Abstract
Background: MiR-23a function as an oncogene in several human cancers, however, its clinical value has not been investigated in NSCLC. Methods: Tissue samples were obtained from 127 NSCLC patients who underwent complete resection at Yantaishan Hospital from March 2008 to January 2014. The expression level of miR-23a was detected in NSCLC tissues and the matched adjacent lung tissues by qRT-PCR. The survival analysis was estimated by the Kaplan-Meier method and was compared by using the log-rank test. Multivariate analysis was performed using the Cox proportional hazard model. Results: The expression level of miR-23a was significantly up-regulated in NSCLC tissues compared with matched adjacent lung tissues (P<0.001). The expression of miR-23a in NSCLC tissues was significantly associated with the smoking status (P=0.001), tumor size (P=0.002), lymphnode metastasis (P<0.001), TNM stage (P=0.001), and tumor differentiation (P=0.004). The overall survival was significantly lower in patients with higher miR-23a expression than in patients with lower miR-23a expression (P=0.02). In addition, multivariate analysis demonstrated that high miR-23a expression (HR=3.558, 95% CI: 2.982-6.635, P=0.011) was significant prognostic factor for NSCLC patients. Conclusions: miR-23a might play an oncogenic role in NSCLC and is a poor prognostic factor. Our results must be verified by large-scale prospective studies with standardized methodology.
Keywords: MicroRNA-23a, expression, NSCLC, prognosis, biomarker
Introduction
Lung cancer is the most common cause of death among all cancers. Non-small cell lung cancer (NSCLC) accounts for approximately 85% of all lung cancers [1]. Despite extensive research and significant improvement in early detection and treatment options, the disease is still difficult to treat and many patients develop recurrent diseases after surgery. Therefore, recent studies focused on finding new prognosis biomarker and pivotal molecular associated with development and metastases of NSCLC [2].
MicroRNAs (miRNAs) represent a class of endogenous, highly conserved, small nonprotein-coding RNAs that are approximately 22 nucleotides in length [3]. miRNAs bind to specific sequences in the 3’ UTR of target genes, resulting in either modulation of translation efficiency or degradation of the mRNA [4,5]. miRNAs have been shown to regulate the expression of a variety of genes involved in the carcinogenesis, metastasis, invasion, and recurrence of lung cancer [6-9].
MiR-23a is a miRNA cluster located in chromosome 19p13.12, which can function as an oncogene in several human cancers [10-12]. Previously, Cao et al found that miR-23a regulated TGF-β-induced EMT by targeting E-cadherin in lung cancer cells and might be useful as a new therapeutic target in NSCLC [13]. However, its clinical value has not been investigated in NSCLC.
Materials and methods
Ethics statement
The project protocol was approved by the Institutional Ethics Committee of Yantaishan Hospital prior to the initiation of the study. All patients provided written informed consent for the use of the tumor tissues for clinical research.
Patients and tissue samples
Tissue samples were obtained from 127 NSCLC patients who underwent complete resection at Yantaishan Hospital from March 2008 to January 2014. None of the patients had received preoperative radiotherapy or chemotherapy prior to surgical resection. Demographic and clinicopathological data were collected from medical records and histopathological reports. Clinical stage was reassessed according to the 7th edition of the American Joint Committee on Cancer TNM staging system. Follow-up data were included until December 2014 or until death. The clinicopathological characteristics of NSCLC patients are summarized in Table 1.
Table 1.
