Abstract
MicroRNAs are implicated in the regulation of gene expression via various mechanisms in health and disease, including fibrotic processes. Pterygium is an ocular surface condition characterized by abnormal fibroblast proliferation and matrix deposition. We aimed to investigate the role of microRNAs in pterygium and understand the relevant cellular and molecular mechanisms. To achieve this objective, a combination of approaches using surgically excised paired human pterygium and conjunctival tissues as well as cultured primary fibroblast cells from tissue explants were evaluated. Fibroblast dysfunction has been shown to play a central role in pterygium pathology. Here we show that miR-215, among a few others, was down-regulated (2-fold) in pterygium compared to control, and this was consistent in microarray, real-time PCR and fluorescent in-situ hybridization. The effects of increased miR-215 were investigated by adding exogenous miR-215 to fibroblasts, and this showed a decrease in cell proliferation but no significant apoptosis compared to control. Further cell cycle analysis showed that miR-215 depressed progression of cells at G1/S as well as G2/M. A few cell cycle related transcripts were downregulated (2.2–4.5-fold) on addition of miR-215: Mcm3, Dicer1, Cdc25A, Ick, Trip13 and Mcm10. Theoretic binding energies were used to predict miR-215 binding targets and luciferase reporter studies confirmed Mcm10 and Cdc25A as direct targets. In summary, mir-215 could play a role in inhibiting fibroblast proliferation in ocular surface conjunctiva. Dampening of this mir-215 could result in increased fibroblast cell cycling and proliferation, with possibly increased fibroblastic production of matrix, inducing pterygium formation.
Keywords: cell cycle, fibroblast, microRNA, ocular surface, pterygium
Introduction
Endogenously produced microRNAs are small, single-stranded, non-coding RNA molecules that regulate post-transcriptional gene expression. Each microRNA forms complementary base pairs within the 3′ untranslated region (UTR) of multiple target messenger(m)RNAs, promoting mRNA degradation or translational repression.1 Most microRNAs are located in the intergenic non-coding regions of the genome where they require a unique RNA polymerase and transcriptional regulators for independent tissue-specific and developmental stage-specific transcription.2 Alternatively, microRNAs may be found within gene sequences where they are generally transcribed together with their host genes.2,3
MicroRNAs participate in a vast range of developmental and physiological processes, including fibrosis,4,5 cell proliferation,6 apoptosis,7 differentiation,8 metabolism,9 and angiogenesis.10 Because of their extensive involvement in cellular processes, derangement of microRNA is associated with disease formation such as in tumorigenesis,11,12 inflammatory diseases13,14 and defective neuronal development.15
Nucleotide arrays16-20 and other studies have detected microRNAs in the eye,16,17 and elucidated their roles in development and differentiation16-18 of the retina,7,19,21 lens20 and cornea.16 In humans, microRNAs have been implicated in various diseases,11,22-24 including neovascularization25 and fibrosis. However, the involvement of microRNAs in ocular surface fibrosis have been limited to descriptive studies of microRNA levels in diseases,26 or induction by laser,27 exogenous growth factor28,29 or immunosuppressive drugs30,31 in non-disease derived cultured ocular surface fibroblasts.
Pterygium is a relatively common ocular surface disease characterized by a wedge-shaped lesion growing centripetally from the conjunctiva to the cornea, affecting up to 200 million people globally.32 The pathogenesis of pterygium is largely unknown, although it is thought to be associated with overexposure to ultraviolet light.33 Studies have linked aberrant cell proliferation, defects in wound healing,34 cell transformation35 and matrix remodeling,36,37 angiogenesis,38 oxidative stress mechanisms39 and genetic susceptibility40 to the formation of pterygium. The definitive treatment for pterygium is surgical resection combined with conjunctival autograft, mitomycin C administration or other adjunctive forms of treatment, as bare sclera excision is associated with high recurrence rates.41
Pterygium is a good and relevant model for dissecting molecular control of fibrosis. The clinical severity of pterygium is normally categorized into T1–3, based on the fleshiness or amount of fibrous tissue.42 The clinical staging is related to biological activity as higher grade pterygium is associated with higher recurrence rates.42 Fibroblast proliferation in pterygium has been previously documented,33 which leads to increased matrix production, resulting in a ′fleshy lesion′ to varying extents. A molecular pathway that has been shown to result in fibroblast proliferation is the up-regulation of the stromal cell-derived factor 1 (SDF-1) and its interaction with the chemokine receptor CXCR4.43
There is good accessibility of human pterygium tissues through surgical excision by virtue of its anatomical position, compared to other organs affected by fibrosis such as the liver and kidney. Due to accessibility of the lesion, there is greater potential for findings to be translated to medicine using local injection of microRNA intervention, limiting issues of systemic toxicity.44 MicroRNAs have been implicated in the development of fibrosis via extracellular matrix and proliferative changes especially in liver, kidney and lung fibrotic diseases.5,45,46
In this study, we aimed to ascertain the role of microRNAs in fibroblast function, using human pterygium as a model. Here we found miR-215, a conserved microRNA previously unreported in pterygium, to regulate cell cycling in pterygium-derived fibroblasts.
Results
Seven microRNAs were significantly dysregulated in pterygium
Analysis of the Exiqon miRCURY LNATM microRNA Array found 6 microRNAs to be up-regulated and one down-regulated significantly in pterygium compared to conjunctival control tissue (Table 1). For validation, microRNAs with greater than 1.5-fold differences (P < 0.05) between pterygium and conjunctival samples obtained in microarray analyses were selected. Among the validated microRNA changes, miR-215 was selected for further studies.
Table 1.
MicroRNAs dysregulated in pterygium, presented as fold change over conjunctiva levels (microarray data).
| Annotation | Fold change | p-value |
|---|---|---|
| hsa-miR-138 | 3.024437 | 0.019 |
| hsa-miRPlus-E1233 | 2.3784142 | 0.019 |
| hsa-miR-215 | −1.8617594 | 0.028 |
| hsa-miR-518b | 1.7451285 | 0.032 |
| hsa-miR-1236 | 1.6934906 | 0.047 |
| hsa-miRPlus-E1258 | 1.6548113 | 0.044 |
| hsa-miR-766 | 1.5764362 | 0.017 |
Expression of miR-215 in pterygium
MiR-215 was found to be downregulated (0.54-fold change) in pterygium compared to control conjunctiva in the Exiqon microRNA array (Fig. 1A). To confirm the dysregulation in pterygium, qRT-PCR (Fig. 1B) was performed using 3 samples of paired human pterygium and normal conjunctival tissues from new donors. qRT-PCR results showed that 100% of the pterygium samples displayed down-regulation ranging from 0.30 to 0.80-fold, averaging 0.49-fold in pterygium relative to uninvolved conjunctiva (P < 0.05).
Figure 1.
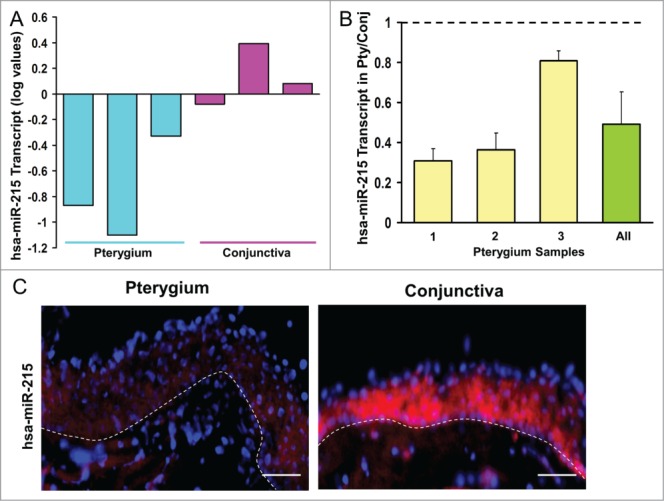
(A) Bar chart showing results of the Exiqon microRNA Array. Height of the bars shows the mean normalized log values of miR-215 levels in conjunctiva and pterygium tissues from different patients. (B) Bar chart showing individual and overall results from qRT-PCR assay performed on 3 separate pairs of human pterygium and conjunctival tissues. Height of the bars represents the relative fold change of miR-215 levels in pterygium with respect to conjunctival control. Error bars represent standard error of mean. Values are normalized to the 5S rRNA control values. (C) miR-215 in situ staining of human pterygium and conjunctival tissues. Diagram on the right showing conjunctival epithelium and some stroma. Pterygium and conjunctival sections hybridized with DIG-labeled miR-215 LNA probes (red). DNA was counterstained with DAPI (blue). Scale bar represents 100 μm. Dashed lines represent position of basement membrane separating epithelium from stroma.
