Abstract
We distinguished the four alleles of OXA subgroups from 339 strains of Acinetobacter baumannii using Polymerase Chain Reaction, and investigated distributions of OXA subgroups in clinical isolated strains. A total of 196 Acinetobacter baumannii were isolated from the Central Hospital of Zhumadian between 2010 and 2014. Amplification of OXA genes, blaOXA-23, blaOXA-24, blaOXA-51 and blaOXA-58, were performed by PCR. Patients with Acinetobacter baumannii were selected from ICU, pneumology, emergency and cerebral surgery, accounting for 33.67%, 17.86%, 16.33% and 32.14%, respectively. Most strains showed resistance to different classes of agents, especially in ceftazidime, piperacillin, cefepime, nitrofurantoin and ertapenem. Multiplex PCR results showed, out of the 339 isolated strains, 164 (48.38%) were blaOXA-51, 157 (46.31%) were blaOXA-23, 18 (5.31%) were blaOXA-58, and no strain for blaOXA-24. 143 (47.67%) strains of blaOXA-51, 143 (47.67%) strains of blaOXA-23, and 14 (4.66%) strains of blaOXA-58 showed multidrug-resistant. In conclusion, our study found that OXA-51 and OXA-23 were the main mechanisms of resistant or sensitivity to carbapenems.
Keywords: Genomic diversity, OXA, Acinetobacter baumannii
Introduction
Acinetobacter baumannii is an aerobic non-motile gram-negative coccobacillus, and it has become an important hospital-acuired pathogen worldwide, such as pneumonia, septicemia and urinary tract as well as wound infections. Polymorphic bacterial pathogen of Acinetobacter baumannii is frequently involved in outbreaks, which can persist in the environment for several days [1]. Previous studies reported that Acinetobacter baumannii is a most common pathogenic bacteria isolated from hospitalized patients with pneumonia [2,3]. It is well known that Acinetobacter is a common nosocomial pathogen, and is widely found in intensive care units (ICUs) and can cause severe infections. However, Acinetobacter baumannii usually found to have multidrug resistant to many drugs, such as third generation cephalosporins, aminoglycosides and fluoroquinolone [4].
It is reported that multidrug-resistant Acinetobacter baumannii strains are gained their antibiotic-resistant genes through class 1 integrons that carry single or multiple gene cassettes [5]. Integrons are genetic elements to encode antibiotic resistance and integrate or mobilize their inherent gene cassettes [6]. Aminoglycoside resistance genes could influence the enzymatic inactivation of aminoglycoside antibiotics, including acetyltransferase, phosphotransferase and adenylyltransferase. Carbapenemase genes are important part of class I integrins, and they could cause carbapenem resistance, such as blaIMP, blaVIM, blaGIM, blaSIM, or blaOXA-like [7,8]. Carbapenemase production is the most well-described resistance mechanism to carbapenems [9]. The mechanisms of drug resistance are usually reported to be correlated with hydrolyzing β-lactamases of metallo-β-lactamases (Ambler class B) and oxacillinases (Ambler class D). Four subgroups of acquired CHDLs in Acinetobacter baumannii, including blaOXA-23, blaOXA-24, blaOXA-51 and blaOXA-58. In our study, we collected 196 strains of Acinetobacter baumannii to detect drug-resistant genes of OXA subgroups by Polymerase Chain Reaction (PCR) analysis, and conduct homology analysis.
Methods and materials
A total of 339 strains of Acinetobacter baumannii were isolated from 196 patients in the Central Hospital of Zhumadian between 2010 and 2014. All the isolated strains were identified as Acinetobacter baumannii using multiple Polymerase Chain Reaction (PCR) test.
Antibiotic susceptibility testing
General antimicrobial susceptibilities for A. baumannii identification were performed using Vitek 2 Compact system (bioMérieux, Inc., Marcy-l’Etoile, France). Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 27853), and Staphylococcus aureus (ATCC 29213) were taken as quality control strains. Susceptibility results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines.
