Abstract
Hepatocellular carcinoma (HCC) is one of the most common lethal malignancies in the world, and the current knowledge on the molecular and genetic basis of HCC is still limited. Previous study has shown miR-149 plays a tumor suppressive role in HCC, here we aimed to investigate the association between rs71428439 polymorphism, which located in the pre-miR-149, and the risk of HCC in a Chinese Han population. A total of 177 HCC patients and 103 healthy controls were genotyped, by a multivariate logistic regression, we found that individuals with GG genotype have significantly higher risk of HCC (adjusted OR=3.397, 95% CI=1.565-7.375, P=0.002) compared with those with AA genotype, similar results were also observed in recessive model (adjusted OR=2.563, 95% CI=1.300-5.054, P=0.007) and dominant model (adjusted OR=2.074, 95% CI=1.147-3.752, P=0.016). We further observed that tumor tissues in patients with GG genotype expressed lower level of miR-149 compared with those with AA or AG genotype, and consequently, AKT1, a pre-validated miR-149 target in vitro, was found to have higher expression level in tumors with GG genotype. In summary, our data indicated that rs71428439 may be a genetic risk factor of HCC in the Chinese Han population, and its mechanism possibly involves downregulated miR-149 expression and upregulated AKT1 expression.
Keywords: HCC, polymorphism, miR-149, AKT1
Introduction
Hepatocellular carcinoma (HCC), the most common type hepatic cancer, is the third leading cause of cancer associated death [1], and is characterized by high invasiveness and poor prognosis. The pathogenesis of HCC is by far considered to be closely related to the interplay between environmental and genetic factors, of which its association with viral infection (HBV and HCV) represents a good exemplification. Despite the several susceptibility locus revealed by recent Genome wide association studies on HBV- and HCV-related HCC [2-4], the genetic basis of HCC is still incompletely understood.
MicroRNAs are a class of short non-coding RNAs that act endogenously to post-transcriptionally modify gene expression [5]. It shows a broad regulatory effect on many diseases by tethering critical biological progresses such as proliferation, apoptosis and differentiation [6,7]. A large amount of evidence has suggested the vital functions of microRNA in the pathogenesis of HCC [8]. MiR-149, which is transcribed from chromosome 2, has been identified as tumor suppressor microRNA in several cancers [9-11]. Thus far, numbers of its target genes that involved in cell proliferation have been experimentally validated. Notably, a recent study has implicated the tumor suppressive role of miR-149 by targeting AKT1 in HCC [12].
Single nucleotide polymorphisms (SNPs) are widely existed in the human genome. Genetic variants caused by exonic SNPs have been functionally linked to the risk of several cancers, besides, a substantial amount of intronic polymorphisms or exonic polymorphisms that result in synonymous mutation are also relevant to the disease status with largely unknown mechanisms. Despite the extensive studies of polymorphisms of protein-coding genes, the role of polymorphisms in microRNAs has received little attention. Although previous studies have suggested the association of miR-149 C/T polymorphism with the carcinogenesis of HCC [13,14], the mechanism underlying this association is still unknown. Nonetheless, the role of rs71428439 A/G, another polymorphism within the pre-microRNA-149, has never been discovered clinically in the scenario of HCC.
The aim of this case-control study is to test whether rs71428439 is associated with the risk of HCC in ethnically Han Chinese, 177 HCC cases and 103 healthy controls were genotyped, and the results were analyzed by multivariate logistic regression. Our data suggest a previously unreported relationship between this polymorphism and the susceptibility of HCC, and may thereby provide insights into the genetic basis of oncogenesis of HCC.
Materials and methods
Study subjects
In this study, we consecutively recruited a total of newly diagnosed 177 HCC patients from gastroenterology department of The Second Affiliated Hospital of Xi’an University. All the diagnoses of HCC were based on the histological evidences, and the histological examination was confirmed by two experienced pathologists. The healthy controls were selected from the individuals who were from the same geographic location with patients and underwent physical examination in the outpatient department of the same hospital. Individuals with other severe complications such as cardiovascular and cerebral disorder and diabetes were excluded from this study. We obtained an informed consent from each individual upon recruitment, and this study was approved by the ethics review committee of The Second Affiliated Hospital of Xi’an university.
