Abstract
Artificial selection has played a critical role in animal breeding. Detection of artificial selection footprints in genomic regions can provide insights for understanding the function of specific phenotypic traits and better guide animal breeding. To more fully understand the relationship between genomic composition and phenotypic diversity arising from breed development, a genome-wide scan was conducted using an OvineSNP50 BeadChip and integrated haplotype score and fixation index analyses to detect selection signatures on the X chromosome in three sheep breeds. We identified 49, 34, and 55 candidate selection regions with lengths of 27.49, 16.47, and 25.42 Mb in German Mutton, Dorper, and Sunit sheep, respectively. Bioinformatics analysis showed that some of the genes in these regions with selection signatures, such as BMP15, were relevant to reproduction. We also identified some selection regions harboring genes that had human orthologs, including BKT, CENPI, GUCY2F, MSN, PCDH11X, PLP1, VSIG4, PAK3, WAS, PCDH19, PDHA1, and SRPX2. The VSIG4 and PCDH11X genes are associated with the immune system and disease, PDHA1 is associated with biosynthetic related pathways, and PCDH19 is expressed in the nervous system and skin. These genes may be useful as candidate genes for molecular breeding.
Keywords: selection signature, X chromosome, sheep
1. Introduction
Artificial selection has played a significant role in the domestication of livestock. Approximately 11,000 years ago, domesticated animals began to appear in the Fertile Crescent [1]. Sheep, the first livestock known to be domesticated, were initially reared for their meat before their breeding became specialized for secondary products, such as wool, which occurred between 4000 and 5000 years ago [2]. Domestication reshaped the behavior, morphology, and genetics of the involved livestock. For example, two studies found that artificial selection changed sheep coat pigmentation, horn morphology and growth developmental traits [3,4] using a genome scan of recent positive selection signatures in three sheep populations.
Selection has many effects on the genome. Positive selection can improve advantageous allele frequencies and fix them within a population [5]. As a result, polymorphism at a selection site is reduced in the population. With the development of high-density single nucleotide polymorphism (SNP) chips and high-throughput genotyping technology, a number of statistics have been used to explore selection signatures on genes and the genome [6]. For instance, signatures indicating diversified selection among breeds are found in genomic regions associated with traits related to the standard criteria for breeds, such as coat color and ear morphology. Other selection signals are found in genomic regions such as quantitative trait loci and genes associated with production traits, including reproduction, growth, and fat deposition. Some selection signatures in sheep are associated with regions showing evidence of introgression from Asian breeds. When European sheep breeds were compared with the wild boar, genomic regions with high levels of differentiation were found to harbor genes related to bone formation, growth, and fat deposition [7].
Selection signatures can be detected through the variation of allele frequency and decay of linkage disequilibrium. To date, some related methods have been put forward and can be classified into categories of linkage disequilibrium and site-frequency spectrum [8]. The integrated haplotype score (iHS) [9] and F-statistics (FST) [10] are extensively used in identifying selection signatures. The iHS has been used mainly to reveal the selection signatures (within-population methods) using information from a single population, whereas FST has been used primarily to detect selection signatures between populations. The iHS is based on linkage disequilibrium theory and can detect regions with a rapidly increased frequency of the derived allele at selected sites [9]. The FST, first used to evaluate population variation through the DNA polymorphism of a population, is currently based on a Bayesian hierarchical model [11]. McRae et al. [12] used FST to identify 14 novel regions associated with resistance or susceptibility to gastrointestinal nematodes in sheep.
The X chromosome has a high density of genes and thus may be a good target for detecting selection signatures. Rubin et al. [13] pointed out that the X chromosome should be solely analyzed for the identification of selection signatures, and only sows should be used because sex chromosomes and autosomes, even between genders, are subjected to different selective pressures and have different effective population sizes. The X chromosome undergoes more drift than autosomes, as its effective population size is three-quarters that of autosomes [14]. The X chromosome is more specialized than an autosome and plays an important role in evolution of human and animals. Studies have shown that selection on the X chromosome reduces the genomic diversity to a greater extent than that on autosomes (19%–26%) [15]. Ma et al. [6] studied selection footprints on the X chromosome in pigs and determined that genes relevant to meat quality, reproduction, and the immune system were found in potential selection regions. Moradi et al. [16] used the OvineSNP50 BeadChip to scan selective sweeps in thin and fat tail sheep and found increased homozygosity in selection regions in favor of fat tail breeds on chromosomes 5 and X. Taking into account the sex-specific dosage compensation, the selection pressure on the X chromosome is higher than that on the autosomes, indicating that genes on the X chromosome are under more direct and effective selection [17,18]. The X chromosome in sheep contains several genes relevant to desirable breeding traits, including those related to tail fat deposition [19], further indicating that this is a good target chromosome for examining selection signatures in sheep.
Therefore, in the present study, within-population (iHS) and between-population (FST) methods were used to search the whole X chromosome in three breeds of sheep for signatures of positive selection using the OvineSNP50 BeadChip array, followed by candidate gene enrichment analysis and gene annotation to elucidate the biological functions of the selection signature.