PCR amplification
Two amplification bands were determined as Acinetobacter baumannii. Amplification of OXA genes, blaOXA-23, blaOXA-24, blaOXA-51 and blaOXA-58, were performed by Taq PCR Master Mix (Shanghai Lifefeng Biotech Co., Ltd, Shanghai, China). The PCR analysis were performed using the primers as follows: for blaOXA-23, the primer sequences were 5’-GATCGGATTGGAGAACCAGA-3’ (forwards) and 5’-ATTTCTGACCGCATTTCCAT-3’ (reverse); The primers for blaOXA-24 were 5’-TTCCCCTAACATGAATTTGT-3’ (forwards) and 5’-GTACTAATCAAAGTTGTGAA-3’ (reverse); The primers for for blaOXA-51 were 5’-TAATGCTTTGATCGGCCTTG-3’ (forwards) and 5’-TGGATTGCACTTCATCTTGG-3’ (reverse); The primers for blaOXA-58 were 5’-TGGCACGCATTTAGACCG-3’ (forwards) and 5’-AAACCCACATACCAACCC-3’ (reverse). The amplicon sizes for blaOXA-23, blaOXA-24, blaOXA-51 and blaOXA-58 were 501 bp, 1024 bp, 353 bp and 507 bp.
Culturing of strains
The isolated strains of Acinetobacter baumannii were stored at -70°C until use. The genomic DNA of Acinetobacter baumannii was extracted using TIANamp Bacteria DNA Kit (Tiangen, Beijing, China). PCR was performed using Taq PCR Master Mix (TaKaRa Bio, Dalian, China). Each PCR reaction mix was conducted in a 50 µL reaction solution, which contained 50 ng genomic DNA, 200 µM dNTP, 2.5 U Taq DNA polymerase (Promega, Madison, WI, USA), and 200 µM primers, in a total volume of 20 µL.OXA-51 and 16SrRNA was used as the internal control. The cycling conditions were performed with a preliminary denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 25 s, and annealing at 53°C for 40 s, with a final extension at 72°C for 6 min. The PCR products were verified by 1.0% agarose gel electrophoresis and visualized using ethidium bromide staining and UV light. Samples were coded for case-control status, and at least 10% of the samples were randomly selected and subjected to repeat analysis as quality control for verification of genotyping procedures. Two researchers independently reviewed all genotyping results.
Statistical analysis
Statistical analyses were conducted using the SPSS® statistical package, version 16.0 (SPSS Inc., Chicago, IL, USA) for Windows®. Categorical variables were expressed by frequency and percentage, and differences between categorical variables were compared by χ2 test or Fisher’s exact test. All P-values were two sided, and a P-values less than 0.05 are considered statistically significant.
Results
The characteristics of patients infected with Acinetobacter baumannii
The 349 strains of Acinetobacter baumannii were isolated from 196 patients, and the mean age of the 196 patients were 67.3 ± 11.5 years old (ranged from 31 to 81 years old). There were 115 (58.67%) males and 81 (41.33%) females from 196 selected patients (Table 1). Patients with Acinetobacter baumannii were selected from ICU, pneumology, emergency and cerebral surgery, accounting for 33.67%, 17.86%, 16.33% and 32.14%, respectively. 123 strains (86.62%) of clinical specimens were isolated from sputum, and 6 (4.23%) were isolated from cerebrospinal fluid.
Table 1.
Characteristics of included patients
| Variables | Number N = 196 | % |
|---|---|---|
| Age | ||
| < 60 | 38 | 19.39 |
| ≥ 60 | 158 | 80.61 |
| Gender | ||
| Male | 115 | 58.67 |
| Female | 81 | 41.33 |
| Location of acinetobacter baumannii | ||
| ICU | 66 | 33.67 |
| Emergency | 35 | 17.86 |
| Cerebral surgery | 32 | 16.33 |
| Pneumology | 63 | 32.14 |
Antimicrobial susceptibility of Acinetobacter baumannii
The antimicrobial susceptibility of 196 patients with Acinetobacter baumannii strain was shown in Table 2. Among the 196 patients with drug resistant strains, most strains showed resistance to different classes of agents, especially in ceftazidime, piperacillin, cefepime, nitrofurantoin and ertapenem.
Table 2.
