Abstract
Background: Notch signaling is indicated as novel therapeutic targets to prevent recurrence of breast cancer. LncRNAs were identified as downstream target of Notch pathway. However, the exact mechanisms involved in Notch signaling, lncRNAs and breast cancer remain to be explained. Objective: This original research aimed to determine the prognostic implications of Notch-1 for breast cancer, and explain mechanisms involved in regulation of lnRNA GAS5 by Notch-1, and identify the function of this mechanism on breast cancer. Method: Thirty breast cancer patients were included from The First Affiliated Hospital of Anhui Medical University (China) since January 2006 in this study. The mRNA level by RT-PCR and protein level of Notch-1 by western blot in tumor tissues and adjacent normal tissues were evaluated and 5-year survival analysis was applied to examine the significance of Notch-1. The levels of ten reported lncRNAs were determined by RT-PCR, and subsequently linear analysis was applied to analyze the relationship between these four unique lncRNAs and protein level of Notch-1, which identified the most relevant lncRNA GAS5 with Notch-1 in breast cancer. Subsequently, Notch1-siRNA was applied to influence the expression of Notch-1 in T47D, then the level of RSA5 was measured by RT-PCR, and CCK-8 assay was applied to measure the proliferation of T47D cells. Results: High level of Notch-1 provided a poor prognosis in breast cancer. Interference of Notch-1 significantly suppressed proliferation of T47D cell (P < 0.05), and significantly increased the level of GAS5. Conclusion: Notch-1 promotes breast cancer cells proliferation by regulating LncRNA GAS5.
Keywords: Breast cancer patients, 5-year survival analysis, Notch-1, LncRNAs, GAS5, T47D cell, proliferation assay
Introduction
Breast cancer is the leading cause of cancer death in women aged 20 to 59 years, which is expected to be account for 29% of all new cancers in women [1]. Recently, radiotherapy, chemotherapy and surgery have been applied as common therapeutic modalities for patients with breast cancer [2]. These traditional chemo-radio therapeutic approaches could provide consequent disease remission, however, are toxic and of limited efficacy in the treatment of breast cancer [3]. Therefore, developing new and effective prognostic markers and therapeutic strategies is an urgent issue to improve treatment of breast cancer.
Notch signaling was indicated as novel therapeutic targets to prevent recurrence of pre-invasive and invasive breast cancer [4]. Notch signaling, which included four receptors (Notch1-Notch4) [5], regulated cell fate during development, stem cell renewal and differentiation in postnatal tissues [6]. Notch-1 was reported to be intimately linked to the behaviors of breast cancer stem cells (BCSCs), and blocking Notch-1 signaling could inhibit the malignant behaviors of BCSCs [7]. Furthermore, Qun et al. suggested that Notch-1 promoted gastric cancer formation through up-regulating the expression of lncRNA AK022798 expression, which indicated that notch-1 signaling system might involve in the regulation of lncRNAs [8]. However, the exact system has not been well documented.
The development of high resolution microarray and genome wide sequencing technologies have revealed that human genomes with protein coding mRNAs constitute only a minor fraction of the transcribed sequences, while more than 70% is transcribed as noncoding RNAs (ncRNAs) [9]. These ncRNAs function as regulators of gene expression, which identified a novel RNA-based gene regulation mechanism that complements the central dogma [10].
Long noncoding RNAs (lncRNAs, > 200 nt) belong to ncRNAs [11], which still remain poorly understood, however, evidence for their importance and functionality is mounting. Emerging studies suggest that lncRNAs play critical roles in numerous biological processes, including tumorigenesis [12], cell growth [13] and transcriptional regulation [14], and are associated with a number of diseases involved in cancer [15], cardiovascular disease [16] and neurodegeneration disease [17]. According to the lncRNA Disease Database [18], there are 17 lncRNAs known to play a role in breast cancer (Table 1) including BCAR4, PVT1, MALAT1 and GAS5. Recently, other lncRNAs were also reported to be associated with breast cancer, including UCA1 [19], LUNAR1 [20], POU3F3 [21] and HIF1A-AS1 [22].
Table 1.