Genotyping
Each study subject was kindly asked to provide 5 ml blood sample in an EDTA-anticoagulate tube. Genomic DNA was isolated using a QIAamp DNA Blood Midi Kit (Qiagen, Valencia, CA). The DNA samples were stored in -20°C before genotyping. The genotype of each individual was determined by Sanger sequencing as previously described [15].
Realtime PCR
Fresh tumor tissues from the surgical resection were obtained from at least 25 HCC patients for each genotype. The tissues were immediately stored in liquid nitrogen and then transferred to -80°C prior to final analysis. The total RNA was isolated using Trizol (Invitrogen, Carlsbad, CA, USA) and reverse transcribed using an NCode™ miRNA First-Strand cDNA Synthesis Kit (Invitrogen, Carlsbad, CA, USA) according to the protocols of the manufacturer. Taqman assay kit for has-miR-149 (002255) was from applied biosystems (Applied Biosystems, Foster City, CA, USA). U6 was amplified by the SYBR green method and served as internal control. The primers for U6 were forward AAAGCAAATCATCGGACGACC and reverse GTACAACACATTGTTTCCTCGGA.
Western blot
Total proteins were extracted from the fresh tissues described above by RIPA lysis buffer, and the proteins were mixed with appropriate amount of SDS loading buffer and heated at 95°C for 5 min. The denatured protein was then subjected to electrophoresis in a 10% SDS-PAGE gel, followed by transferring onto a PVDF membrane. The membranes were incubated with primary antibodies at 4°C overnight and added with HRP-conjugated secondary antibodies the following day and then visualized by BeyoECL Plus chemiluminescence detection system (P0018, Beyotime, Shanghai China). The expression of β-actin was used as loading control. Antibodies for Cyclin D1 AKT1 and β-actin were all purchased from Cell Signaling Technology Inc. (Beverly, MA, USA). The band density was analyzed by ImageJ software (version 1.44, NIH).
Statistical analysis
The baseline characteristics were presented as means or medians, comparison of age was performed by independent sample t test. The association between HCC status and smoking, drinking, family history or viral infection (HBV and HCV) was analyzed by chi-square test. A multivariate logistic analysis was conducted in order to study whether the genotype of rs71428439 is associated with HCC risk. The differences of miR-149 mRNA level and protein level of AKT1 and cyclin D1 were compared by one-way ANOVA followed by Bonferroni post hoc test. A two sided p value below 0.05 was considered as statistical significance.
Results
Baseline clinical characteristics
The baseline clinical characteristics were summarized in Table 1. No significant difference regarding age (P=0.17), sex (P=0.746), family history (P=0.929) and smoking status (P=0.344) between healthy controls and HCC patients was found. By contrast, the frequency of drinkers in HCC patients was markedly higher than that in healthy population (P<0.001). HBV and HCV infection, which were previously thought to be closely related to the etiology of HCC, were also more frequently occurred in HCC population (P<0.001 and P=0.036, respectively).
Table 1.
Baseline characteristics
| Control | HCC | P | |
|---|---|---|---|
| Age | 53.5±9.3 | 53.9±10.0 | 0.17 |
| Sex | |||
| Male | 82 | 138 | 0.746 |
| Female | 21 | 39 | |
| Smoking status | |||
| No | 63 | 98 | |
| Yes | 40 | 79 | 0.344 |
| Drinking status | |||
| No | 75 | 91 | |
| Yes | 28 | 86 | <0.001 |
| HBV infection | |||
| No | 83 | 98 | |
| Yes | 20 | 79 | <0.001 |
| HCV infection | |||
| No | 100 | 160 | |
| Yes | 3 | 17 | 0.036 |
| Family history | |||
| No | 91 | 157 | |
| Yes | 12 | 20 | 0.929 |
The association between rs71428439 SNP and risk of HCC
We next assessed the genotype distribution in both the case and the control group. As displayed in Table 2, the distribution of each genotype of rs71428439 was in line with Hardy-Weinberg equilibrium in both the control group (P=0.798) and HCC group (P=0.423), indicating the lack of selection bias in our study. To test whether rs71428439 is associated with the susceptibility of HCC, we employed a multivariate logistic analysis to compute the adjusted odds ratio of each genotype in additive, recessive and dominant models, respectively. We found that the frequency of GG genotype was significantly increased in HCC group, and individuals with this genotype showed an increased risk of HCC (adjusted OR=3.397, 95% CI=1.565-7.375, P=0.002). Similar result was observed when we combined AA and AG genotypes to construct a recessive model (adjusted OR=2.563, 95% CI=1.300-5.054, P=0.007). Moreover, the adjusted OR for HCC risk in recessive model was higher than that in dominant model (adjusted OR=2.074, 95% CI=1.147-3.752, P=0.016). Allele frequencies were also compared, individuals with G allele had a 1.858-fold risk of HCC compared with those carrying A allele (adjusted OR=1.858, 95% CI=1.270-2.716, P=0.001).