2. Results
2.1. Markers and Core Haplotypes
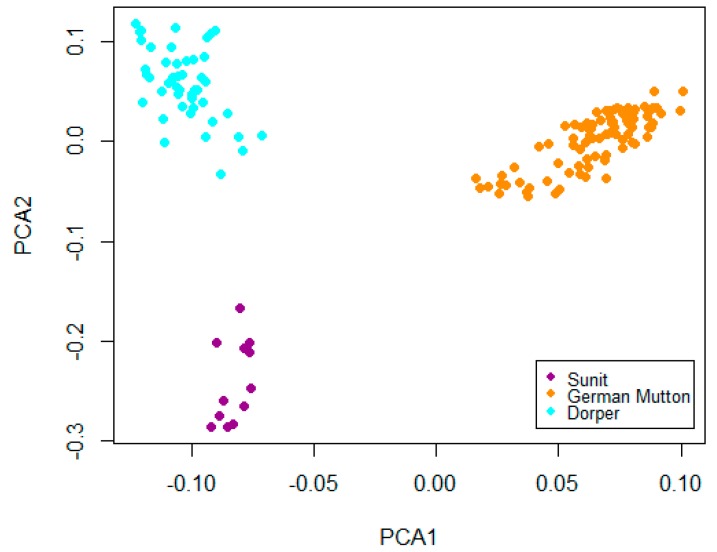
After quality control and PCA analyses (Figure 1), 89 German Mutton, 47 Dorper, and 12 Sunit ewes were used in the final analyses. A total of 1226 SNPs were obtained per breed. The average distance between two SNPs was 110.11 kb.
Figure 1.
Animal clusters based on principal component analysis (PCA) using individuals. Plots for the first (PCA1) and second (PCA2) components reveal the clustering of 148 animals from German Mutton, Dorper and Sunit.
2.2. Empirical Distribution of Test Statistics
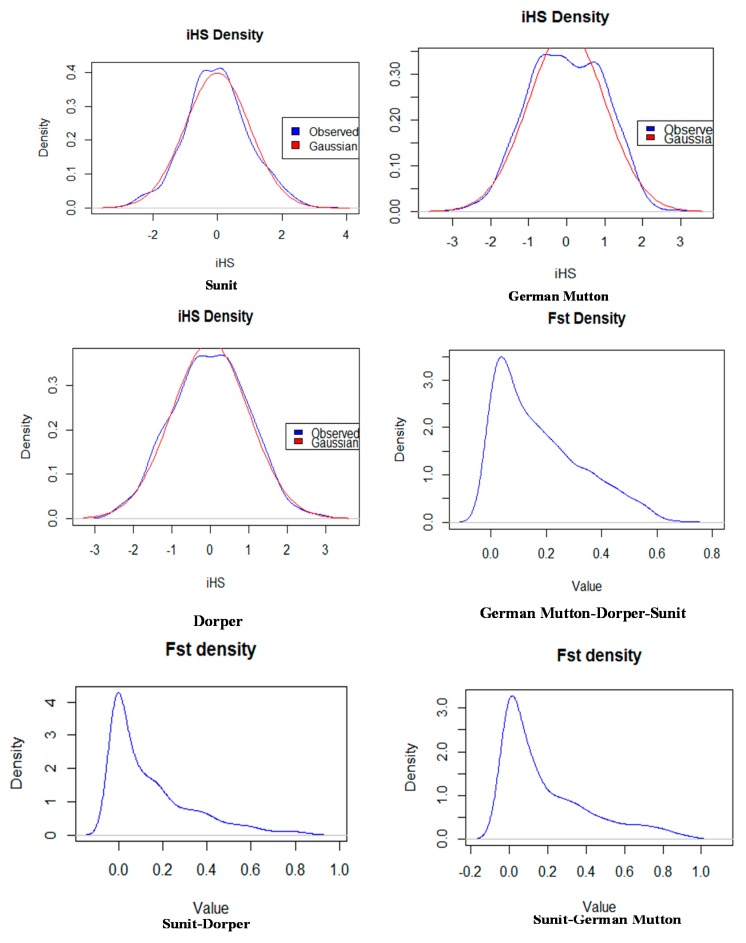
The empirical distributions of the two test statistics for each breed and breed pair were clearly observed. Figure 2 shows the distributions of the iHS and FST values on the X chromosome of German Mutton sheep and pairwise for German Mutton, Dorper, or Sunit. Table S1, Table S2, Table S3 and Table S4 show the true values for each SNP in every breed. The standardized iHS and FST values approximately followed a standard normal distribution, as pointed out by Sabeti et al. [20]. Moreover, the distributions of the iHS test statistics indicated that the other individual breeds and the comparison of breed pairs with one another showed similar results.
Figure 2.
Empirical distribution of test statistics within the three individual populations (iHS) and between (FST) the German Mutton, Dorper and Sunit ewe populations.