The lncRNAs reported to be associated with breast cancer
| Index | LncRNA name | Dysfunction type | Species |
|---|---|---|---|
| 2 | BCYRN1 | Expression | Human |
| 3 | CDKN2B-AS1 | Expression | Human |
| 4 | DSCAM-AS1 | Expression/Locus/Mutation | Human |
| 5 | GAS5 | Expression | Human |
| 6 | H19 | Expression/regulation | Human |
| 7 | HOTAIR | Expression/Mutation/regulation | Human |
| 8 | Loc554202 | Regulation/Expression | Human |
| 9 | LSINCT5 | Regulation | Human |
| 10 | MALAT1 | Expression | Human |
| Regulation | Human | ||
| 11 | MEG3 | Expression | Human |
| 12 | MIR31HG | Epigenetics | Human |
| 13 | PINC | N/A | Mouse |
| 14 | PVT1 | Expression | Human |
| 15 | SRA1 | Regulation/Locus | Human |
| 16 | XIST | Mutation | Human |
| 17 | ZNFX1-AS1 | Expression/Locus | Human |
| 18 | UCA1 | Regulation | Human |
| 19 | LUNAR1 | Regulation | Human |
| 20 | POU3F3 | Regulation | Human |
| 21 | HIF1A-AS1 | Regulation | Human |
| 22 | 7SK | Regulation | Human |
In this study, we examined the prognostic role of Notch-1 in breast cancer, which indicated that high expression of Notch-1 would provide poor prognosis. And we aimed to investigate the most relevant lncRNA (GAS5) with Notch-1 in breast cancer, and further explored the clinical significance and biological functions of Notch-1 and GAS5 in cancer cells.
Materials and methods
Patients and specimens
The original research was approved by the Medical ethics committee of “The First Affiliated Hospital of Anhui Medical University” and the informed consent letters were obtained from patients. A total of 30 breast cancer patients were included from The First Affiliated Hospital of Anhui Medical University (China) since January 2006 in this study. All patients recruited in this study were not subjected to preoperative radiotherapy and/or chemotherapy. All patients were regularly followed up, with up to 100 months after the surgery. Tumor specimens and corresponding adjacent normal tissues were collected and stored in liquid nitrogen until use.
Cell culture
The human breast cancer cell lines were purchased from the American Type Culture Collection (ATCC, USA) including MDA-MB-453, MDA-MB-231, MCF-7 and T47D. All the cells were maintained in DMEM/F-12 (1:1) medium (HyClone, USA) containing 10% fetal bovine serum (FBS, Hyclone, USA) and 1% penicillin-streptomycin solution (100×, Beyotime, China) at 37°C in a humidified atmosphere containing 5% CO2.
SiRNA transfection
Small interfering RNA that targeted Notch1-RNA (Notch1-siRNA) and a scrambled negative control (Scrambled-SiRNA) were generously provided by Life Technologies. Human breast cancer cells were transfected with either 50 nmol Notch1-siRNA or Scrambled-SiRNA using Lipofectamine 2000 transfection reagent according to the manufacturer’s protocol (Life Technologies). The sequences were as follows:
Notch1, Forward 5’-GTCTCCATTGCTAGCCAC-3’ and Reverse 5’-ATGCAGCTGCAGGTCTTAAGAG-3’; GAPDH, Forward 5’-ACAGGGGAGGTGATAGCATT-3’ and Reverse 5’-GACCAAAAGCCTTCATACATCTC-3’.
Notch1-SiRNA: GUC CAG GAA ACA ACU GCA ATT (sense) and UUG CAG UUG UUU CCU GGA CTT (antisense).
Scrambled-SiRNA: UUC UCC GAA CGU GUC ACG UTT (sense) and ACG UGA CAC GUU CGG AGA ATT (antisense).
Quantitative real-time PCR assay
Total RNA was isolated from tissues using TRIZOL reagent (Invitrogen) or cells using Trizol plus kit (TaKaRa, Japan) according to the manufacturer’s protocol. RNA was reverse transcribed using SuperScript First Strand cDNA System (Invitrogen) according to the manufacturer’s instructions. The PCR amplification were performed for 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, on a Applied Biosystems 7900HT (Applied Biosystems) with 1.0 μl of cDNA and SYBR Green Real-time PCR Master Mix (Takara). Data was collected and analyzed by SDS2.3 Software (Applied Biosystems). The expression level of each sample was internally normalized against that of the GAPDH. The relative quantitative value was expressed by the 2-ΔΔCt method. Each experiment was performed in triplicates.