The microRNA results were further confirmed by fluorescent in situ hybridization (FISH) on the tissue sections from another 3 donors (Fig. 1C). MiR-215 was localized to the epithelial as well as the stromal layer of pterygium with a less intense red fluorescent staining in pterygium compared to conjunctival tissue. The presence of miR-215 in the stromal layer, in addition to epithelial cells, suggests the involvement of this microRNA in fibroblasts. We accept that it is difficult to determine differential expression levels from the staining in the stroma. Based on these results we proceeded to evaluate the effects of mir-215 by using a mir-215 mimic in a cell-based assay.
MiR-215 reduced cell impedance
When miR-215 mimic was added to cultured primary pterygium fibroblast cells and observed over 48 h, fibroblast cells showed a markedly reduced cell index, which is a measure of cell impedance (Fig. 2). At 24 h, fibroblast cells treated with miR-215 mimic had a 0.63-fold reduction in cell index (p = 0 .02) compared to random oligonucleotide control-transfected cells, and 0.29-fold at 48 h (p = 0 .01). An increase in cell impedance can be attributed to a few properties, mainly involving proliferative and/or adhesive changes, but at time intervals of 24–48 h, the impedance is mainly a measure of cell numbers or proliferation rates.47
Figure 2.
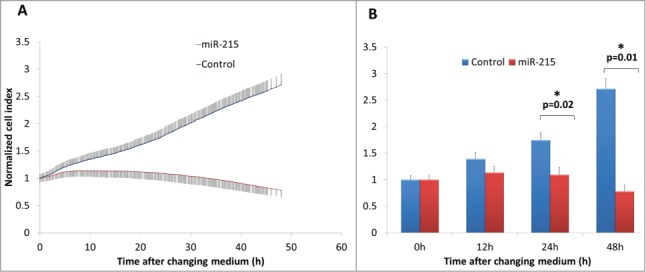
(A) Real-time cell impedance index measured using the xCELLigence cell impedance system. Cells are transfected with 100 nM miR-215 mimic (red) or non-specific oligonucleotide control (blue) and medium changed at 6 h. Error bars show standard deviation across 3 biological samples in 3 separate experimental replicates. (B) Bar chart representing cell indices of miR-215 or control-transfected cells as described above at various time points after changing medium. Error bars show standard deviation across 3 biological samples in 3 separate experimental replicates.
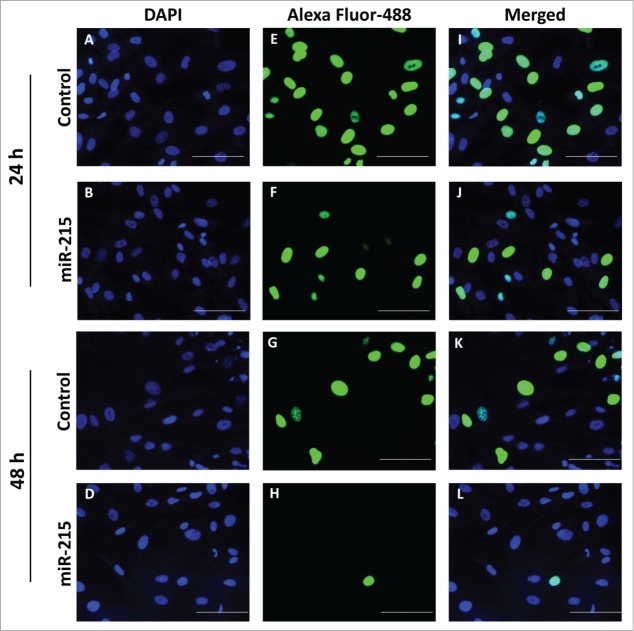
To confirm that the impedance findings were mainly due to cell proliferation, we corroborated the findings using the 5-ethynyl-2'-deoxyuridine (EdU) cell proliferation assay. Cultured primary pterygium fibroblast cells transfected with miR-215 mimic showed significantly reduced replication activity, measured as the ratio of replicating nuclei (Alexa Fluor 488-labeled) over total number of nuclei (DAPI-labeled) (Fig. 3). The number of replicating nuclei is significantly reduced at 48 h of transfection with miR-215 mimic (0.05-fold, p = 0 .007) (Fig. 3K and L) compared to 24 h (0.57-fold, p = 0 .048) (Fig. 3I and J).
Figure 3.
Cell proliferation was analyzed using the Click-it EdU Alexa Fluor 488 (Life Technologies, Carlsbad, CA, USA) assay according to the manufacturer′s protocol. Pterygium fibroblast cells reverse-transfected with miR-215 mimic or non-specific oligonucleotide control and 10 uM of 5-ethynyl-2′-deoxyuridine (EdU) added and incubated for 10 h. (I), Cells transfected with miR-215 for 24 h, (J) cells transfected with control for 24 h, (K), cells transfected with miR-215 for 48 h, (L), cells transfected with control for 48 h; (E), (F), (G), (H), DAPI fluorescence only, for the same cells in (I)-(L), respectively; (A), (B), (C), (D), Alexa-Fluor 488 fluorescence only. Images shown are representative of 3 experiments. Scale bar represents 100 uM.
MiR-215 induced cell cycling arrest
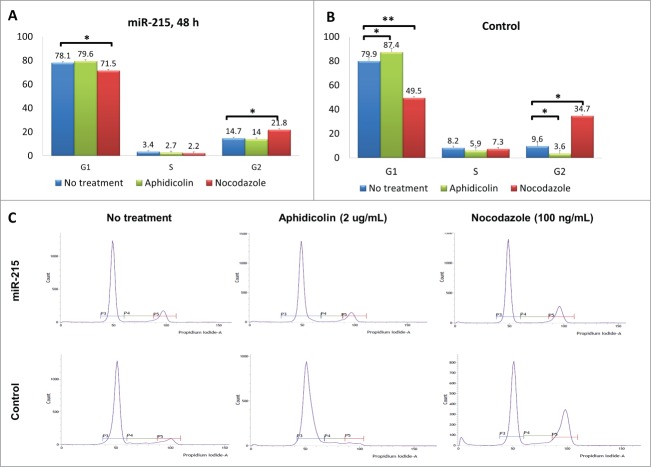
The effects of mir-215 on cell proliferation may be due to increased cell cycling. To investigate whether miR-215 has an effect on cell cycling, we performed cell cycle analysis by synchronizing cells at G1 with aphidicolin and G2 with nocodazole for 18 h after transfection with miR-215 mimic or random oligonucleotide control (Fig. 4).
Figure 4.
Results of the cell cycle analysis: Cultured primary pterygium fibroblast cells were reverse-transfected with 100 nM miR-215 mimic or non-specific control oligonucleotide for 48 h, and arrested at G1 and G2 phases by blocking with 2 ug/mL aphidicolin or 100 ng/mL nocodazole respectively. Eighteen hours after drug treatment, cells were collected for cell cycle analysis using flow cytometry. (A and B) Cluster bar chart showing percentage of cells arrested at G1, S and G2, for miR-215 (A) or random oligonucleotide control (B) transfected cells treated with aphidicolin (green), or nocodazole (red). Non-treated controls are shown in blue. Numbers represent mean of 3 experiments ± standard deviation C. Histograms illustrate proportions of cells observed at different phases: the first peak corresponds to G1 and second peak G2. The histogram is extracted from one representative experiment from a series of 3 biological replicates.
In the control set (random oligonucleotide, Fig. 4B), 7.5% of cells (87.4 − 79.9) could be arrested at G1 phase (difference between green and blue bars, p = 0 .03), but with addition of miR-215 mimic (Fig. 4A), only 1.5% of cells (79.6 − 78.1) were arrested (difference between green and blue bars, p = 0 .42).
An additional observation (Fig. 4B) was that 25.1% of the cells (34.7 − 9.6) were arrested at G2 phase in control (difference between red and blue bars, p = 0 .001), while only 7.1% (21.8 − 14.7) could be thus arrested when miR-215 mimic was added (difference between red and blue bars, p = 0 .05) (Fig. 4A).
These experiments suggest that miR-215 had arrested a proportion of cells at G1 and G2 and therefore further effects by aphidicolin and nocodazole were less evident. To investigate how G1/S progression and G2/M progression can be affected by miR-215, we are interested to determine the effects of adding miR-215 in cell cycle related transcripts.
MiR-215 controlled a group of genes including cell cycling regulators
The addition of miR-215 mimic in cultured primary fibroblast cells was performed for 24 h and global transcript analysis investigated using microarray. The microarray data has been deposited in NCBI′s Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) with GEO series accession number GSE57296. Gene ontology analysis was performed using the GeneSpring platform (Agilent Technologies, Santa Clara, CA, USA) and interestingly, the genes described as “cell cycle” related were over-represented in the overall list of dysregulated genes compared to control (p = 0 .008). Cell cycling genes were dysregulated by miR-215 with a 12.03% representation among all dysregulated genes compared to 6.20% representation of all cell cycling genes with valid signals on the microarray chip (Table S1). Table S2 shows the breakdown of the 16 cell cycling genes and their levels in the microarray.