Antibiotics resistance of multidrug-resistant Acinetobacter baumannii isolates
| Antibiotics | Patients with drug resistant strains | |
|---|---|---|
|
| ||
| Number | % | |
| Ampicillin | 162 | 82.65 |
| Piperacillin | 180 | 91.84 |
| Cefotetan | 146 | 74.49 |
| Cefazolin | 154 | 78.57 |
| Ceftriaxone | 143 | 72.96 |
| Ceftazidime | 183 | 93.37 |
| Cefepime | 178 | 90.82 |
| Cefoperazone | 42 | 21.43 |
| Imipenem | 147 | 75.00 |
| Aztreonam | 162 | 82.65 |
| Amikacin | 102 | 52.04 |
| Gentamicin | 129 | 65.82 |
| Tobramycin | 114 | 58.16 |
| Levofloxacin | 128 | 65.31 |
| Ciprofloxacin | 141 | 71.94 |
| Nitrofurantoin | 196 | 100.00 |
| Ertapenem | 196 | 100.00 |
| Meropenem | 150 | 76.53 |
Detection of OXA genes in Acinetobacter baumannii
The detection of blaOXA-51, blaOXA-23, blaOXA-24 and blaOXA-58 were shown in Table 3 and Figure 1. Of 339 isolated strains, 164 (48.38%) were blaOXA-51, 157 (46.31%) were blaOXA-23, 18 (5.31%) were blaOXA-58, and no strain for blaOXA-24. 143 (47.67%) strains of blaOXA-51, 143 (47.67%) strains of blaOXA-23, and 14 (4.66%) strains of blaOXA-58 showed multidrug-resistant. However, 21 (53.85%) stains of blaOXA-51, 14 (35.90%) strains of blaOXA-23, and 4 (10.26%) strains of blaOXA-58 showed non-multidrug resistant.
Table 3.
The PCR results of drug resistant genes
| Genes | Total | % | Multidrug-resistant N = 339 | % | Non-multidrug resistant N = 39 | % |
|---|---|---|---|---|---|---|
| blaOXA-51 | 164 | 48.38 | 143 | 47.67 | 21 | 53.85 |
| blaOXA-23 | 157 | 46.31 | 143 | 47.67 | 14 | 35.90 |
| blaOXA-24 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 |
| blaOXA-58 | 18 | 5.31 | 14 | 4.66 | 4 | 10.26 |
Figure 1.
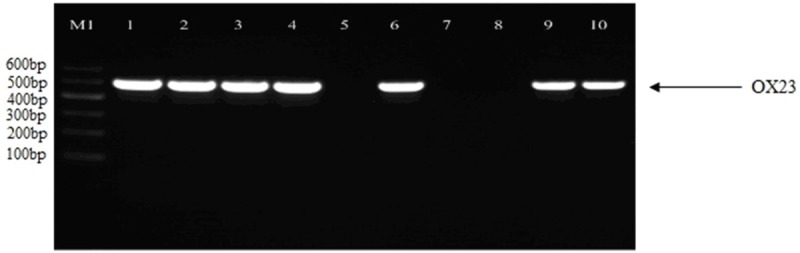
Detection of genes encoding blaOXA-23 gene PCR products. Moreover, we detected whether the Acinetobacter baumannii showed multidrug-resistant (Table 3). Of 339 isolated strains, 164 (48.38%) were blaOXA-51, 157 (46.31%) were blaOXA-23, 18 (5.31%) were blaOXA-58, and no strain for blaOXA-24. 143 (47.67%) strains of blaOXA-51, 143 (47.67%) strains of blaOXA-23, and 14 (4.66%) strains of blaOXA-58 showed multidrug-resistant.
Discussion
Acinetobacter baumannii causes a significant number of nosocomial outbreaks worldwide, which commonly occur in settings with high antibiotic selective pressures, such as ICUs. Most outbreak strains are highly resistant to antibiotics, and therefore therapeutic options are becoming increasingly limited [10]. Previous studies reported that blaOXA-23 and blaOXA-51 are the most common detected genes in Acinetobacter baumannii [11]. blaOXA-24 in Acinetobacter baumannii was reported to be detected in Spain and Iran [12,13]. blaOXA-58 was reported to be sequential outbreaks in a Saudia Arabia [14]. The present study finds that the main resistant genes in Acinetobacter baumannii were blaOXA-51 and blaOXA-23, and the main multidrug-resistant genes were found in ICU and pneumology departments.
Our study showed that the Acinetobacter baumannii presented multidrug resistant to different classes of agents, especially in ceftazidime, piperacillin, cefepime, nitrofurantoin and ertapenem. The main mechanism of drug resistant of Acinetobacter baumannii is due to four carbapenemases. One of the four carbapenemases includes blaOXA-23, blaOXA-24, blaOXA-51 and blaOXA-58. The OXA-51 is naturally existed and different with other acinetobacters [15].