Correlation of miR-23a expression with clinicopathological characteristics in NSCLC
| Characteristic | Number of patients | miR-23a high-expression | miR-23a low-expression | P value | ||
|---|---|---|---|---|---|---|
|
| ||||||
| N | % | N | % | |||
| Age (years) | ||||||
| ≤65 | 65 | 34 | 52.31 | 31 | 47.69 | 0.467 |
| >65 | 62 | 31 | 50.00 | 31 | 50.00 | |
| Gender | ||||||
| Male | 77 | 35 | 45.45 | 42 | 54.55 | 0.396 |
| Female | 50 | 30 | 60.00 | 20 | 40.00 | |
| Smoking | ||||||
| Yes | 89 | 54 | 60.67 | 35 | 39.33 | 0.001 |
| No | 38 | 11 | 28.95 | 27 | 71.05 | |
| Tumor size (cm) | ||||||
| ≤3 | 41 | 13 | 31.71 | 28 | 68.29 | 0.002 |
| >3 | 86 | 52 | 60.47 | 34 | 39.53 | |
| Lymphnode metastasis | ||||||
| Yes | 37 | 29 | 78.38 | 8 | 21.62 | <0.001 |
| No | 90 | 36 | 40.00 | 54 | 60.00 | |
| Stage | ||||||
| I/II | 85 | 35 | 41.18 | 50 | 58.82 | 0.001 |
| III/IV | 42 | 30 | 71.43 | 12 | 28.57 | |
| Histologicaltype | ||||||
| Squamous cell carcinoma | 64 | 31 | 48.44 | 33 | 51.56 | 0.596 |
| Adenocarcinoma | 63 | 34 | 53.97 | 29 | 46.03 | |
| Differentiation | ||||||
| Well/Moderate | 96 | 42 | 43.75 | 54 | 56.25 | 0.004 |
| Poor | 31 | 23 | 74.19 | 8 | 25.81 | |
RNA extraction and quantitative RT-PCR
Total RNA was extracted from the corresponding tissues using a miRcute miRNA isolation kit (Tiangen, Bejing, China) according to the manufacturer’s instructions. Reverse transcription was performed using the One Step Prime Script miRNA cDNA Synthesis Kit (Takara, Dalian, China) following the manufacturer’s instructions. Quantitative RT-PCR was performed using SYBR® Premix Ex Taq™ II (Takara, Dalian, China) according to the manufacturer’s instructions with the 7500 real-time RT-PCR system (Applied Biosystems, Foster City). U6B was used as the normalization control. Each detection was performed in triplicate.
Statistical analysis
To evaluate the possible relationships between the expression levels of miR-23a and various clinicopathological parameters, we used either independent sample t test or one-way ANOVA. The survival analysis was estimated by the Kaplan-Meier method and was compared by using the log-rank test. Multivariate analysis was performed using the Cox proportional hazard model. A difference was considered significant when P<0.05. All statistical analyses were performed with SPSS version 18.0 (SPSS Inc., Chicago, IL, USA).
Results
miR-23a expression in 127 NSCLC tissues samples and corresponding normal tissues
The expression level of miR-23a was detected in 127 pairs of NSCLC tissues and the matched adjacent lung tissues by qRT-PCR. As showed in Figure 1, the expression level of miR-23a was significantly up-regulated in NSCLC tissues compared with matched adjacent lung tissues (P<0.001), suggesting that miR-23a takes part in the genesis of NSCLC. The median value of miR-23a expression in NSCLC tissues were used as the cut-off point for the classification of miR-23a-low/high. NSCLC tissues with the relative expression levels of miR-23a exceeding median value were deemed to be miR-23a-high group; all other tissues were considered to be miR-23a-low group.
Figure 1.
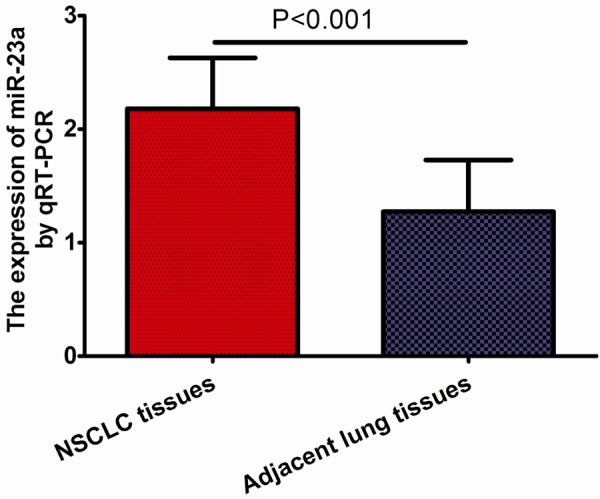
miR-23a expression in 127 NSCLC tissues samples and corresponding normal tissues.
Correlation of miR-23a expression with clinicopathological characteristics in NSCLC
Of the 127 NSCLC patients, 65 patients were high expression of miR-23a, and 62 patients were low expression of miR-23a. Table 1 summarized the association of miR-23a upregulation with the clinicopathological features of NSCLC patients. As a result, the expression of miR-23a in NSCLC tissues was significantly associated with smoking status (P=0.001), tumor size (P=0.002), lymph node metastasis (P<0.001), TNM stage (P=0.001), and tumor differentiation (P=0.004).
High miR-23a level contributes to poor prognosis in patients with NSCLC
Patients with higher miR-23a expression had poorer prognosis. The overall survival was significantly lower in patients with higher miR-23a expression than in patients with lower miR-23a expression (P=0.02, Figure 2). In addition, multivariate analysis demonstrated that lymph node metastasis (HR=7.833, 95% CI: 2.522-10.047, P=0.002), tumor stage (HR=7.912, 95% CI: 3.016-10.726, P=0.001), tumor differentiation (HR=3.447, 95% CI: 1.281-7.883, P=0.037), and miR-23a expression (HR=3.558, 95% CI: 2.982-6.635, P=0.011) were significant prognostic factors for NSCLC patients (Table 2).