Western blot analysis
Tissues or cells were rinsed with cold PBS and harvested in lysis buffer. Then, the extractions were obtained and then centrifuged at 14,000 rpm for 30 min. Twenty-five micrograms of protein was loaded per lane and separated by 10% SDS-PAGE, then transferred to nitrocellulose membranes and blocked overnight in Trisbuffered saline containing 0.1% Tween and 5% skim milk. Then, the membrane was incubated with primary antibodies (Notch 1, and β-actin) at 4°C and subsequently incubated with a secondary antibody for 2 h at room temperature. The detection of specific proteins was carried out using an ECL western blotting kit (Amersham Biosciences, Piscataway, New Jersey, USA) according to the recommended procedure.
Cell proliferation assay
The CCK-8 assay kit (Beyotime Biotech Company) was used to determine the proliferation of T47D cells. In each 96-well plate, the cells (1×105) were seeded and cultured for 24 h, then transfected with Scrambled-SiRNA or Notch1-siRNA, and further incubated for 0 h, 12 h, 24 h, 36 h and 48 h respectively. After 10 μL CCK-8 reagents had been added to each well, the plate was re-incubated for 1 h and the absorbance at 450 nm was subsequently detected. In each group, five wells were measured for cell proliferation, and all of the independent groups were performed in triplicate.
Statistical analysis
All computations were carried out using the software of SPSS version 18.0 for Windows. Data were expressed as mean ± SD. The data were analyzed using independent two-tailed t test. Categorical data were analyzed using the two-side chi-square test. Overall survival was estimated by using Kaplan-Meier method, and univariate analysis was conducted by log-rank test. The Cox proportional hazards model was used in the multivariate analysis. Values of P < 0.05 were considered statistically significant.
Results
Breast cancer tumors highly expressed Notch-1
The mRNA and protein level of Notch-1 in tumors and adjacent normal tissues were measured, which revealed that the level of Notch-1 increased significantly (about 3 folds) than adjacent normal tissues (Figure 1). This indicated that Notch-1 might be a potential biomarker for breast cancer. Subsequently, according to the protein level of Notch-1, we divided these patients into two groups, including Notch1-high group and Notch1-low group. After the follow-up of about 100 months, the 5-year survival analysis showed that Notch1-high group has a significantly poorer prognosis (28% of survival rate) than Notch1-low group (57% of survival rate), which indicated that Notch-1 might play a pivotal role in breast cancer.
Figure 1.
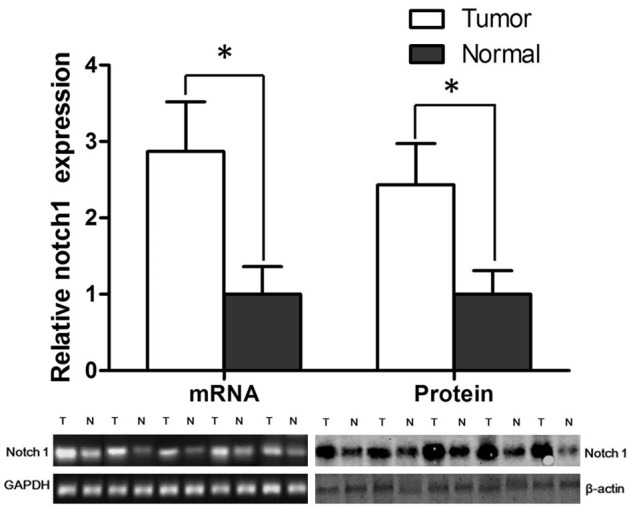
The mRNA level and protein level of Notch-1 in breast tumor tissues and their adjacent normal tissues. *P < 0.05.