Table 2.
Genotype distribution of rs71428439 in healthy control and HCC patients
| Control (n, %) | HCC (n, %) | P | Adjusted ORa | 95% CI | |
|---|---|---|---|---|---|
| Genotype | |||||
| AA | 41 (39.8%) | 37 (20.9%) | - | 1 | - |
| AG | 47 (45.6%) | 82 (46.3%) | 0.144 | 1.607 | 0.851-3.037 |
| GG | 15 (14.6%) | 58 (32.8%) | 0.002 | 3.397 | 1.565-7.375 |
| HWE (P) | 0.798 | 0.423 | |||
| Recessive model | |||||
| AA+AG | 88 (85.4%) | 119 (67.2%) | - | 1 | - |
| GG | 15 (14.6%) | 58 (32.8%) | 0.007 | 2.563 | 1.300-5.054 |
| Dominant model | |||||
| AA | 41 (39.8%) | 37 (20.9%) | - | 1 | - |
| AG+GG | 62 (60.2%) | 140 (79.1%) | 0.016 | 2.074 | 1.147-3.752 |
| Allele | |||||
| A | 129 (62.6%) | 156 (44.1%) | - | 1 | - |
| G | 77 (37.4%) | 198 (55.9%) | 0.001 | 1.858 | 1.270-2.716 |
OR adjusted for age, sex, smoking status, drinking status, family history, HBV infection and HCV infection.
The association between rs71428439 SNP and miR-149 expression in HCC patients
As previous study has suggested that rs71428439 is involved in the maturation of miR-149 and thus affects its expression [15], we then tested whether the deceased miR-149 expression is also presented in the tissue sample of HCC patients with rs71428439 genetic variants. Realtime PCR was used to quantitatively assess the miR-149 expression in HCC tumor tissues. As shown in Figure 1, miR-149 expression in patients with GG genotype was significantly lower than those with AG or AA genotype.
Figure 1.
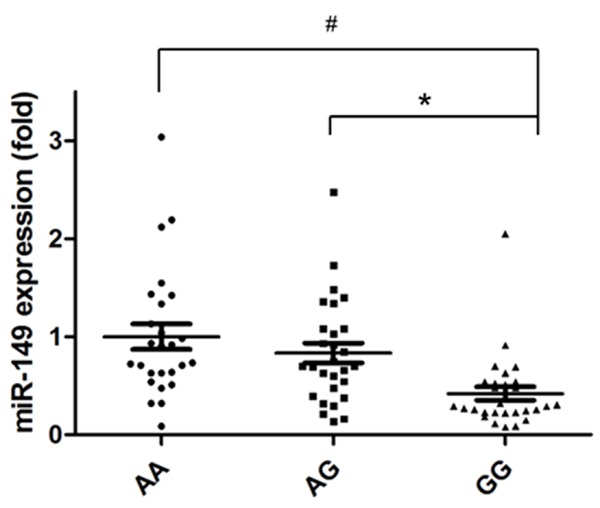
The expression level of miR-149 in tumors with different rs71428349 genotypes. *P<0.05 compared with AG group, #P<0.05 compared with AA group.
Rs71428439 SNP affects AKT1 and cyclin D1 expression in HCC
Recent study has implicated AKT1 as a direct target of miR-149 in neuroblastoma cell and HeLa cell [16]. In order to test whether rs71428439 SNP shows effect on the expression of genes that involved in cell proliferation and cycle progression, we next performed western blot to detect the expression of AKT1 and Cyclin D1. As shown in Figure 2, tumor tissues from patients with GG genotype showed increased AKT1 and Cyclin D1 expression as compared with those in patients with AA and AG genotypes.