Figure 2.
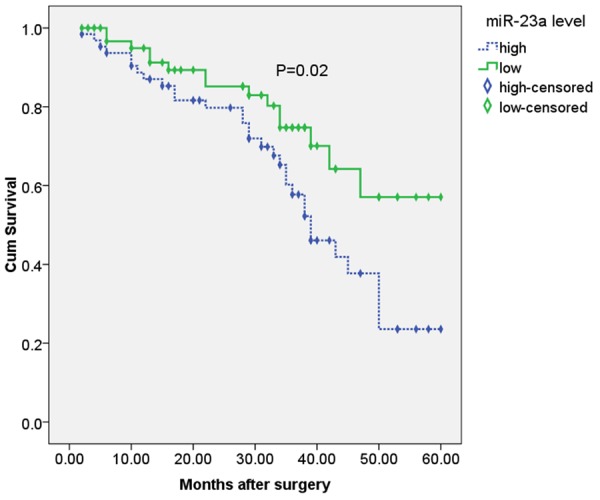
Overall survival of 127 NSCLC patients in relation to mir-23a expression levels.
Table 2.
COX regression analysis in the relationship of clinicopathologic characteristics and prognosis
| Characteristics | HR | 95% CI | P value |
|---|---|---|---|
| Age (years) | 1.371 | 0.526-3.197 | 0.381 |
| Gender | 0.561 | 0.228-2.916 | 0.493 |
| Smoking | 2.918 | 0.816-4.178 | 0.165 |
| Tumor size (cm) | 2.762 | 0.745-5.197 | 0.142 |
| Lymphnode metastasis | 7.883 | 2.522-10.047 | 0.002 |
| Stage | 7.912 | 3.016-10.726 | 0.001 |
| Histological type | 0.273 | 0.198-3.182 | 0.372 |
| Differentiation | 3.447 | 1.281-7.883 | 0.037 |
| miR-23a expression | 3.558 | 2.982-6.635 | 0.011 |
Discussion
Optimal confrontation of NSCLC remains a largely unachieved objective, as the 5-year survival rate struggles to surpass 15% [14]. Most decisions throughout the clinical management of NSCLC are dictated by TNM staging. Surgical removal of tumors, where feasible, is the gold standard of treatment [15]. However, even early-stage resected patients present an extremely variable oncologic outcome; this is a reflection of the large biological heterogeneity between patients who are currently erroneously categorized in the same prognostic group [14]. For example, adjuvant chemotherapy has not been proven to prolong the survival of stage I patients (especially stage IA) [16]. Meanwhile, ~30-40% of stage I NSCLC patients are expected to relapse within 5 years, and this illustrates a severe inability, intrinsic to TNM staging, in defining high-risk patients who should receive adjuvant chemotherapy [17]. In advanced disease stages, an accurate prognostic indicator could guide the administration of maintenance therapy, a clinical decision that currently remains under serious controversy [18]. Consequently, there is a necessity for novel molecular prognostic indicators that would enhance the conventional staging system and improve the decision-making process.
MicroRNAs suppress gene expression by binding to the complementary regions of mRNAs, which either blocks translation or facilitates mRNA degradation through the RNA-induced silencing complex [4]. Recent reports have summarized that aberrant expression of miRNAs has been associated with carcinogenesis and tumor progression [1,15,18]. Numerous lines of evidence have demonstrated that miR-23a is involved in the development of various cancers, including colon cancer, hepatocarcinoma, glioma, breast cancer, colorectal cancer, gastric adenocarcinoma, and haematological malignancies [13,19-25]. Most of them identified miR-23a as an oncogene due to its stimulatory role in cell migration and invasion. For example, miR-23a is regulated by c-MYC and promotes mammary carcinoma cell invasion and hepatic metastasis by targeting Sprouty-2 [23].
The prognostic value of miR-23a has been investigated in several types of cancers. For example, Zhang et al found that miR-23a was significantly up-regulated in laryngeal cancer tissues compared to normal adjacent laryngeal tissues (P<0.01). Tumors with high miR-23a expression had significantly greater extent of lymph node metastasis (P<0.01), worse clinical stage (P<0.05) and shorter overall five-year survival (P<0.01) than those with low miR-23a expression. Both univariate and multivariate Cox hazard regression analysis results showed that clinical stage and miR-23a expression were significantly correlated with patient five-year survival (P<0.01) [12]. He et al identified miR-23a as a tumor suppressor in osteosarcoma. Their data clarify the mechanism of osteosarcoma progression and demonstrated the potential for exploiting miR-23a as a diagnostic marker for osteosarcoma [10]. Ma et al found that the dysregulation of miR-23a and miR-23b might be implicated in the progression of human gastric cancer. Combined expression of miR-23a and miR-23b appeared to be a valuable marker for prognosis of gastric cancer [11]. Previously, Cao et al found that miR-23a regulated TGF-β-induced EMT by targeting E-cadherin in lung cancer cells and might be useful as a new therapeutic target in NSCLC [13]. However, its clinical value has not been investigated in NSCLC.