LncRNA GAS5 was identified as the most relevant one with Notch-1 in tumor tissues
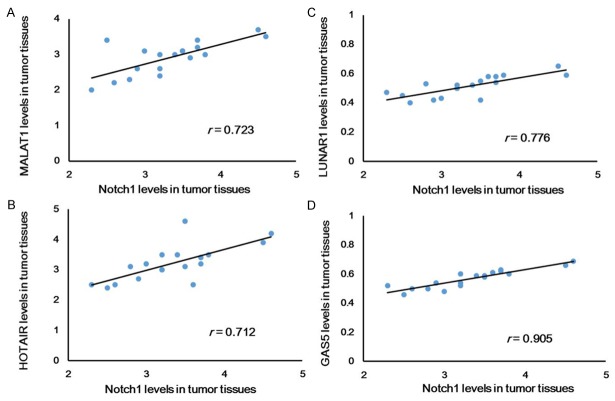
Ten different lncRNAs were applied in this study, which was reported to be associated with Notch-1. The level of these ten lncRNAs in breast tumors and adjacent normal tissues were evaluated (Figure 2), which suggested that HOTAIR and MALAT1 increased most significantly, and LUNAR1 and GAS5 decreased most significantly. This indicated that these four lncRNAs might be most relevant with breast cancer. Subsequently, the mRNA level of these four lncRNAs and the protein level of Notch-1 in tumor tissues was analyzed based on linear analysis, which suggested that GAS5 and Notch-1 mostly conformed (r = 0.905) to the linear relationship (Figure 3). This indicated that there might be a strong association between Notch-1 and GAS5 in breast cancer.
Figure 2.
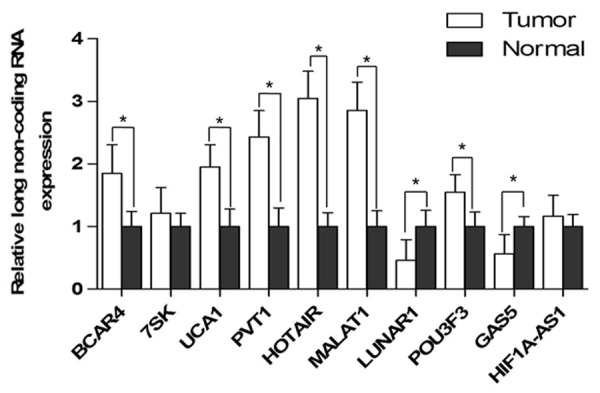
The mRNA level of LncRNAs in tumors and adjacent normal tissues. *P < 0.05.
Figure 3.
Linear analyses of lncRNAs and Notch1 in breast cancer tumors. *P < 0.05.
Interference of Notch1 up-regulates the expression of GAS5 in T47D cells and suppresses the proliferation of T47D cell lines
Further investigation in cell lines was carried out. The expression of Notch-1 and GAS5 was investigated in four breast cancer cell lines including MDA-MB-453, MDA-MB-231, MCF-7 and T47D, which showed that T47D was a suitable cell line model for further investigation. The reasons were as follows: (i) the expression in MDA-MD-453 was regarded as the comparable object, then the expressions of Notch-1 in MCF7 and T47D was expressed lower than MDA-MB-453 (Figure 4A); (ii) but only the mRNA level of GAS5 in T47D cell lines was higher than that in MDA-MB-453 (Figure 4B).
Figure 4.
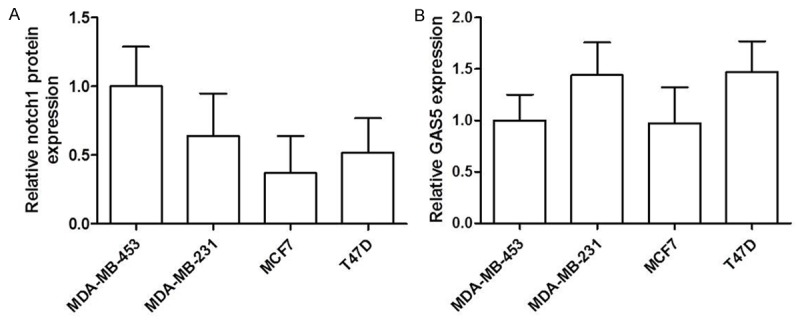
The protein level of Notch-1 (A) and the mRNA level of GAS5 (B) in four breast cancer cell lines including MDA-MB-453, MDA-MB-231, MCF-7 and T47D. *P < 0.05.