In our study, 164 (48.38%) were blaOXA-51, 157 (46.31%) were blaOXA-23, 18 (5.31%) were blaOXA-58, and no strain for blaOXA-24. In a recent study, they detected the blaOXA-23 and blaOXA-58 in bacterial chromonsomes, but they also did not isolated strains of blaOXA-24 from patients [16]. The drug resistant rate of Acinetobacter baumannii to ceftazidime, piperacillin, cefepime, nitrofurantoin and ertapenem is above 90%. In a previous study, Vakili et al. reported that 95% of isolated strains showed multidrug resistant and 76.6% were high resistant, which were similar with the drug resistant with ours [17].
One previous study reported that the bla blaOXA-51-like and bla blaOXA-23-like were the main mechanisms of resistance to imipenem in Acinetobacter baumannii [12]. Another study in an Indian population reported that blaOXA-51-like and bla blaOXA-23-like are the main pathogen for carbapenem-resistant Acinetobacter [18]. However, some studies did not reported similar results with ours. One study in a Spanish population reported that all isolates of multidrug-resistant Acinetobacter baumannii contained the blaOXA-51-like and blaOXA-58-like genes [19]. Another study in a Chinese population reported that blaOXA-23-like are the most frequent carbapenem-resistant Acinetobacter baumannii in China, and blaOXA-24 and blaOXA-58 gene have become the potential threats of hospital outbreaks of multidrug-resistant Acinetobacter baumannii [20]. The discrepancies between studies may be due to differences in samples selection and gene variations in different ethnicities as well as sample size.
In conclusion, our study found that blaOXA-51 and blaOXA-23 were the main mechanisms of resistant or sensitivity to carbapenems. Drug resistance is increasing in Acinetobacter baumannii, and thus the resistance surveillance has become increasingly important to prevent the spread of carbapenem resistant Acinetobacter baumannii.
Disclosure of conflict of interest
None.
References
- 1.Metan G, Sariguzel F, Sumerkan B, Reijden TV, Dijkshoorn L. Clonal diversity and high prevalence of OXA-58 among Acinetobacter baumannii isolates from blood cultures in a tertiary care centre in Turkey. Infect Genet Evol. 2013;14:92–97. doi: 10.1016/j.meegid.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Lin YC, Sheng WH, Chen YC, Chang SC, Hsia KC, Li SY. Differences in carbapenem resistance genes among Acinetobacterbaumannii, Acinetobacter genospecies 3 and Acinetobacter genospecies 13TU in Taiwan. Int J Antimicrob Agents. 2010;35:439–443. doi: 10.1016/j.ijantimicag.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 3.Cefai C, Richards J, Gould FK, McPeake P. An outbreak of Acinetobacter respiratory tract infection resulting from incomplete disinfection of ventilatory equipment. J Hosp Infect. 1990;15:177–182. doi: 10.1016/0195-6701(90)90128-b. [DOI] [PubMed] [Google Scholar]
- 4.Sinha M, Srinivasa H. Mechanism of resistance to crabapenem-resistant Acinetobacter isolates from clinical sample. Ind J Med Microbiol. 2008;25:121–125. doi: 10.4103/0255-0857.32717. [DOI] [PubMed] [Google Scholar]
- 5.Petersen A, Guardabassi L, Dalsgaard A, Olsen JE. Class I integrons containing a dhfrI trimethoprim resistance gene cassette in aquatic Acinetobacter spp. FEMS Microbiol Lett. 2000;182:73–76. doi: 10.1111/j.1574-6968.2000.tb08876.x. [DOI] [PubMed] [Google Scholar]