Figure 2.
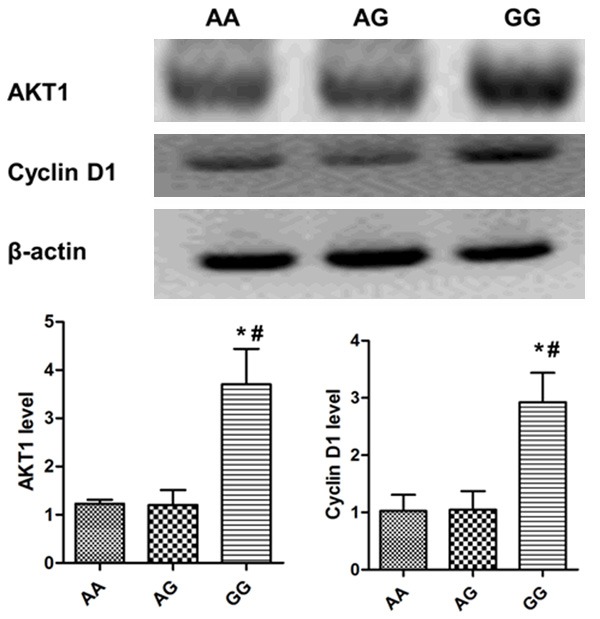
AKT1 and cyclin D1 expression in tumors with different rs71428349 genotypes. *P<0.05 compared with AG group, #P<0.05 compared with AA group.
Discussion
We demonstrate here in the present case-control study that rs71428439 genetic variants may serve as a potential genetic risk factor of HCC. By multivariate logistic regression analysis, we put forward the first ever clinical evidence of the association between this SNP site and the risk of HCC. Our further analysis, which confirmed the association between rs71428439 polymorphism and miR-149 expression as well as proteins that associated with cell cycle regulation, not only corroborates the mechanisms proposed by the recent study [15] but also provided insights into the possible mechanisms by which this pre-miR-149 polymorphism is involved. Hence, rs71428439 pre-miR-149 polymorphism is of considerable importance in the clinical setting.
MicroRNAs are a class of evolutionarily conserved, endogenous and short non-coding RNA strands that are able to repress gene expression by binding to the 3’-UTR of target mRNA. A substantial amount of microRNAs have emerged as key regulatory molecules in the development and progression of tumors [17]. Aberrant expression of microRNAs has been reported in the pathogenesis of HCC [8]. Human miR-149 is mapped to chromosome 2, and it reportedly functions to inhibit tumor development and metastasis. Recent study has revealed an epigenetic silencing of miR-149 by methylation in colorectal cancer tissues [18], which suggested its prognostic and therapeutic potentials. Particularly, miR-149 has been identified as a direct target of AKT1 in glioma cells [10], which has been replicated in a comprehensive study in HCC recently [12]. Although these studies support that miR-149 plays a favorable role in the prognosis of HCC, how the expression of miR-149 is regulated in the HCC remains unknown. Ding et al. have reported the relationship between rs71428439 SNP and miR-149 expression in HEK cells [15], and indeed miR-149 expression was decreased in GG genotype of clear cell renal cell carcinoma [19], as reported by Wang et al. However, whether rs71428439 genetic variant is associated with decreased miR-149 expression and the cancer risk in HCC is still undetermined. In the current study, we found that rs71428439 GG genotype was significantly associated with increased risk of HCC. We demonstrate that patients with GG genotype exhibited lower level of miR-149 expression, which is consistent with the works by Ding et al. and Wang et al. Intriguingly, a C/T polymorphism within miR-149 has been established as a risk factor of HCC [13,14]. However, the exact mechanism is still unknown, whether miR-149 C/T polymorphism also affects its expression and whether it is in linkage equilibrium with rs71429439 in HCC warrant future investigation.