In the present study, we found that the expression level of miR-23a was significantly up-regulated in NSCLC tissues compared with matched adjacent lung tissues, suggesting that miR-23a takes part in the genesis of NSCLC. Of the 127 NSCLC patients, 65 patients were high expression of miR-23a, and 62 patients were low expression of miR-23a. We found that the expression of miR-23a in NSCLC tissues was significantly associated with the smoking status, tumor size, lymph node metastasis, TNM stage, and tumor differentiation. Patients with higher miR-23a expression had poorer prognosis. The overall survival was significantly lower in patients with higher miR-23a expression than in patients with lower miR-23a expression. In addition, multivariate analysis demonstrated that lymph node metastasis, tumor stage, tumor differentiation, and miR-23a expression were significant prognostic factors for NSCLC patients. In conclusion, miR-23a might play an oncogenic role in NSCLC and is a poor prognostic factor. Our results must be verified by large-scale prospective studies with standardized methodology.
Disclosure of conflict of interest
None.
References
- 1.Subramaniam S, Thakur RK, Yadav VK, Nanda R, Chowdhury S, Agrawal A. Lung cancer biomarkers: State of the art. J Carcinog. 2013;12:3. doi: 10.4103/1477-3163.107958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soes S, Daugaard IL, Sorensen BS, Carus A, Mattheisen M, Alsner J, Overgaard J, Hager H, Hansen LL, Kristensen LS. Hypomethylation and increased expression of the putative oncogene ELMO3 are associated with lung cancer development and metastases formation. Oncoscience. 2014;1:367–374. doi: 10.18632/oncoscience.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barshack I, Meiri E, Rosenwald S, Lebanony D, Bronfeld M, Aviel-Ronen S, Rosenblatt K, Polak-Charcon S, Leizerman I, Ezagouri M, Zepeniuk M, Shabes N, Cohen L, Tabak S, Cohen D, Bentwich Z, Rosenfeld N. Differential diagnosis of hepatocellular carcinoma from metastatic tumors in the liver using microRNA expression. Int J Biochem Cell Biol. 2010;42:1355–1362. doi: 10.1016/j.biocel.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 4.Wu Y, Crawford M, Yu B, Mao Y, Nana-Sinkam SP, Lee LJ. MicroRNA delivery by cationic lipoplexes for lung cancer therapy. Mol Pharm. 2011;8:1381–1389. doi: 10.1021/mp2002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang QZ, Xu W, Habib N, Xu R. Potential uses of microRNA in lung cancer diagnosis, prognosis, and therapy. Curr Cancer Drug Targets. 2009;9:572–594. doi: 10.2174/156800909788486731. [DOI] [PubMed] [Google Scholar]
- 6.He D, Wang J, Zhang C, Shan B, Deng X, Li B, Zhou Y, Chen W, Hong J, Gao Y, Chen Z, Duan C. Down-regulation of miR-675-5p contributes to tumor progression and development by targeting pro-tumorigenic GPR55 in non-small cell lung cancer. Mol Cancer. 2015;14:73. doi: 10.1186/s12943-015-0342-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang T, Thakur A, Chen T, Yang L, Lei G, Liang Y, Zhang S, Ren H, Chen M. MicroRNA-15a induces cell apoptosis and inhibits metastasis by targeting BCL2L2 in non-small cell lung cancer. Tumour Biol. 2015;36:4357–4365. doi: 10.1007/s13277-015-3075-1. [DOI] [PubMed] [Google Scholar]
- 8.Chen D, Guo W, Qiu Z, Wang Q, Li Y, Liang L, Liu L, Huang S, Zhao Y, He X. MicroRNA-30d-5p inhibits tumour cell proliferation and motility by directly targeting CCNE2 in non-small cell lung cancer. Cancer Lett. 2015;362:208–217. doi: 10.1016/j.canlet.2015.03.041. [DOI] [PubMed] [Google Scholar]
- 9.Liu B, Qu J, Xu F, Guo Y, Wang Y, Yu H, Qian B. MiR-195 suppresses non-small cell lung cancer by targeting CHEK1. Oncotarget. 2015;6:9445–9456. doi: 10.18632/oncotarget.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He Y, Meng C, Shao Z, Wang H, Yang S. MiR-23a functions as a tumor suppressor in osteosarcoma. Cell Physiol Biochem. 2014;34:1485–1496. doi: 10.1159/000366353. [DOI] [PubMed] [Google Scholar]