- 6.Recchia GD, Hall RM. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 7.Srinivasan VB, Rajamohan G, Pancholi P, Stevenson K, Tadesse D, Patchanee P, Marcon M, Gebreyes WA. Genetic relatedness and molecular characterization of multidrug resistant Acinetobacter baumannii isolated in central Ohio, USA. Ann Clin Microbiol Antimicrob. 2009;8:21. doi: 10.1186/1476-0711-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sung JY, Kwon KC, Park JW, Kim YS, Kim JM, Shin KS, Kim JW, Ko CS, Shin SY, Song JH, Koo SH. Dissemination of IMP-1 and OXA type b-lactamase in carbapenemresistant Acinetobacter baumannii. Korean J Lab Med. 2008;28:16–23. doi: 10.3343/kjlm.2008.28.1.16. [DOI] [PubMed] [Google Scholar]
- 9.Poirel L, Pitout JD, Nordmann P. Carbapenemases: molecular diversity and clinical consequences. Future Microbiol. 2007;2:501–512. doi: 10.2217/17460913.2.5.501. [DOI] [PubMed] [Google Scholar]
- 10.Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, Pitt TL. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett. 2006;258:72–77. doi: 10.1111/j.1574-6968.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- 11.Kusradze Ia, Diene SM, Goderdzishvili M, Rolain JM. Molecular detection of OXA carbapenemases genes in multidrug-resistant Acinetobacter baumannii isolates from Iraq and Georgia. Int J Antimicrob Agents. 2011;38:164–168. doi: 10.1016/j.ijantimicag.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 12.Mohajeri P, Farahani A, Feizabadi MM, Ketabi H, Abiri R, Najafi F. Antimicrobial susceptibility profiling and genomic diversity of Acinetobacter baumannii isolates: a study in western Iran. Iran J Microbiol. 2013;5:195–202. [PMC free article] [PubMed] [Google Scholar]
- 13.Cayô R, Merino M, Ruiz Del Castillo B, Cano ME, Calvo J, Bou G, Martínez-Martínez L. OXA-207, a novel OXA-24 variant with reduced catalytic efficiency against carbapenems in Acinetobacter pittii from Spain. Antimicrob Agents Chemother. 2014;58:4944–4948. doi: 10.1128/AAC.02633-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aly M, Tayeb HT, Al Johani SM, Alyamani EJ, Aldughaishem F, Alabdulkarim I, Balkhy HH. Genetic diversity of OXA-51-like genes among multidrug-resistant Acinetobacter baumannii in Riyadh, Saudi Arabia. Eur J Clin Microbiol Infect Dis. 2014;33:1223–1228. doi: 10.1007/s10096-014-2068-0. [DOI] [PubMed] [Google Scholar]
- 15.Durante-Mangoni E, Zarrilli R. Global spread of drug-resistant Acinetobacter baumannii: molecular epidemiology and management of antimicrobial resistance. Future Microbiol. 2011;6:407–422. doi: 10.2217/fmb.11.23. [DOI] [PubMed] [Google Scholar]
- 16.Li C, Wang ZX, Shen JL. Detection of OXA carbapenemases and molecular mechanism in nosocomial outbreak caused by multi-drug resistant Acinetobacter baumannii strains. Chin J Clin Lab Sci. 2010;28:383–385. [Google Scholar]
- 17.Vakili B, Fazeli H, Shoaei P, Yaran M, Ataei B, Khorvash F, Khaleghi M. Detection of colistin sensitivity in clinical isolates of Acinetobacter baumannii in Iran. J Res Med Sci. 2014;19(Suppl 1):S67–70. [PMC free article] [PubMed] [Google Scholar]
- 18.Bali NK, Fomda BA, Bashir H, Zahoor D, Lone S, Koul PA. Emergence of carbapenem-resistant Acinetobacter in a temperate north Indian State. Br J Biomed Sci. 2013;70:156–160. doi: 10.1080/09674845.2013.11669950. [DOI] [PubMed] [Google Scholar]
- 19.Alvargonzalez JJ, Vindel Hernando A, Martín MD, Casas CM, Iglesias JO, Marín MF, Alvarez ML, Sanchez VB, Marí JM. Sequential outbreaks in a Spanish hospital caused by multiresistant OXA-58-producing Acinetobacter baumannii ST92. J Med Microbiol. 2014;63:1093–1098. doi: 10.1099/jmm.0.067280-0. [DOI] [PubMed] [Google Scholar]
- 20.Ji S, Chen Y, Ruan Z, Fu Y, Ji J, Fu Y, Wang H, Yu Y. Prevalence of carbapenem-hydrolyzing class D β-lactamase genes in Acinetobacter spp. isolates in China. Eur J Clin Microbiol Infect Dis. 2014;33:989–97. doi: 10.1007/s10096-013-2037-z. [DOI] [PubMed] [Google Scholar]