We further studied a list of cell cycle regulators (Table S3) that would be dysregulated had mir-215 targeted them. The functions of these regulators in different phases of the cell cycle are shown, and interestingly all these are positive regulators.
To validate the effects of miR-215 on the change in expression levels in the cell cycling genes, qRT-PCR was employed. Out of the 10 cell cycling genes which were down-regulated by miR-215 in the microarray, 6 were confirmed by qRT-PCR: Cell division cycle 25 homolog A (Cdc25A), thyroid hormone receptor interactor 13 (Trip13), minichromosome maintenance complex component 10 (Mcm10), minichromosome maintenance complex component 3 (Mcm3), dicer 1 (Dicer1) and intestinal cell (MAK-like) kinase (Ick) were found to be downregulated by miR-215 by 4.64 (p = 0.005), 4.52 (p = 0.013), 4.04 (p = 0.014), 3.59 (p = 0.019), 3.01 (p = 0.001) and 2.64 (p = 0.030) folds respectively using qRT-PCR.
CDC25A, MCM10, ICK AND TRIP13 were upregulated in pterygium
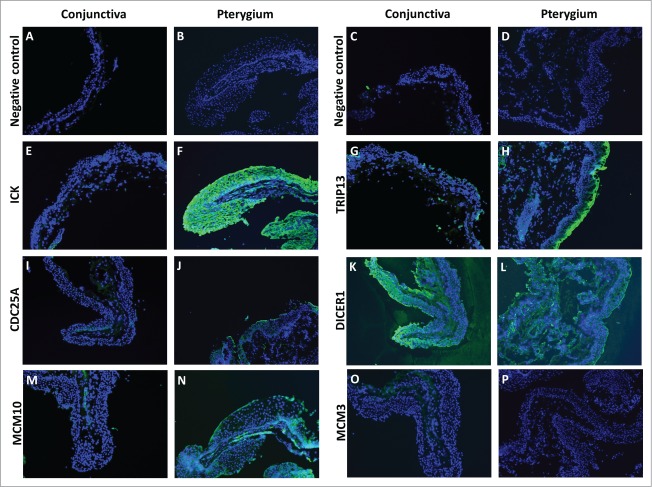
For the cell cycle regulators, we were interested to know if the transcripts down-regulated by miR-215 were up-regulated at the protein level in pterygium. To address that issue, we used immunofluorescent staining for cell cycle regulators ICK, TRIP13, CDC25A, DICER1, MCM10 AND MCM3. We found that compared to conjunctiva, pterygium tissues stained more strongly for ICK, TRIP13, CDC25A and MCM10. While TRIP13, CDC25A and MCM10 were more localized in the epithelial layer, ICK and DICER1 were found abundantly in both the epithelium and the fibroblast-containing stroma. DICER1 stained strongly in both pterygium and conjunctiva, while MCM3 stained weakly in both tissue types (Fig. 5A–T).
Figure 5.
Fluorescent immunostaining images of conjunctiva (first and third column) and pterygium (second and fourth column) tissues using primary antibodies against ICK (E and F), TRIP13 (G and H), CDC25A (I and J), DICER1 (K and L), MCM10 (M and N), MCM3 (O and P), followed by Alexa Fluor-488 conjugated secondary antibodies. Conjunctiva and pterygium tissues which were stained with only secondary antibodies (A–D) were used as negative controls for each primary antibody used. Dicer1: dicer 1, ribonuclease type III, Mcm3: minichromosome maintenance complex component 3, Cdc25A: cell division cycle 25A, Mcm10: minichromosome maintenance complex component 10, Trip13: thyroid hormone receptor interactor 13, Ick: intestinal cell (MAK-like) kinase.
MiR-215 bound directly to Cdc25A and Mcm10
Specific miRNAs can interfere with cell cycle regulators in more than one way. The most direct and obvious mechanism will be the binding of miR-215 to the transcript of the cell cycle regulator. To assess the possible binding of miR-215 to the 3′UTR of the target transcripts that encode cell cycle regulators, we performed the dual-luciferase 3′UTR assays with wildtype sequences as well as mutated sequences at the potential miR-215 binding sites. Of the 4 cell cycling genes tested, transfection of the reporters for 3′UTR of Cdc25A and Mcm10 caused a 0.65-fold (p = 0 .002) and 0.72-fold (p = 0 .002) reduction in luciferase activity compared to their respective mutated 3′ UTR controls, while Trip13 and Ick did not show any significant changes at 0.96- and 1.06-folds (p>0.05) respectively (Fig. 6). When miR-215 mimic is substituted for control oligonucleotide, the reduction in luciferase activity in wild type 3′UTR of Cdc25A and Mcm10 compared to mutant 3′UTR was abrogated. These suggest that endogenous miR-215 binds to and inhibits expression of Cdc25A and Mcm10 transcripts directly, but may not affect the Trip13 and Ick transcripts.
Figure 6.
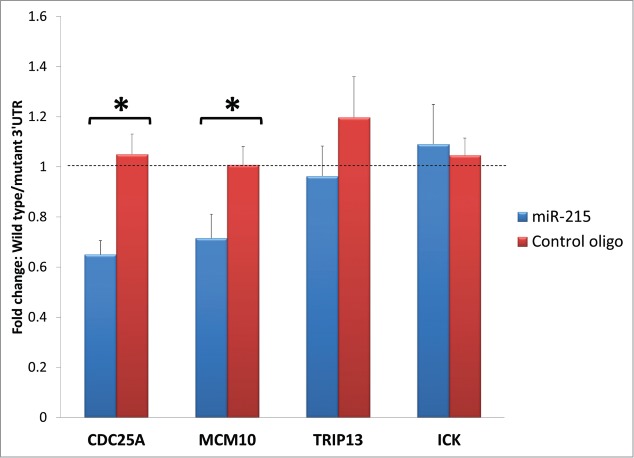
Bar charts represent fold change difference in luciferase activity when HEK293T cells are transfected with wild type 3′ UTR or mutant 3′ UTR. Wild type Cdc25A and Mcm10 show a marked reduction in luciferase activity compared to their mutant control (illustrated as fold change with blue bars) but this reduction is abolished when miR-215 mimic is substituted for random oligonucleotide control (illustrated as fold change with red bars), highlighting specificity of miR-215 to binding sites on Cdc25A and Mcm10. No significant changes were observed for Trip13 and Ick. Cdc25A: cell division cycle 25A, Mcm10: minichromosome maintenance complex component 10, Trip13: thyroid hormone receptor interactor 13, Ick: intestinal cell (MAK-like) kinase.
Discussion
We showed miR-215, one of the downregulated microRNAs in pterygium, suppresses proliferation in primary fibroblast cells and this is likely mediated by reduced G1/S and G2/M cell cycling. We found a panel of cell cycling genes to be inhibited by miR-215, out of which Cdc25A and Mcm10 were likely direct targets of miR-215. Cdc25A and Mcm10 were also upregulated in pterygium.
As far as we are aware, this is the first report of a microRNA, i.e. miR-215, being down-regulated in a fibrotic ocular surface condition and exerting an effect on fibroblast proliferative function, through its direct cell cycling targets. Our work is consistent with previously reported findings, which were not related to fibroblasts or fibrosis. It was reported that miR-192, an analog of miR-215, is up-regulated in HCT116 DICERex5 cells upon p53 induction, bringing about cell cycling arrest via a panel of cell cycling regulators.48 While both Georges et al. and our studies confirmed Mcm10 transcripts to be targets of miR-215 using the luciferase assay, we found another transcript, Cdc25A, to be a target as well.
Previous studies have implicated the role of microRNAs in fibrosis in the context of ocular wound healing. One study reported an upregulation of miR-200b in human Tenon′s fibroblasts (HTF) upon stimulation with transforming growth factor (TGF)-ß1,28 via proliferative effects of p27/kip1 and Rho Family GTPase (RND) 3. Meanwhile, Li et al. found miR-29b to be downregulated by TGF-ß1 in HTF by targeting collagen type 1 variant A1 (Col1A1) and facilitating extracellular matrix remodelling.29 Another study found miR-216b to regulate apoptosis and autophagy effects of hydroxycamptothecin treatment in HTF after glaucoma surgery via beclin (Bcl)-2.30 These studies on ocular fibroblasts have implications for glaucoma surgery with regard to wound healing and scarring, however the specific miRNAs involved have not been shown to be dysregulated in any ocular surface fibrotic disease.
In addition, Liu et al. profiled the expression of microRNAs on human corneal fibroblasts after dexamethasone treatment and found miR-16, -21, and -29C to be upregulated and miR-100 down-regulated.31 Robinson et al. focused on the expression of miR-133b following laser ablation in mouse cornea. The authors found miR-133b to affect cell migration by targeting connective tissue growth factor, smooth muscle actin, and COL1A1.27 Finally, in a paper on trachomatous scarring, Derrick et al. found miR-147 and -1285 to be up-regulated in inflammatory trachomatous conjunctiva.26 However, functions have not been attributed to these microRNAs in the papers.