- 11.Ma G, Dai W, Sang A, Yang X, Gao C. Upregulation of microRNA-23a/b promotes tumor progression and confers poor prognosis in patients with gastric cancer. Int J Clin Exp Pathol. 2014;7:8833–8840. [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang XW, Liu N, Chen S, Wang Y, Zhang ZX, Sun YY, Qiu GB, Fu WN. High microRNA-23a expression in laryngeal squamous cell carcinoma is associated with poor patient prognosis. Diagn Pathol. 2015;10:22. doi: 10.1186/s13000-015-0256-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao M, Seike M, Soeno C, Mizutani H, Kitamura K, Minegishi Y, Noro R, Yoshimura A, Cai L, Gemma A. MiR-23a regulates TGF-beta-induced epithelial-mesenchymal transition by targeting E-cadherin in lung cancer cells. Int J Oncol. 2012;41:869–875. doi: 10.3892/ijo.2012.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powell CA, Halmos B, Nana-Sinkam SP. Update in lung cancer and mesothelioma 2012. Am J Respir Crit Care Med. 2013;188:157–166. doi: 10.1164/rccm.201304-0716UP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson DH, Schiller JH, Bunn PA Jr. Recent clinical advances in lung cancer management. J. Clin. Oncol. 2014;32:973–982. doi: 10.1200/JCO.2013.53.1228. [DOI] [PubMed] [Google Scholar]
- 16.Reck M, Heigener DF, Mok T, Soria JC, Rabe KF. Management of non-small-cell lung cancer: recent developments. Lancet. 2013;382:709–719. doi: 10.1016/S0140-6736(13)61502-0. [DOI] [PubMed] [Google Scholar]
- 17.Goodgame B, Viswanathan A, Miller CR, Gao F, Meyers B, Battafarano RJ, Patterson A, Cooper J, Guthrie TJ, Bradley J, Pillot G, Govindan R. A clinical model to estimate recurrence risk in resected stage I non-small cell lung cancer. Am J Clin Oncol. 2008;31:22–28. doi: 10.1097/COC.0b013e3180ca77d1. [DOI] [PubMed] [Google Scholar]
- 18.Gadgeel SM, Ramalingam SS, Kalemkerian GP. Treatment of lung cancer. Radiol Clin North Am. 2012;50:961–974. doi: 10.1016/j.rcl.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, Watanabe M, Nakagama H, Yokota J, Kohno T, Tsuchiya N. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9:e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bao L, Zhao J, Dai X, Wang Y, Ma R, Su Y, Cui H, Niu J, Bai S, Xiao Z, Yuan H, Yang Z, Li C, Cheng R, Ren X. Correlation between miR-23a and onset of hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2014;38:318–330. doi: 10.1016/j.clinre.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Wang WL, Yang C, Han XL, Wang R, Huang Y, Zi YM, Li JD. MicroRNA-23a expression in paraffin-embedded specimen correlates with overall survival of diffuse large B-cell lymphoma. Med Oncol. 2014;31:919. doi: 10.1007/s12032-014-0919-2. [DOI] [PubMed] [Google Scholar]
- 22.Hu X, Chen D, Cui Y, Li Z, Huang J. Targeting microRNA-23a to inhibit glioma cell invasion via HOXD10. Sci Rep. 2013;3:3423. doi: 10.1038/srep03423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Liu X, Xu W, Zhou P, Gao P, Jiang S, Lobie PE, Zhu T. c-MYC-regulated miR-23a/24-2/27a cluster promotes mammary carcinoma cell invasion and hepatic metastasis by targeting Sprouty2. J Biol Chem. 2013;288:18121–18133. doi: 10.1074/jbc.M113.478560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Wei W, Sarkar FH. miR-23a, a critical regulator of “migR” ation and metastasis in colorectal cancer. Cancer Discov. 2012;2:489–491. doi: 10.1158/2159-8290.CD-12-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu LH, Liu T, Tang H, Tian RQ, Su C, Liu M, Li X. MicroRNA-23a promotes the growth of gastric adenocarcinoma cell line MGC803 and downregulates interleukin-6 receptor. FEBS J. 2010;277:3726–3734. doi: 10.1111/j.1742-4658.2010.07773.x. [DOI] [PubMed] [Google Scholar]