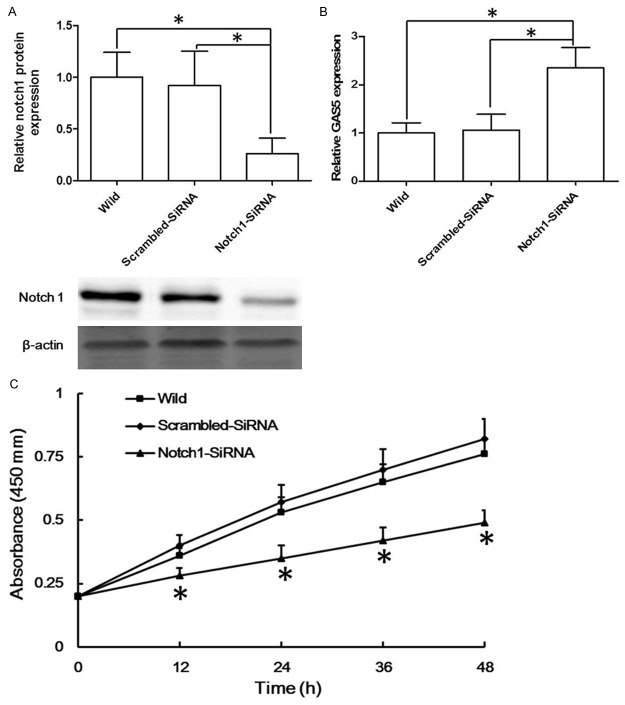
Subsequently, Notch1-siRNA was applied to influence the expression of Notch1 in T47D cell lines. As Figure 5A shown, Notch1-siRNA significantly decreased the protein level of Notch1 in T47D cell lines. Then the mRNA level of GAS5 in Notch1-siRNA significantly increased (more than 2 folds) compared with that in scrambled-siRNA group or that in wild group (Figure 5B).
Figure 5.
After interference of the expression of Notch1 in T47D cell lines, the expression of Notch-1 (A), the level of GAS5 (B) was measured, *P < 0.05. Then the proliferation assay was applied to identify the proliferation of T47D cell lines (C), *P < 0.05 compared with the wild group.
The cell proliferation assay was applied to further confirm the role of Notch-1 in breast cancer cells. These cell lines were incubated for 48 h following the interference of Notch-1 in T47D cell lines, which showed that the interference of Notch-1 suppressed T47D cell proliferation significantly (Figure 5C).
Discussion
The function of Notch-1 on cancers has been investigated for decades, but the exact mechanisms remained to be explained. Co-expression of Notch-1 and vascular endothelial growth factor-A were reported to predict poor survival in nonsmall cell lung cancer [23]. However, Jiayuan et al. showed that patients with positive Notch-1 expression had a prolonged progression of overall survival compared with those with negative Notch-1 expression [24]. We demonstrate that the expression of Notch-1 was significant higher in tumor tissues than the adjacent normal tissues. And 5-year survival analysis in our study based on Western Bot has been applied to confirmed that high level of Notch-1 provide poor prognosis for breast cancer. Additional studied has confirmed the role of Notch-1 in breast cancer. The mRNA expression of Notch-1 based on in-situ hybridization showed that patients with high level of Notch-1 had a 5-year survival rate of 42% compared with patients with low level of Notch-1 with a 5-year survival rate of 65% [25]. And Notch-1 signaling was proven to promote the malignant behaviors of breast cancer stem cells (BCSCs) [7]. Although many studies have investigated the mechanisms of Notch-1 signaling, most of them focused on the factors regulating Notch-1 signaling, including Silibinin [26], Genistein [27] and p53 [28,29]. Our study examined the strong association between lncRNAs and Notch-1, and suggested that GAS5 identified as downstream target of Notch-1 signaling in breast cancer.
Only a few studies investigated the mechanisms involved in lncRNAs and Notch-1. In 2007, Tsutsumi and Itoh suggested that lncRNAs might be novel downstream target genes of Notch pathway by conducting a microarray screen in zebrafish [30]. Tang et al. identified the cross talk between Notch signaling and lncRNAs on the fate of stem cells [31]. Recently, Trimarchi et al. confirmed that lncRNAs are important downstream targets of the Notch signaling pathway, and additionally they are key regulators of the oncogenic state in acute leukemia [32]. Furthermore, Hang et al. indicated that Notch-1 promoted the expression lncRNA AK022798 and subsequently leaded to the formation of SGC7901/DDP and BGC823/DDP cells in gastric cancer [8]. Our group suggested the possibility that Notch-1 played a role in breast cancer by regulating lncRNAs, and we found the high correlation of lncRNA GAS5 (r = 0.907) with Notch-1. Mourtada et al. suggested that GAS5 transcript levels were significantly reduced in breast cancer samples relative to adjacent unaffected normal breast epithelial tissues [33]. Unfortunately, the emerging functional role of GAS5 in breast cancer remains largely unknown. Our results revealed that GAS5 was regulated by Notch-1 involved in the proliferation of breast cancer cells. The level of GAS5 was increased significantly by the down-regulation of Notch-1 in T74D cells, and consequently the proliferation of these cells was significantly inhibited. This regulation might explain the prognostic role of Notch-1 in breast cancer patients, however, the exact mechanisms remained to be identified. Breast cancer remains to be one of the leading causes of death, the crosstalk between Notch signaling and lncRNAs might provide new therapeutic method.