Potential mechanisms
Mcm10 is known to be involved in DNA replication and initiation and elongation. In replication initiation, Mcm10 binds to single-stranded DNA (ssDNA) and promotes helicase activation,49 and may recruit DNA polymerase-α primase to aid DNA elongation.50 In view of its function in DNA replication, defects of Mcm10 may result in defective G1/S progression. Increase in replication has been correlated to proliferative state.51 An overexpression of Mcm10 has been linked to cervical cancer.52
Cdc25A is reported to be an important regulator of cell proliferation53 and promotes cell cycle progression at both G1/S phase and G2/M by dephosphorylation of CDK1 and CDK2.54-58 An overexpression of Cdc25A results in uncontrolled cell proliferation and genomic instability; upregulation of Cdc25A has been notably reported in several cancers including breast, gastric and colorectal cancer.59,60 In addition, it was reported that Cdc25A has anti-apoptotic properties through inhibiting apoptosis signal-regulating kinase 1 (ASK1), a positive regulator of the mitogen-activated protein kinase kinase 4 (MKK4)-c-Jun N-terminal kinases (JNK) cascade.61
We postulate that miR-215 plays an important role in propagating pterygium by increasing fibroblast proliferation via Mcm10 and Cdc25A. Cdc25A and Mcm10 are in turn interacting with a network of other cell cycling regulators such as cyclin dependent kinases which will have further amplifying effects on cell cycling. Although increased numbers of fibroblasts could result from increased cell cycling and proliferation, we did not show that the amount of matrix proteins was consequently increased. We agree that matrix regulation is an important issue and we have a separate report that focused on the spectrum of protein secreted by pterygium fibroblasts.
The connection between UV radiation and microRNA disturbance may possibly be via the processing of endogenous microRNAs. For instance, enzymes that catalyze the assembly of the RNA-Induced Signaling Complex (RISC) may be affected by ultraviolet radiation.62 Presently, 1872 different human microRNAs have been discovered and predicted to control over 60% of all genes in the human genome.63,64 Therefore only a small number of microRNAs are required to cause a widespread disorder of gene expression to result in fibrosis. Although we investigated only one human disease, the findings may be applicable to other fibrotic diseases.
During the preparation of this manuscript, 2 papers have been published on the dysregulation of microRNAs in pterygium. Chien et al. reported the correlation of mir-145 with pterygium severity.65 The authors examined the expression of miR-145 in 253 patient samples, and clinically classified pterygium into 3 grades of severity without comparing to uninvolved tissue controls. Engelsvold et al. reported the dysregulation of microRNAs including the downregulation of miR-200b, by comparing 8 pterygium tissues to matched conjunctiva controls. Using clustering analysis, functional analysis and pathway analysis on the dysregulated microRNAs and a list of dysregulated mRNAs, the authors hypothesized on the role of the miR-200 family in epithelial mesenchymal transition in pterygium. In this hypothesis, epithelial cells transform into abnormal fibroblasts which then mediate the changes that result in pterygium.35
It is surprising that neither Chien et al.65 nor Engelsvold et al.66 reported miR-215 to be of importance. One reason would be the heterogeneity of tissues involved and the lack of same eye controls in other studies. This may cause confounding by differences in ethnicity, hours of sun exposure, and genetic factors such as expression of G6PD and redox genes.
Strengths
The pterygium and conjunctival tissue specimens were harvested in a single eye center, which handles many cases of such a disease. Samples were obtained from patients using uniform diagnostic criteria and surgical techniques and processed in a standardized manner. Use of controls from the same eye eliminates issues concerning inter-individual differences such as age and genetics.
Limitations
We acknowledge a few limitations to our studies. One limitation is that the number of tissues used is relatively small. The miRCURY LNA™ microRNA Array featured 6 slides for microRNA comparison, therefore only 3 pairs of pterygium and conjunctiva tissues were used for microRNA profiling. Small sample size and tissue heterogeneity could result in a smaller number of targets identified. However, we used 3 other pairs of tissues for qRT-PCR and another 3 for FISH experiments. These showed that dysregulation of miR-215 was found in all 9 patients that we evaluated.
We accept that studies on cell culture would not necessarily yield similar results from a tissue study due to the absence of cell matrix or matrix influences in cell culture. A further limitation is that the extent or severity of the pterygium lesion was not documented. In theory, it is possible that the level of microRNA may be different at different stages of pterygium or between different parts of the lesion (e.g. head versus body) and therefore poses the important question of whether derangements in microRNA are the cause of pterygium formation or merely an effect of pterygium.
There are other potential binding sites on the 3′ UTR of Trip13 and Ick which we did not mutate, so we cannot exclude them out as direct targets of miR-215. Mcm3 and Ppp1ca were potentially targets of miR-215 as the complementary sequences were found on the 3′UTR, but we did not validate this specificity by luciferase assay.
The current manuscript focused on miR-215 among the dysregulated microRNAs based on the relatively well known cellular function in other studies as well as the function of predicted targets of this miR-215. Our data cannot exclude the possible role of other microRNAs
Future studies
It would be useful to perform binding specificity of miR-215 to the other cell cycling regulators found in microarray screening (Table S2). Phenotypic studies involving the knock-down or rescue of target genes will validate the functions of the target proteins in association with miR-215. The in vivo role of microRNA and its target transcripts in other models of fibrosis will be studied.
Clinical application
Our findings on microRNA and fibroblast function may be applied to a human disease. It may be possible to arrest pterygium formation by non-surgical treatment using supplementation of miR-215. The elevation of microRNA expression can be achieved via the use of short interfering RNAs in therapy.67,68 In fleshy pterygium lesions, these strategies may even play a role in the prevention of post-surgical recurrence after excision.
Material and Methods
Specimen and patient samples
The method for patient specimen collection was previously described.69 All protocols complied with the tenets of the Declaration of Helsinki and were approved by the Central Institutional Review Board of Singhealth. Written informed consents were obtained from patient donors. Pterygium tissue was excised from a patient diagnosed with primary pterygium undergoing an elective surgical procedure. An upper bulbar free conjunctival graft from the same eye was removed and placed over the site of the original lesion. The whole pterygium tissue and a small portion of uninvolved conjunctiva located adjacent to the graft site were collected separately.
Primary pterygium fibroblast cell culture
Primary pterygium fibroblasts were cultured as previously reported70,71 with slight modifications: Fresh pterygium specimen were transferred immediately into 1 ml of Dulbecco′s modified eagle′s medium (DMEM)/F-12 culture medium (Life Technologies, Carlsbad, CA, USA) containing 1x antibiotic and antimycotic (1x A/A) (Life Technologies, Carlsbad, CA, USA) and transferred to lab on ice. Each specimen was cut into explant pieces of approximately 1–2 mm2 and placed onto 60 mm culture plates coated with 1 ml fetal bovine serum (FBS) (Life Technologies, Carlsbad, CA, USA) for 10 min at 37°C. Sixty microliters of FBS with 1X A/A was dropped onto the explants and incubated overnight at 37°C with 5% CO2. On the second morning, 120 ul of DMEM/F-12 with 10% FBS and 1× A/A was dropped over each explant, and an additional 400 ul topped up on the same evening. About 3–5 d later upon first observations of cell migration, the volume of medium was increased to 2 ml and replaced on alternative days. Cells were subcultured at 80–90% confluency using TrypLE™ Express (Life Technologies, Carlsbad, CA, USA). After Passage 0, cells were cultured in DMEM/F-12 with 10% FBS.
RNA extraction
Fresh tissue specimens were immediately transferred to RNAlater RNA stabilization reagent (Qiagen, Hilden, Germany) upon surgical excision and stored at −20°C until RNA extraction. Tissues were minced using surgical scissors and ground using pestle and mortar with liquid nitrogen, transferred to TRIzol (Life Technologies, Carlsbad, CA, USA) and passed through 22G needles (Becton Dickenson). Chloroform was added and the RNA layer extracted after separation into organic phases. The RNeasy RNA extraction kit (Qiagen, Hilden, Germany) was used to purify RNA obtained, performed according to manufacturer′s protocol. RNA concentration and quality were assessed using Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA) for microarray use or the spectrophotometer (Thermo Scientific, Waltham, MA, USA) for qRT-PCR use.
MicroRNA and mRNA microarray profiling
Comprehensive microRNA profiling was outsourced to Exiqon′s ISO 9001:2000 certified microRNA profiling service. Briefly, RNA from 3 pterygium and 3 conjunctival samples were labeled using miRCURY LNATM microRNA Power Labeling Kit after removal of 5′-phosphatases from the terminal ends and hybridized to miRCURY LNATM microRNA Array (Exiqon, Vedbaek, Denmark). The samples are incubated for 16 h at 56°C, washed and images acquired.