If rs71428439 did affect the expression of miR-149, then its target genes should be downregulated. To test this idea in the clinical setting of HCC, we assayed the protein level of AKT1 in the samples of each genotype. To our expectation, AKT1 level in tumor tissues of GG genotype was the highest compared with the other two groups. Therefore, our study is in consistent with the in vitro data of the previous study [12], which identified AKT1 as a direct target of miR-149. More importantly, cyclin D1, a key component of cell cycle regulation that controls G1/S transition, was also expressed at a higher level in GG genotype. Given the well documented involvement of Cyclin D1 in AKT-mTOR transcriptional axis in early studies [20], it is conceivable that a regulating apparatus that comprises rs71428439 GG genotype, increased AKT1 expression and increased cyclin D1 expression is involved in the development of HCC. This result indicated an increased proliferative activity in tumors with GG genotype.
It should be noted that this study is still limited due to the facts of several aspects. Firstly, as our study is a retrospective study, no causal link between rs71428439 polymorphism and the onset of cancer has been established, experimental animal studies are warrant to better understand its role in the carcinogenesis of HCC. Secondly, as the study subjects in this study are limited to the Han population from a single hospital, our conclusion may not necessarily accurately represent the whole population. Our evidence may as well be confirmed in other ethnic groups. Thirdly, we failed to follow up these patients with different genotypes, and thus it is not able to figure out whether this polymorphism is associated with the prognosis of HCC; earlier study that propose cell cycle regulatory proteins as the prognostic markers of HCC should shed some light on this issue [21].
Conclusion
Our study provided the first compelling clinical evidences which support GG genotype of rs71428439 SNP within pre-miR-149 as a risk factor for HCC susceptibility. Our study further implicated the link between rs71428439 polymorphism, AKT1 expression and Cyclin D1 expression, which suggested a new biomarker for developing new therapeutic approaches for HCC.
Disclosure of conflict of interest
None.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Kumar V, Kato N, Urabe Y, Takahashi A, Muroyama R, Hosono N, Otsuka M, Tateishi R, Omata M, Nakagawa H, Koike K, Kamatani N, Kubo M, Nakamura Y, Matsuda K. Genome-wide association study identifies a susceptibility locus for HCV-induced hepatocellular carcinoma. Nat Genet. 2011;43:455–458. doi: 10.1038/ng.809. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H, Zhai Y, Hu Z, Wu C, Qian J, Jia W, Ma F, Huang W, Yu L, Yue W, Wang Z, Li P, Zhang Y, Liang R, Wei Z, Cui Y, Xie W, Cai M, Yu X, Yuan Y, Xia X, Zhang X, Yang H, Qiu W, Yang J, Gong F, Chen M, Shen H, Lin D, Zeng YX, He F, Zhou G. Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat Genet. 2010;42:755–758. doi: 10.1038/ng.638. [DOI] [PubMed] [Google Scholar]
- 4.Jiang DK, Sun J, Cao G, Liu Y, Lin D, Gao YZ, Ren WH, Long XD, Zhang H, Ma XP, Wang Z, Jiang W, Chen TY, Gao Y, Sun LD, Long JR, Huang HX, Wang D, Yu H, Zhang P, Tang LS, Peng B, Cai H, Liu TT, Zhou P, Liu F, Lin X, Tao S, Wan B, Sai-Yin HX, Qin LX, Yin J, Liu L, Wu C, Pei Y, Zhou YF, Zhai Y, Lu PX, Tan A, Zuo XB, Fan J, Chang J, Gu X, Wang NJ, Li Y, Liu YK, Zhai K, Zhang H, Hu Z, Liu J, Yi Q, Xiang Y, Shi R, Ding Q, Zheng W, Shu XO, Mo Z, Shugart YY, Zhang XJ, Zhou G, Shen H, Zheng SL, Xu J, Yu L. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet. 2013;45:72–75. doi: 10.1038/ng.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaravinos A. The Regulatory Role of MicroRNAs in EMT and Cancer. J Oncol. 