Acknowledgements
We acknowledge the support from patients and their family, and the financial support from The First Affiliated Hospital of Anhui Medical University (China).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Hart CD, Migliaccio I, Malorni L, Guarducci C, Biganzoli L, Leo AD. Challenges in the management of advanced, ER-positive, HER2-negative breast cancer. Nat Rev Clin Oncol. 2015;12:541–52. doi: 10.1038/nrclinonc.2015.99. [DOI] [PubMed] [Google Scholar]
- 3.Yallapu MM, Jaggi M, Chauhan SC. Curcumin nanomedicine: a road to cancer therapeutics. Curr Pharm Des. 2013;19:1994–2010. doi: 10.2174/138161213805289219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farnie G, Clarke RB. Mammary stem cells and breast cancer--role of Notch signalling. Stem Cell Rev. 2007;3:169–175. doi: 10.1007/s12015-007-0023-5. [DOI] [PubMed] [Google Scholar]
- 5.Miele L. Notch signaling. Clin Cancer Res. 2006;12:1074–1079. doi: 10.1158/1078-0432.CCR-05-2570. [DOI] [PubMed] [Google Scholar]
- 6.Espinoza I, Miele L. Notch inhibitors for cancer treatment. Pharmacol Ther. 2013;139:95–110. doi: 10.1016/j.pharmthera.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng GL, Tian Y, Lu C, Guo H, Zhao XW, Guo YW, Wang LQ, Du QL, Liu CP. Effects of Notch-1 down-regulation on malignant behaviors of breast cancer stem cells. J Huazhong Univ Sci Technolog Med Sci. 2014;34:195–200. doi: 10.1007/s11596-014-1258-4. [DOI] [PubMed] [Google Scholar]
- 8.Hang Q, Sun R, Jiang C, Li Y. Notch 1 promotes cisplatin-resistant gastric cancer formation by upregulating lncRNA AK022798 expression. Anticancer Drugs. 2015;26:632–640. doi: 10.1097/CAD.0000000000000227. [DOI] [PubMed] [Google Scholar]
- 9.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Zhao J, Zhang W, Gan J, Hu C, Huang G, Zhang Y. lncRNA GAS5 enhances G1 cell cycle arrest via binding to YBX1 to regulate p21 expression in stomach cancer. Sci Rep. 2015;5:10159. doi: 10.1038/srep10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yarmishyn AA, Kurochkin IV. Long noncoding RNAs: a potential novel class of cancer biomarkers. Front Genet. 2015;6:145. doi: 10.3389/fgene.2015.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M, Wu F, Mo YY. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013;20:1558–1568. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiyomaru T, Yamamura S, Fukuhara S, Yoshino H, Kinoshita T, Majid S, Saini S, Chang I, Tanaka Y, Enokida H, Seki N, Nakagawa M, Dahiya R. Genistein inhibits prostate cancer cell growth by targeting miR-34a and oncogenic HOTAIR. PLoS One. 2013;8:e70372. doi: 10.1371/journal.pone.0070372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA. 2010;16:324–337. doi: 10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Wan L, Lu K, Sun M, Pan X, Zhang P, Lu B, Liu G, Wang Z. The Long Noncoding RNA MEG3 Contributes to Cisplatin Resistance of Human Lung Adenocarcinoma. PLoS One. 2015;10:e0114586. doi: 10.1371/journal.pone.0114586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma Y, Ma W, Huang L, Feng D, Cai B. Long non-coding RNAs, a new important regulator of cardiovascular physiology and pathology. Int J Cardiol. 2015;188:105–110. doi: 10.1016/j.ijcard.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 17.Roberts TC, Morris KV, Wood MJ. The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos Trans R Soc Lond B Biol Sci. 2014:369. doi: 10.1098/rstb.2013.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X, Zhang Q, Yan G, Cui Q. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M, Mo YY. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1) Cell Death Dis. 2014;5:e1008. doi: 10.1038/cddis.