For mRNA expression profiling, the microarray experiment was performed on the Agilent Sureprint G3 GE 8 × 60K (ID: G6851–60510) single-color chip (Agilent Technologies, Santa Clara, CA, USA) containing 27,958 entrez gene RNA sequences. Four pairs of matched pterygium and conjunctiva tissue samples were evaluated. Briefly, total RNA was labeled with Low Input Quick Amp Labeling Kit. Total RNA was converted to double-stranded cDNA using oligo-dT primers and transcribed to produce cyanine 3-CTP labeled cRNA. Labeled cRNA was hybridized onto Agilent SurePrint G3 human GE 8 × 60K microarray for 17h at 65°C in Agilent hybridization oven. After hybridization, the microarray slide is washed and scanned on Agilent High Resolution Microarray scanner. Raw signal data was extracted from the TIFF image with Agilent Feature Extraction Software (V10.7.1.1). The workflow was performed by Molecular Genomics Pte Ltd, Singapore.
To screen for potential targets of miR-215, 4 pairs of primary pterygium fibroblast cells that have been treated with 100 nM miR-215 mimic or non-specific control oligonucleotide (Thermo Scientific, Waltham, MA, USA) for 24 h were analyzed for gene expression using the same chip and procedure described above.
Analysis of microarray data
The threshold of the raw signals was set to 1.0. Data were transformed to logbase 2 values. Per chip normalization was performed by percentile shift to 75th percentile. Per gene normalization was performed by baseline transformation to the median of all samples. A box plot of normalized data for the 8 samples showed a similar distribution. Principal component analysis (PCA) of the normalized data was performed and the 3D scatter plot showed separation of the 4 points representing pterygium tissue from the 4 points representing conjunctival tissue. Subsequent quality control on the probeset to remove faulty or suspicious probes was done by filtering on flags (probes flagged as ′not detected′ and ′compromised′) and filtering on expression (removal of probes with very low raw signal intensity (<20 )).
Quantitative Real-time Polymerase Chain Reaction (qRT-PCR)
Primary pterygium and conjunctival tissues from 3 pairs of samples were used in a 2-step qRT-PCR experiment for the analyses of both microRNA and target mRNAs. For quantification of microRNA expression, qRT-PCR was performed using the miRCURY LNA™ PCR system (Exiqon, Vedbaek, Denmark) in accordance with the manufacturer′s instructions. Total RNA was diluted to a concentration of 10 ng in 4.5 μl using nuclease-free water and added to reagents from the miRCURY LNA™ First-strand cDNA Synthesis Kit. Resultant complementary cDNA products were diluted 2-fold and 4 μl was added to reagents in the miRCURY LNA™ SYBR® Green Master Mix. Twenty microlitres of the final product was amplified in triplicates for qRT-PCR on a LightCycler® 480 real-time PCR System. Control reactions using primers against 5S rRNA (rRNA) were used to normalize microRNA expression levels.
In mRNA expression analysis, SuperScript™ II First-Strand Synthesis System for RT-PCR (Life Technologies, Carlsbad, CA, USA) was used to reverse transcribe 1 μg of each sample′s total RNA preparation. Gene-specific forward and reverse primers (Table S4) were designed using Primer3 (http://bioinfo.ut.ee/primer3/) and synthesized by FBCO (Singapore). Quantification was performed using the comparative ΔCt method as described earlier.72 Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal control.
Fluorescent In Situ hybridization (FISH) for microRNA
FISH was carried out as described in the protocol reported earlier,73 with adjustments made to the incubation time with anti-DIG alkaline phosphatase antibody so as to avoid non-specific binding. Three pairs of pterygium and conjunctival specimens were embedded in OCT, cryosectioned to 8 μm-thick and mounted on microscope slides. Sections were fixed in 4% paraformaldehyde (PFA) and washed in 1X PBS. They were immersed in acetylation solution (590 ml DEPC-treated water, 8 ml triethanolamine, 1050 μl HCl and 1.5 ml acetic anhydride), washed in 1X PBS and treated with Proteinase K. The slides were prehybridized for 6 h in hybridization solution (50% formamide, 5X sodium chloride/sodium citrate (SSC), 5X Denhardt′s, 200 μg/ml yeast RNA, 500 μg/ml salmon sperm DNA, 0.4 g Roche blocking reagent and 1.75 ml DEPC-treated water) at room temperature. Tissues were hybridized at 50–60oC overnight in the presence of a probe mix consisting of 0.1 μl of LNA DIG-labeled probe and 150 μl of denaturating hybridization solution (prepared as hybridization solution but 500 μl 10% CHAPS, 100 μl 20% Tween-20 and 1150 μl DEPC-treated water was added instead of 1.75 ml DEPC-treated water). Hybridized sections were soaked in 60oC 5X SSC followed by a high stringency wash in 0.2X SSC at 60oC for 1 h. Slides were incubated at room temperature for 10 min in B1 solution (0.1 M Tris pH 7.5 and 0.15 M NaCl) and 1 h in blocking solution (2 ml FBS, 18 ml B1 solution and 100 μl 10% Tween-20). Diluted anti-DIG-alkaline phosphatase antibody 1:2000 was incubated with the sections for another hour at room temperature. Slides were washed in B1 solution, equilibrated 10 min with 1 M Tris pH 8.2, and incubated 30 min with Fast Red solution. Images were taken at 200X magnification using Axioplan2 fluorescence microscope equipped with an AxioCam MR camera (Carl Zeiss AG, Jena, Germany).
Gene ontology classification
Microarray data was analyzed by Genespring 12.5 (Agilent Technology, Redwood City, CA). A differential expression gene list was generated based on the following criteria: fold change of miR-215 treated pterygium fibroblast over control > 1.5. General gene ontology analysis was performed on the generated gene list to identify involved cellular processes.
Immunofluorescent staining and microscopy
Three pairs of human pterygium and control conjunctival specimens were embedded in Optimal Cutting Temperature (OCT) (Sakura, USA) compound. Eight microlitres-thick sections were made using a Microm HM550 cryostat (Microm, Walldorf, Germany). Sections were fixed in ice-cold acetone, washed in 1X PBS, blocked with 4% BSA in 1X PBS containing 0.1% Triton X-100 for 45 min and incubated with primary antibodies (Table S5) at 4°C overnight. Sections were washed in 1X PBS and incubated with Alexa Fluor 488-conjugated secondary antibody 1:800 in PBST (1X PBS, 0.1% Tween-20) for 30 min at room temperature. UltraCruz™ Mounting Medium was added to stain the nuclei. Images were captured at 200X magnification using Axioplan2 fluorescence microscope equipped with an AxioCam MR camera (Carl Zeiss AG, Jena, Germany).
Impedance assay for cell proliferation
This was performed using a continuous non-labeling method as previously described.74 The xCELLigence system (Roche, Basel, Switzerland) allows cellular dynamics involving cell viability, adhesion, migration and proliferation to be monitored real-time, using microelectronic biosensors to track cell impedance. Briefly, 3,000 cultured primary pterygium fibroblast cells were reverse transfected with 100 nM of either miR-215 mimic or non-specific control oligonucleotide (Thermo Scientific, Waltham, MA, USA) in each well of a 96-well plate. Transfection medium is changed to complete culture medium at 6 h and cell impedance is monitored. The experiments were repeated using 3 biologically different (different donors) cultured primary pterygium fibroblast cells.
Flow cytometry analysis for cell cycle
Approximately 250,000 cultured pterygium fibroblast cells were transfected in each well of a 6-well plate with 100 nM of either miR-215 or non-specific control oligonucleotide (Thermo Scientific, Waltham, MA, USA). At 48 h, cells are treated with 100 ng/ml nocodazole (Sigma Aldrich) or 2 ug/ml aphidicolin (Sigma Aldrich, St. Louis, MO, USA) for 18 h, after which cells are collected. After one wash using PBS, cells are resuspended in 500 uL PBS and 70% cold ethanol is added drop-wise under gentle vortexing. Cells are fixed overnight in 4°C, collected by centrifugation and washed once in PBS. Cells are collected again and stained with PI Staining Solution composed of 20 ug/ml propidium iodide (Abcam), 200 ug/ml RNAse A (Thermo Scientific, Waltham, MA, USA) and 0.1% Triton X-100 (Thermo Scientific, Waltham, MA, USA) for 15 min at 37°C. The DNA contents of the stained cells are analyzed using a flow cytometer, FACSVerse (Becton Dickenson, Franklin Lakes, NJ, USA). The experiments were repeated using 3 biologically different (different donors) cultured primary pterygium fibroblast cells. The highest non-lethal dosage of aphidicolin and nocodazole were used.
