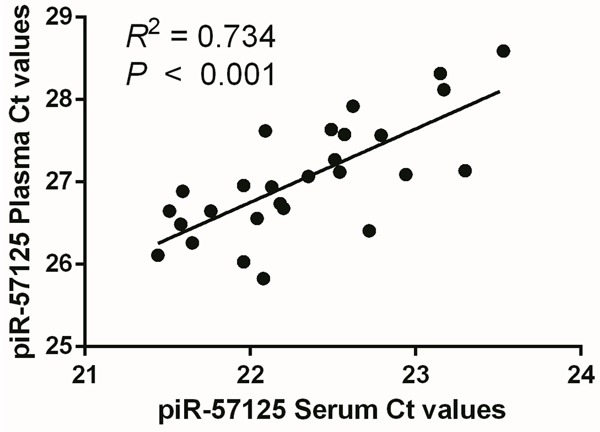
Abundance of piRNAs in serum and plasma samples collected from the same individual. Observed piRNA levels highly correlated (R 2 = 0.734, P < 0.001). Each plot presents the average Ct values in triplicate.
Discussion
Recent reports have revealed that piRNAs play roles in the epigenetic regulation of cancers and other diseases, suggesting that piRNAs can serve as potential diagnostic markers for cancer and other diseases [10]. Tissue-based diagnosis remains invasive and time-intensive; thus, blood-based diagnosis has become a trend for disease screening in high-risk patients. To the best of our knowledge, this study is the first to show that piRNAs are stably expressed in blood serum and plasma.
Previous assumption about the role of piRNAs focused on germline development [6]. However, recent studies have indicated that piRNAs are involved in disease development, especially carcinogenesis. For instance, piR-594040 reportedly participates in the development of bladder cancer [11]. Up-regulated piR-4987 expression is associated with lymph node positivity in breast cancer [12]. piRNA-823 acts as an oncogenic inducer in multiple myelomas [9] but as a tumour suppressor in gastric cancer [13]. By contrast, piR-651 might be involved in the development of gastric cancer [14].
ncRNAs are important epigenetic regulators which contribute to the genesis and progression of human disorders, especially cancer. Studies on the relationship of ncRNAs to human disease mainly focused on the expression and function of miRNAs [5]. However, recent findings on piRNAs suggested that these novel ncRNAs are also implicated in human diseases, indicating that piRNAs are potential diagnostic biomarkers for disease detection. For instance, piRNAs are reportedly potential biomarkers for the diagnosis and prognosis of gastric cancer [13], breast cancer [12,15], hepatocellular carcinoma [8], bladder cancer [11] and multiple myelomas [9].
Interestingly, circulating miRNAs have attracted increasing attention in recent years [16]. As a distinct class of small ncRNAs, piRNAs can become novel circulating biomarkers similar to miRNAs. This study is the first to determine through clone sequencing that piRNAs are stably expressed in blood serum and plasma. We applied qRT-PCR to measure the level of piR-57125 in blood serum and plasma; melting curve analysis (Figure 1D) showed that piR-57125 was efficiently amplified without non-specific interference. Considering that stability is an important prerequisite for utility as a biomarker, we explored the stability of the piRNAs in blood serum and plasma. We observed that the piRNAs remained stable regardless of repetitive freeze–thawing or long-term incubation at room temperature (Figure 2). In addition, the piR-57125 levels detected from the plasma or serum samples strongly correlated (Figure 3). A previous study showed the stability of circulating miRNAs and the highly correlated expression levels of miRNAs between serum and plasma samples [3]. One possible explanation is that miRNAs are packaged inside exosomes, which are secreted from cells [17]. The protection of miRNAs by the formation of RNA-protein complexes could be an alternative explanation. As another type of small RNAs, circulating piRNAs can remain stable in the same manner as circulating miRNAs. However, further studies must be conducted to validate this hypothesis.
In conclusion, this study is the first to report that piRNAs are stably expressed in human serum or plasma samples. Therefore, piRNAs can serve as valuable blood-based biomarkers for disease detection and monitoring.
Disclosure of conflict of interest
None.
References
- 1.Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, Radich J, Anderson G, Hartwell L. The case for early detection. Nat Rev Cancer. 2003;3:243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 2.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: The long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating micrornas as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou H, Hu H, Lai M. Non-coding rnas and their epigenetic regulatory mechanisms. Biol Cell. 2010;102:645–655. doi: 10.1042/BC20100029. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M. Non-coding rnas in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 6.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small rnas binds mammalian piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 7.Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A pirna pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Law PT, Qin H, Ching AK, Lai KP, Co NN, He M, Lung RW, Chan AW, Chan TF, Wong N. Deep sequencing of small rna transcriptome reveals novel non-coding rnas in hepatocellular carcinoma. J Hepatol. 2013;58:1165–1173. doi: 10.1016/j.jhep.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 9.Yan H, Wu QL, Sun CY, Ai LS, Deng J, Zhang L, Chen L, Chu ZB, Tang B, Wang K, Wu XF, Xu J, Hu Y. Pirna-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia. 2015;29:196–206. doi: 10.1038/leu.2014.135. [DOI] [PubMed] [Google Scholar]
- 10.Mei Y, Clark D, Mao L. Novel dimensions of pirnas in cancer. Cancer Lett. 2013;336:46–52. doi: 10.1016/j.canlet.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu H, Hui G, Yuan L, Shi D, Wang Y, Du M, Zhong D, Ma L, Tong N, Qin C, Yin C, Zhang Z, Wang M. Identification of novel pirnas in bladder cancer. Cancer Lett. 2015;356:561–7. doi: 10.1016/j.canlet.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Huang G, Hu H, Xue X, Shen S, Gao E, Guo G, Shen X, Zhang X. Altered expression of pirnas and their relation with clinicopathologic features of breast cancer. Clin Transl Oncol. 2013;15:563–568. doi: 10.1007/s12094-012-0966-0. [DOI] [PubMed] [Google Scholar]
- 13.Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z, Guo J. Pir-823, a novel non-coding small rna, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012;315:12–17. doi: 10.1016/j.canlet.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z, Zhou H, Li QN. Pirna, the new non-coding rna, is aberrantly expressed in human cancer cells. Clin Chim Acta. 2011;412:1621–1625. doi: 10.1016/j.cca.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Ren Y, Xu H, Pang D, Duan C, Liu C. The expression of stem cell protein piwil2 and pir-932 in breast cancer. Surg Oncol. 2013;22:217–223. doi: 10.1016/j.suronc.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Cheng G. Circulating mirnas: Roles in cancer diagnosis, prognosis and therapy. Adv Drug Deliv Rev. 2015;81:75–93. doi: 10.1016/j.addr.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mrnas and micrornas is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]