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Zang C, Liu XS, Aster JC. The role of Notch receptors in transcriptional regulation. J Cell Physiol. 2015;230:982–988. doi: 10.1002/jcp.24872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plourde KV, Labrie Y, Desjardins S, Belleau P, Ouellette G, Durocher F, BRCAs I. Analysis of ZNF350/ZBRK1 promoter variants and breast cancer susceptibility in non-BRCA1/2 French Canadian breast cancer families. J Hum Genet. 2013;58:59–66. doi: 10.1038/jhg.2012.127. [DOI] [PubMed] [Google Scholar]
- 22.Zhang QQ, Xu MY, Qu Y, Hu JJ, Li ZH, Zhang QD, Lu LG. TET3 mediates the activation of human hepatic stellate cells via modulating the expression of long non-coding RNA HIF1A-AS1. Int J Clin Exp Pathol. 2014;7:7744–7751. [PMC free article] [PubMed] [Google Scholar]
- 23.Donnem T, Andersen S, Al-Shibli K, Al-Saad S, Busund LT, Bremnes RM. Prognostic impact of Notch ligands and receptors in nonsmall cell lung cancer: coexpression of Notch-1 and vascular endothelial growth factor-A predicts poor survival. Cancer. 2010;116:5676–5685. doi: 10.1002/cncr.25551. [DOI] [PubMed] [Google Scholar]
- 24.Huang J, Song H, Liu B, Yu B, Wang R, Chen L. Expression of Notch-1 and its clinical significance in different histological subtypes of human lung adenocarcinoma. J Exp Clin Cancer Res. 2013;32:84. doi: 10.1186/1756-9966-32-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reedijk M, Odorcic S, Chang L, Zhang H, Miller N, McCready DR, Lockwood G, Egan SE. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 26.Kim TH, Woo JS, Kim YK, Kim KH. Silibinin induces cell death through reactive oxygen species-dependent downregulation of notch-1/ERK/Akt signaling in human breast cancer cells. J Pharmacol Exp Ther. 2014;349:268–278. doi: 10.1124/jpet.113.207563. [DOI] [PubMed] [Google Scholar]
- 27.Pan H, Zhou W, He W, Liu X, Ding Q, Ling L, Zha X, Wang S. Genistein inhibits MDA-MB-231 triple-negative breast cancer cell growth by inhibiting NF-kappaB activity via the Notch-1 pathway. Int J Mol Med. 2012;30:337–343. doi: 10.3892/ijmm.2012.990. [DOI] [PubMed] [Google Scholar]
- 28.Yun J, Espinoza I, Pannuti A, Romero D, Martinez L, Caskey M, Stanculescu A, Bocchetta M, Rizzo P, Band V, Band H, Kim HM, Park SK, Kang KW, Avantaggiati ML, Gomez CR, Golde T, Osborne B, Miele L. p53 Modulates Notch Signaling in MCF-7 Breast Cancer Cells by Associating with the Notch Transcriptional Complex via MAML1. J Cell Physiol. 2015;230:3115–27. doi: 10.1002/jcp.25052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Timani KA, Liu Y, Fan Y, Mohammad KS, He JJ. Tip110 Regulates the Cross Talk between p53 and Hypoxia-Inducible Factor 1alpha under Hypoxia and Promotes Survival of Cancer Cells. Mol Cell Biol. 2015;35:2254–2264. doi: 10.1128/MCB.00001-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsutsumi M, Itoh M. Novel transcript nort is a downstream target gene of the Notch signaling pathway in zebrafish. Gene Expr Patterns. 2007;7:227–232. doi: 10.1016/j.modgep.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Tang Y, Wang Y, Chen L, Pan Y, Weintraub N. Cross talk between the Notch signaling and noncoding RNA on the fate of stem cells. Prog Mol Biol Transl Sci. 2012;111:175–193. doi: 10.1016/B978-0-12-398459-3.00008-3. [DOI] [PubMed] [Google Scholar]
- 32.Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Dalla-Favera R, Tsirigos A, Aifantis I. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158:593–606. doi: 10.1016/j.cell.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]