Cell proliferation assay
Cell proliferation was analyzed using the Click-it EdU Alexa Fluor 488 (Life Technologies, Carlsbad, CA, USA) assay according to the manufacturer′s protocol. EdU is a thymidine analog that is incorporated into the DNA during the S-phase, and its detection is catalyzed by copper (I) using an azide modified fluorescent dye. This click technology allows insight to cell replication without the need to denature DNA as required by the conventional bromodeoxyuridine (BrdU) cell proliferation assay. Briefly, 50,000 cultured pterygium fibroblast cells reverse-transfected with miR-215 mimic or non-specific control oligonucleotide are seeded into a 4-well plate and allowed to attach overnight. Ten micromolar of 5-ethynyl-2′-deoxyuridine (EdU) was added and cells incubated for 10 h at 37°C. The cells were then fixed with 4% PFA, washed with 3% bovine serum albumin (BSA) and permeabilized using 0.5% Triton X-100. Five hundred microlitres of Click-it reaction cocktail comprising 1X Click-it reaction buffer, copper sulfate solution, Alexa Fluor azide, 1X reaction buffer additive prepared according to manufacturer′s protocol, was added and cells incubated for 30 min, washed and stained with Hoechst 33342 for 30 min, protected from light. The images were viewed using the C2 Confocal microscope (Nikon , Chiyoda, Tokyo, Japan) under 100 x magnification. Experiments were performed 3 times with biological replicates. The number of stained nuclei was measured as a percentage of total number of nuclei in each image for quantification of staining.
Prediction of target binding sites by calculation of binding energies
Analysis of results was performed using a web-based bioinformatics prediction platform. Statistically significant changes (P < 0.05) in mRNA levels after miR-215 treatment in pterygium fibroblast cells were checked for miR-215 seed sequence-mRNA 3′UTR complementarity using RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/). A minimum binding energy, defined arbitrarily, was used to determine potential binding by microRNA. Using this definition, the potential sites had at least 6 base pairs of contiguous nucleotide complementarity.
Dual-luciferase assay
The pEZX-MT01 dual-luciferase reporter vector from Genecopoeia (Genecopoeia, Rockville, MD, USA) was used for target validation. The vectors comprised of 3′ UTR sequences of target transcripts inserted downstream of the firefly luciferase reporter gene driven by SV40 and a renilla luciferase gene driven by CMV. RNAhybrid was used to determine possible binding sites in silico and 3–5 short nucleotide sequences of about 10–15 base pairs which were determined to be potential microRNA binding sites were randomly mutated. The 3′ UTR sequences as well as mutant 3′ UTR sequences are listed in Table S6. Mutation and cloning services were performed by Genecopoeia and validated by IDT (Singapore) sequencing services using the following primers: 3′UTR Forward 5′-GATCCGCGAGATCCTGAT-3′, 3′UTR Reverse 5′-CCTATTGGCGTTACTATG-3′.
Seventy-five thousand HEK293T cells were seeded onto 96-well white walled, clear-bottom plates and reverse transfected with 100 nM miR-215 mimic and 200 ng 3′ UTR plasmid or mutated 3′ UTR plasmid. Medium was changed 24 h later and cells lysed 48 h after transfection using 1x passive lysis buffer (Promega, Madison, WI, USA). Addition of Luciferase Assay Reagent (firefly luciferase substrate) and Stop-and-Glow (renilla luciferase substrate) were performed according to manufacturer′s protocol (Promega, Madison, WI, USA) and luminescence measured using the Infinite M200 luminometer (Tecan, Maennedorf, Switzerland). Each experiment was repeated with 3 different biological samples.
Statistical analysis
Data was presented as mean fold change ± standard error of mean and analyzed using Student′s 2-tailed t test. P values less than 0.05 were considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank Dr. Sebastian Maurer-Stroh for lending his expertise in bioinformatics and help with using the RNAhybrid software.
Funding
This work was supported by the National Medical Research Council (Singapore) [NMRC/CSA/046/2012, NMRC/CSA/013/2009].
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- 1. Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol 2007; 17:118-26; PMID:17197185; http://dx.doi.org/ 10.1016/j.tcb.2006.12.007 [DOI] [PubMed] [Google Scholar]
- 2. Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004; 10:1957-66; PMID:15525708; http://dx.doi.org/ 10.1261/rna.7135204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci U S A 2007; 104:17719-24; PMID:17965236; http://dx.doi.org/ 10.1073/pnas.0703890104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Srivastava SP, Koya D, Kanasaki K. MicroRNAs in kidney fibrosis and diabetic nephropathy: roles on EMT and EndMT. BioMed Res Int 2013; 2013:125469; PMID:24089659; http://dx.doi.org/ 10.1155/2013/125469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tu X, Zhang H, Zhang J, Zhao S, Zheng X, Zhang Z, Zhu J, Chen J, Dong L, Zang Y. MicroRNA-101 suppresses liver fibrosis by targeting TGFbeta signaling pathway. J Pathol 2014; 234(1):46-59; PMID:24817606; http://dx.doi.org/ 10.1002/path.4373 [DOI] [PubMed] [Google Scholar]
- 6. Yao Q, Xu H, Zhang QQ, Zhou H, Qu LH. MicroRNA-21 promotes cell proliferation and down-regulates the expression of programmed cell death 4 (PDCD4) in HeLa cervical carcinoma cells. Biochem Biophys Res Commun 2009; 388(3):539-42; PMID:19682430; http://dx.doi.org/ 10.1016/j.bbrc.2009.08.044 [DOI] [PubMed] [Google Scholar]
- 7. Walker JC, Harland RM. microRNA-24a is required to repress apoptosis in the developing neural retina. Genes Dev 2009; 23:1046-51; PMID:19372388; http://dx.doi.org/ 10.1101/gad.1777709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Friedman JM, Jones PA. MicroRNAs: critical mediators of differentiation, development and disease. Swiss Med Wkly 2009; 139:466-72; PMID:19705306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aumiller V, Forstemann K. Roles of microRNAs beyond development–metabolism and neural plasticity. Biochim Biophys Acta 2008; 1779:692-6; PMID:18498780; http://dx.doi.org/ 10.1016/j.bbagrm.2008.04.008 [DOI] [PubMed] [Google Scholar]
- 10. Kuehbacher A, Urbich C, Dimmeler S. Targeting microRNA expression to regulate angiogenesis. Trends Pharmacol Sci 2008; 29:12-5; PMID:18068232; http://dx.doi.org/ 10.1016/j.tips.2007.10.014 [DOI] [PubMed] [Google Scholar]
- 11. Zhao JJ, Yang J, Lin J, Yao N, Zhu Y, Zheng J, Xu J, Cheng JQ, Lin JY, Ma X. Identification of miRNAs associated with tumorigenesis of retinoblastoma by miRNA microarray analysis. Childs Nerv Syst 2009; 25:13-20; PMID:18818933; http://dx.doi.org/ 10.1007/s00381-008-0701-x [DOI] [PubMed] [Google Scholar]
- 12. Corson TW, Gallie BL. One hit, two hits, three hits, more? Genomic changes in the development of retinoblastoma. Genes Chromosomes Cancer 2007; 46:617-34; PMID:17437278; http://dx.doi.org/ 10.1002/gcc.20457 [DOI] [PubMed] [Google Scholar]
- 13. Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res 2008; 79:581-8; PMID:18550634; http://dx.doi.org/ 10.1093/cvr/cvn156 [DOI] [PubMed] [Google Scholar]
- 14. Sonkoly E, Stahle M, Pivarcsi A. MicroRNAs: novel regulators in skin inflammation. Clin Exp Dermatol 2008; 33:312-5; PMID:18419608; http://dx.doi.org/ 10.1111/j.1365-2230.2008.02804.x [DOI] [PubMed] [Google Scholar]
- 15. Asli NS, Pitulescu ME, Kessel M. MicroRNAs in organogenesis and disease. Curr Mol Med 2008; 8:698-710; PMID:19075669; http://dx.doi.org/ 10.2174/156652408786733739 [DOI] [PubMed] [Google Scholar]
- 16. Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vis 2006; 12:1175-84; PMID:17102797 [PubMed] [Google Scholar]