2015;2015:865816. doi: 10.1155/2015/865816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sidhu K, Kapoor NR, Pandey V, Kumar V. The “Macro” World of microRNAs in Hepatocellular Carcinoma. Front Oncol. 2015;5:68. doi: 10.3389/fonc.2015.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan SH, Huang WC, Chang JW, Chang KJ, Kuo WH, Wang MY, Lin KY, Uen YH, Hou MF, Lin CM, Jang TH, Tu CW, Lee YR, Lee YH, Tien MT, Wang LH. MicroRNA-149 targets GIT1 to suppress integrin signaling and breast cancer metastasis. Oncogene. 2014;33:4496–4507. doi: 10.1038/onc.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan SJ, Zhan SK, Pei BG, Sun QF, Bian LG, Sun BM. MicroRNA-149 inhibits proliferation and invasion of glioma cells via blockade of AKT1 signaling. Int J Immunopathol Pharmacol. 2012;25:871–881. doi: 10.1177/039463201202500405. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Zheng X, Zhang Z, Zhou J, Zhao G, Yang J, Xia L, Wang R, Cai X, Hu H, Zhu C, Nie Y, Wu K, Zhang D, Fan D. MicroRNA-149 inhibits proliferation and cell cycle progression through the targeting of ZBTB2 in human gastric cancer. PLoS One. 2012;7:e41693. doi: 10.1371/journal.pone.0041693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Guo X, Xiong L, Yu L, Li Z, Guo Q, Li Z, Li B, Lin N. Comprehensive analysis of microRNA-regulated protein interaction network reveals the tumor suppressive role of microRNA-149 in human hepatocellular carcinoma via targeting AKT-mTOR pathway. Mol Cancer. 2014;13:253. doi: 10.1186/1476-4598-13-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang XH, Wang FR, Tang YF, Zou HZ, Zhao YQ. Association of miR-149C>T and miR-499A>G polymorphisms with the risk of hepatocellular carcinoma in the Chinese population. Genet Mol Res. 2014;13:5048–5054. doi: 10.4238/2014.July.4.20. [DOI] [PubMed] [Google Scholar]
- 14.Liu MF, Chen WQ, He YZ, Gu YL. Role of miR-149 C>T polymorphisms on the risk of hepatocellular carcinoma in a Chinese population. Genet Mol Res. 2014;13:7184–7189. doi: 10.4238/2014.September.5.4. [DOI] [PubMed] [Google Scholar]
- 15.Ding SL, Wang JX, Jiao JQ, Tu X, Wang Q, Liu F, Li Q, Gao J, Zhou QY, Gu DF, Li PF. A pre-microRNA-149 (miR-149) genetic variation affects miR-149 maturation and its ability to regulate the Puma protein in apoptosis. J Biol Chem. 2013;288:26865–26877. doi: 10.1074/jbc.M112.440453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin RJ, Lin YC, Yu AL. miR-149* induces apoptosis by inhibiting Akt1 and E2F1 in human cancer cells. Mol Carcinog. 2010;49:719–727. doi: 10.1002/mc.20647. [DOI] [PubMed] [Google Scholar]
- 17.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 18.Wang F, Ma YL, Zhang P, Shen TY, Shi CZ, Yang YZ, Moyer MP, Zhang HZ, Chen HQ, Liang Y, Qin HL. SP1 mediates the link between methylation of the tumour suppressor miR-149 and outcome in colorectal cancer. J Pathol. 2013;229:12–24. doi: 10.1002/path.4078. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, Wei M, Ren Y, Liu H, Wang M, Shi K, Jiang H. miR149 rs71428439 polymorphism and risk of clear cell renal cell carcinoma: a case-control study. Tumour Biol. 2014;35:12127–12130. doi: 10.1007/s13277-014-2517-5. [DOI] [PubMed] [Google Scholar]
- 20.Nicholson KM, Anderson NG. The protein kinase B/Akt signalling pathway in human malignancy. Cell Signal. 2002;14:381–395. doi: 10.1016/s0898-6568(01)00271-6. [DOI] [PubMed] [Google Scholar]
- 21.Che Y, Ye F, Xu R, Qing H, Wang X, Yin F, Cui M, Burstein D, Jiang B, Zhang DY. Co-expression of XIAP and cyclin D1 complex correlates with a poor prognosis in patients with hepatocellular carcinoma. Am J Pathol. 2012;180:1798–1807. doi: 10.1016/j.ajpath.2012.01.016. [DOI] [PubMed] [Google Scholar]