- 17. Karali M, Peluso I, Marigo V, Banfi S. Identification and characterization of microRNAs expressed in the mouse eye. Invest Ophthalmol Vis Sci 2007; 48:509-15; PMID:17251443; http://dx.doi.org/ 10.1167/iovs.06-0866 [DOI] [PubMed] [Google Scholar]
- 18. Xu S. microRNA expression in the eyes and their significance in relation to functions. Prog Retin Eye Res 2009; 28:87-116; PMID:19071227; http://dx.doi.org/ 10.1016/j.preteyeres.2008.11.003 [DOI] [PubMed] [Google Scholar]
- 19. Arora A, McKay GJ, Simpson DA. Prediction and verification of miRNA expression in human and rat retinas. Invest Ophthalmol Vis Sci 2007; 48:3962-7; PMID:17724173; http://dx.doi.org/ 10.1167/iovs.06-1221 [DOI] [PubMed] [Google Scholar]
- 20. Makarev E, Spence JR, Del Rio-Tsonis K, Tsonis PA. Identification of microRNAs and other small RNAs from the adult newt eye. Mol Vis 2006; 12:1386-91; PMID:17149364 [PubMed] [Google Scholar]
- 21. Decembrini S, Andreazzoli M, Barsacchi G, Cremisi F. Dicer inactivation causes heterochronic retinogenesis in Xenopus laevis. Int J Dev Biol 2008; 52:1099-103; PMID:18956342; http://dx.doi.org/ 10.1387/ijdb.082646sd [DOI] [PubMed] [Google Scholar]
- 22. Loscher CJ, Hokamp K, Kenna PF, Ivens AC, Humphries P, Palfi A, Farrar GJ. Altered retinal microRNA expression profile in a mouse model of retinitis pigmentosa. Genome Biol 2007; 8:R248; PMID:18034880; http://dx.doi.org/ 10.1186/gb-2007-8-11-r248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Damiani D, Alexander JJ, O'Rourke JR, McManus M, Jadhav AP, Cepko CL, Hauswirth WW, Harfe BD, Strettoi E. Dicer inactivation leads to progressive functional and structural degeneration of the mouse retina. J Neurosci 2008; 28:4878-87; PMID:18463241; http://dx.doi.org/ 10.1523/JNEUROSCI.0828-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang L, Wang T, Wright AF, Suri M, Schwartz CE, Stevenson RE, Valle D. A microdeletion in Xp11.3 accounts for co-segregation of retinitis pigmentosa and mental retardation in a large kindred. Am J Med Genet A 2006; 140:349-57; PMID:16419135; http://dx.doi.org/ 10.1002/ajmg.a.31080 [DOI] [PubMed] [Google Scholar]
- 25. Shen J, Yang X, Xie B, Chen Y, Swaim M, Hackett SF, Campochiaro PA. MicroRNAs regulate ocular neovascularization. Mol Ther 2008; 16:1208-16; PMID:18500251; http://dx.doi.org/ 10.1038/mt.2008.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Derrick T, Roberts C, Rajasekhar M, Burr SE, Joof H, Makalo P, Bailey RL, Mabey DC, Burton MJ, Holland MJ. Conjunctival MicroRNA expression in inflammatory trachomatous scarring. PLoS Negl Trop Dis 2013; 7:e2117; PMID:23516655; http://dx.doi.org/ 10.1371/journal.pntd.0002117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robinson PM, Chuang TD, Sriram S, Pi L, Luo XP, Petersen BE, Schultz GS. MicroRNA signature in wound healing following excimer laser ablation: role of miR-133b on TGFbeta1, CTGF, SMA, and COL1A1 expression levels in rabbit corneal fibroblasts. Invest Ophthalmol Vis Sci 2013; 54:6944-51; PMID:24065814; http://dx.doi.org/ 10.1167/iovs.13-12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tong J, Fu Y, Xu X, Fan S, Sun H, Liang Y, Xu K, Yuan Z, Ge Y. TGF-beta1 stimulates human Tenon's capsule fibroblast proliferation by miR-200b and its targeting of p27/kip1 and RND3. Invest Ophthalmol Vis Sci 2014; 55:2747-56; PMID:24667864; http://dx.doi.org/ 10.1167/iovs.13-13422 [DOI] [PubMed] [Google Scholar]
- 29. Li N, Cui J, Duan X, Chen H, Fan F. Suppression of type I collagen expression by miR-29b via PI3K, Akt, and Sp1 pathway in human Tenon's fibroblasts. Invest Ophthalmol Vis Sci 2012; 53:1670-8; PMID:22297492; http://dx.doi.org/ 10.1167/iovs.11-8670 [DOI] [PubMed] [Google Scholar]
- 30. Xu X, Fu Y, Tong J, Fan S, Xu K, Sun H, Liang Y, Yan C, Yuan Z, Ge Y. MicroRNA-216b/Beclin 1 axis regulates autophagy and apoptosis in human Tenon's capsule fibroblasts upon hydroxycamptothecin exposure. Exp Eye Res 2014; 123C:43-55; http://dx.doi.org/ 10.1016/j.exer.2014.03.008 [DOI] [PubMed] [Google Scholar]
- 31. Liu L, Walker EA, Kissane S, Khan I, Murray PtdIns, Rauz S, Wallace GR. Gene expression and miR profiles of human corneal fibroblasts in response to dexamethasone. Invest Ophthalmol Vis Sci 2011; 52:7282-8; PMID:21666241; http://dx.doi.org/ 10.1167/iovs.11-7463 [DOI] [PubMed] [Google Scholar]
- 32. Lucas RM, McMichael AJ, Armstrong BK, Smith WT. Estimating the global disease burden due to UV radiation exposure. Int J Epidemiol 2008; 37:654-67; PMID:18276627; http://dx.doi.org/ 10.1093/ije/dyn017 [DOI] [PubMed] [Google Scholar]
- 33. Di Girolamo N, Chui J, Coroneo MT, Wakefield D. Pathogenesis of pterygia: role of cytokines, growth factors, and matrix metalloproteinases. Prog Retin Eye Res 2004; 23:195-228; PMID:15094131; http://dx.doi.org/ 10.1016/j.preteyeres.2004.02.002 [DOI] [PubMed] [Google Scholar]
- 34. Tan DT, Liu YP, Sun L. Flow cytometry measurements of DNA content in primary and recurrent pterygia. Invest Ophthalmol Vis Sci 2000; 41:1684-6; PMID:10845586 [PubMed] [Google Scholar]
- 35. Kato N, Shimmura S, Kawakita T, Miyashita H, Ogawa Y, Yoshida S, Higa K, Okano H, Tsubota K. Beta-catenin activation and epithelial-mesenchymal transition in the pathogenesis of pterygium. Invest Ophthalmol Vis Sci 2007; 48:1511-7; PMID:17389479; http://dx.doi.org/ 10.1167/iovs.06-1060 [DOI] [PubMed] [Google Scholar]
- 36. Austin P, Jakobiec FA, Iwamoto T. Elastodysplasia and elastodystrophy as the pathologic bases of ocular pterygia and pinguecula. Ophthalmology 1983; 90:96-109; PMID:6828309; http://dx.doi.org/ 10.1016/S0161-6420(83)34594-2 [DOI] [PubMed] [Google Scholar]
- 37. Westermarck J, Kahari VM. Regulation of matrix metalloproteinase expression in tumor invasion. FASEB J 1999; 13:781-92; PMID:10224222 [PubMed] [Google Scholar]
- 38. Detorakis ET, Zaravinos A, Spandidos DA. Growth factor expression in ophthalmic pterygia and normal conjunctiva. Int J Mol Med 2010; 25:513-6; PMID:20198298; http://dx.doi.org/ 10.3892/ijmm_00000371 [DOI] [PubMed] [Google Scholar]
- 39. Maxia C, Perra MT, Demurtas P, Minerba L, Murtas D, Piras F, Cabrera R, Ribatti D, Sirigu P. Relationship between the expression of cyclooxygenase-2 and survivin in primary pterygium. Mol Vis 2009; 15:458-63; PMID:19247455 [PMC free article] [PubMed] [Google Scholar]
- 40. Tung JN, Wu HH, Chiang CC, Tsai YY, Chou MC, Lee H, Cheng YW. An association between BPDE-like DNA adduct levels and CYP1A1 and GSTM1 polymorphisma in pterygium. Mol Vis 2010; 16:623-9; PMID:20700368 [PMC free article] [PubMed] [Google Scholar]
- 41. Ang LP, Chua JL, Tan DT. Current concepts and techniques in pterygium treatment. Curr Opin Ophthalmol 2007; 18:308-13; PMID:17568207; http://dx.doi.org/ 10.1097/ICU.0b013e3281a7ecbb [DOI] [PubMed] [Google Scholar]
- 42. Tan DT, Chee SP, Dear KB, Lim AS. Effect of pterygium morphology on pterygium recurrence in a controlled trial comparing conjunctival autografting with bare sclera excision. Arch Ophthalmol 1997; 115:1235-40; PMID:9338666; http://dx.doi.org/ 10.1001/archopht.1997.01100160405001 [DOI] [PubMed] [Google Scholar]
- 43. Kim KW, Park SH, Lee SH, Kim JC. Upregulated stromal cell-derived factor 1 (SDF-1) expression and its interaction with CXCR4 contribute to the pathogenesis of severe pterygia. Invest Ophthalmol Vis Sci 2013; 54:7198-206; PMID:24114538; http://dx.doi.org/ 10.1167/iovs.13-13044 [DOI] [PubMed] [Google Scholar]
- 44. Mulik S, Xu J, Reddy PB, Rajasagi NK, Gimenez F, Sharma S, Lu PY, Rouse BT. Role of miR-132 in angiogenesis after ocular infection with herpes simplex virus. Am J Pathol 2012; 181:525-34; PMID:22659469; http://dx.doi.org/ 10.1016/j.ajpath.2012.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li P, Li J, Chen T, Wang H, Chu H, Chang J, Zang W, Wang Y, Ma Y, Du Y, et al. . Expression analysis of serum microRNAs in idiopathic pulmonary fibrosis. Int J Mol Med 2014; 33:1554-62; PMID:24676360 [DOI] [PubMed] [Google Scholar]
- 46. Wang B, Ricardo S. Role of microRNA machinery in kidney fibrosis. Clin Exp Pharmacol Physiol 2014; 41(8):543-50; PMID:24798583; http://dx.doi.org/ 10.1111/1440-1681.12249 [DOI] [PubMed] [Google Scholar]
- 47. Kustermann S, Boess F, Buness A, Schmitz M, Watzele M, Weiser T, Singer T, Suter L, Roth A. A label-free, impedance-based real time assay to identify drug-induced toxicities and differentiate cytostatic from cytotoxic effects. Toxicol In Vitro 2013; 27:1589-95; PMID:22954529; http://dx.doi.org/ 10.1016/j.tiv.2012.08.019 [DOI] [PubMed] [Google Scholar]
- 48. Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, Jackson AL, Carleton MO, Linsley PS, Cleary MA, et al. . Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res 2008; 68:10105-12; PMID:19074876; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-1846 [DOI] [PubMed] [Google Scholar]
- 49. Thu YM, Bielinsky AK. MCM10: One tool for all-Integrity, maintenance and damage control. Semin Cell Dev Biol 2014; 30:121-30; PMID:24662891; http://dx.doi.org/ 10.1016/j.semcdb.2014.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Maiorano D, Lutzmann M, Mechali M. MCM proteins and DNA replication. Curr Opin Cell Biol 2006; 18:130-6; PMID:16495042; http://dx.doi.org/ 10.1016/j.ceb.2006.02.006 [DOI] [PubMed] [Google Scholar]
- 51. Blow JJ, Hodgson B. Replication licensing–defining the proliferative state? Trends Cell Biol 2002; 12:72-8; PMID:11849970; http://dx.doi.org/ 10.1016/S0962-8924(01)02203-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Das M, Prasad SB, Yadav SS, Govardhan HB, Pandey LK, Singh S, Pradhan S, Narayan G. Over expression of minichromosome maintenance genes is clinically correlated to cervical carcinogenesis. PloS One 2013; 8:e69607; PMID:23874974; http://dx.doi.org/ 10.1371/journal.pone.0069607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shen T, Huang S. The role of Cdc25A in the regulation of cell proliferation and apoptosis. AntiCancer Agents Med Chem 2012; 12:631-9; PMID:22263797; http://dx.doi.org/ 10.2174/187152012800617678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lindqvist A, Rodriguez-Bravo V, Medema RH. The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J Cell Biol 2009; 185:193-202; PMID:19364923; http://dx.doi.org/ 10.1083/jcb.200812045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Terada Y, Tatsuka M, Jinno S, Okayama H. Requirement for tyrosine phosphorylation of Cdk4 in G1 arrest induced by ultraviolet irradiation. Nature 1995; 376:358-62; PMID:7630405; http://dx.doi.org/ 10.1038/376358a0 [DOI] [PubMed] [Google Scholar]
- 56. Iavarone A, Massague J. Repression of the CDK activator Cdc25A and cell-cycle arrest by cytokine TGF-β in cells lacking the CDK inhibitor p15. Nature 1997; 387:417-22; PMID:9163429; http://dx.doi.org/ 10.1038/387417a0 [DOI] [PubMed] [Google Scholar]
- 57. Hoffmann I, Draetta G, Karsenti E. Activation of the phosphatase activity of human cdc25A by a cdk2-cyclin E dependent phosphorylation at the G1/S transition. EMBO J 1994; 13:4302-10; PMID:7523110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhao H, Watkins JL, Piwnica-Worms H. Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc Natl Acad Sci U S A 2002; 99:14795-800; PMID:12399544; http://dx.doi.org/ 10.1073/pnas.182557299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Boutros R, Lobjois V, Ducommun B. CDC25 phosphatases in cancer cells: key players? Good targets? Nat Rev Cancer 2007; 7:495-507; PMID:17568790; http://dx.doi.org/ 10.1038/nrc2169 [DOI] [PubMed] [Google Scholar]
- 60. Cangi MG, Cukor B, Soung P, Signoretti S, Moreira G, Jr., Ranashinge M, Cady B, Pagano M, Loda M. Role of the Cdc25A phosphatase in human breast cancer. J Clin Invest 2000; 106:753-61; PMID:10995786; http://dx.doi.org/ 10.1172/JCI9174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zou X, Tsutsui T, Ray D, Blomquist JF, Ichijo H, Ucker DS, Kiyokawa H. The cell cycle-regulatory CDC25A phosphatase inhibits apoptosis signal-regulating kinase 1. Mol Cell Biol 2001; 21:4818-28; PMID:11416155; http://dx.doi.org/ 10.1128/MCB.21.14.4818-4828.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mori MA, Raghavan P, Thomou T, Boucher J, Robida-Stubbs S, Macotela Y, Russell SJ, Kirkland JL, Blackwell TK, Kahn CR. Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metab 2012; 16:336-47; PMID:22958919; http://dx.doi.org/ 10.1016/j.cmet.2012.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. miRBase Homo sapiens miRNAs (1872 sequences). Homo sapiens miRNAs (1872 sequences). University of Manchester, 2014:http://www.mirbase.org/cgi-bin/mirna_summary.pl?org=hsa. [Google Scholar]
- 64. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009; 19:92-105; PMID:18955434; http://dx.doi.org/ 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chien KH, Chen SJ, Liu JH, Woung LC, Chen JT, Liang CM, Chiou SH, Tsai CY, Cheng CK, Hu CC, et al. . Correlation of microRNA-145 levels and clinical severity of pterygia. Ocul Surf 2013; 11:133-8; PMID:23583047; http://dx.doi.org/ 10.1016/j.jtos.2012.12.001 [DOI] [PubMed] [Google Scholar]
- 66. Engelsvold DH, TP U, OK O, P G, JR E, T L, AM T, DA D, S R. miRNA and mRNA expression profiling identifies members of the miR-200 family as potential regulators of epithelial-mesenchymal transition in pterygium. Exp Eye Res 2013; 115C:189-98; http://dx.doi.org/ 10.1016/j.exer.2013.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pawitan JA. The possible use of RNA interference in diagnosis and treatment of various diseases. Int J Clin Pract 2009; 63:1378-85; PMID:19691623; http://dx.doi.org/ 10.1111/j.1742-1241.2008.01940.x [DOI] [PubMed] [Google Scholar]
- 68. Pushparaj PN, Aarthi JJ, Manikandan J, Kumar SD. siRNA, miRNA, and shRNA: in vivo applications. J Dent Res 2008; 87:992-1003; PMID:18946005; http://dx.doi.org/ 10.1177/154405910808701109 [DOI] [PubMed] [Google Scholar]
- 69. Wong YW, Chew J, Yang H, Tan DT, Beuerman R. Expression of insulin-like growth factor binding protein-3 in pterygium tissue. Br J Ophthalmol 2006; 90:769-72; PMID:16488932; http://dx.doi.org/ 10.1136/bjo.2005.087486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Di Girolamo N, Tedla N, Kumar RK, McCluskey P, Lloyd A, Coroneo MT, Wakefield D. Culture and characterisation of epithelial cells from human pterygia. Br J Ophthalmol 1999; 83:1077-82; PMID:10460780; http://dx.doi.org/ 10.1136/bjo.83.9.1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Solomon A, Grueterich M, Li DQ, Meller D, Lee SB, Tseng SC. Overexpression of Insulin-like growth factor-binding protein-2 in pterygium body fibroblasts. Invest Ophthalmol Vis Sci 2003; 44:573-80; PMID:12556385; http://dx.doi.org/ 10.1167/iovs.01-1185 [DOI] [PubMed] [Google Scholar]
- 72. Livak KJ, Flood SJ, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl 1995; 4:357-62; PMID:7580930; http://dx.doi.org/ 10.1101/gr.4.6.357 [DOI] [PubMed] [Google Scholar]
- 73. Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc 2007; 2:1508-14; PMID:17571058; http://dx.doi.org/ 10.1038/nprot.2007.153 [DOI] [PubMed] [Google Scholar]
- 74. Scrace S, O'Neill E, Hammond EM, Pires IM. Use of the xCELLigence System for Real-Time Analysis of Changes in Cellular Motility and Adhesion in Physiological Conditions. Methods Mol Biol 2013; 1046:295-306; PMID:23868595; http://dx.doi.org/ 10.1007/978-1-62703-538-5_17 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.