2.3. Identification of Recent Selection Signatures on the X Chromosome
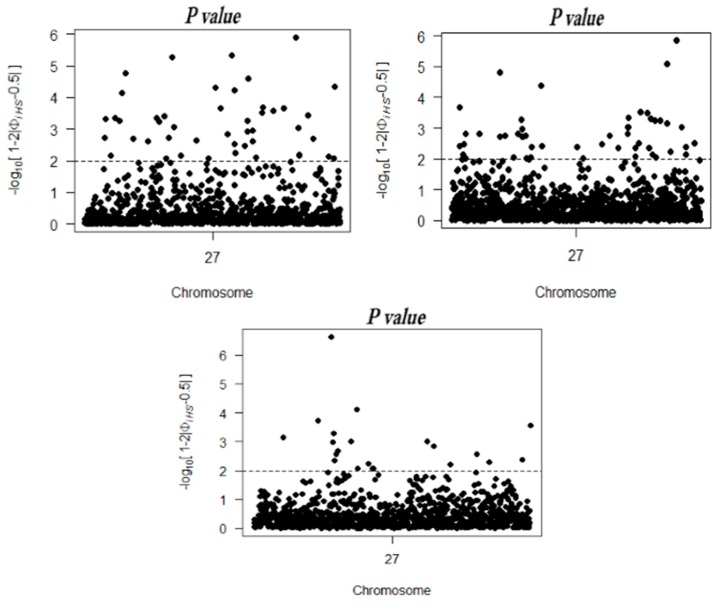
The scanning of the X chromosome for selection signatures in the three sheep breeds was conducted using between- and within-population methods. First, the within-population method iHS was used to look for selection signatures in the three breeds. The iHS scores were computed at each SNP over the whole genome using haplotypes. Figure 3 depicts the distribution of the PiHS value on the X chromosome to visualize the distribution of the selection signatures. As shown in Table 1, there were 51, 21, and 46 outliers identified in German Mutton, Dorper, and Sunit, respectively.
Figure 3.
The X chromosome distribution of PiHS values in the three breeds. The dotted line was denoted as the threshold, indicating significant regions.
Table 1.
Summary of selection signatures detected using iHS and FST in three sheep breeds.
| Items | iHS | FST | |||||
|---|---|---|---|---|---|---|---|
| G M | D | S | G M-D-S | G M-D | S-G M | S-D | |
| Number of SNPs | 51 | 21 | 46 | 12 | 12 | 12 | 12 |
| Number of regions | 35 | 17 | 37 | 10 | 9 | 11 | 9 |
| Average length (Mb) | 0.43 | 0.44 | 0.43 | 0.43 | 0.43 | 0.40 | 0.45 |
| Total length (Mb) | 14.78 | 7.59 | 15.93 | 4.35 | 3.89 | 4.44 | 40.3 |
(G M represents German Mutton; D represents Dorper; S represents Sunit).
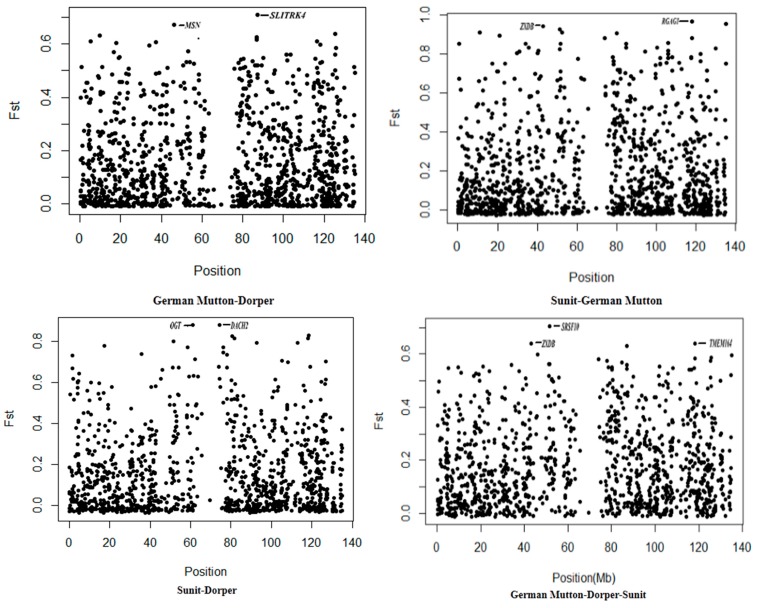
For the FST test, the population genetic differentiation at each genetic marker was detected. Because the FST empirical distribution of a single site was similar to a chi-squared (χ2) distribution with two or three degrees of freedom, the boxplot method was used to identify outliers in the genome-wide selection signal. Figure 4 depicts the distribution of the FST values on the X chromosome. A total of 12 outlier sites was detected in the three breeds.
Figure 4.
The X chromosome distribution of FST values.
2.4. Candidate Selection Regions
A region on the X chromosome having a false discovery rate (FDR) less than 0.1 for both methods, or an FDR less than 0.05 for one method, was considered a candidate selection region. As shown in Table 1, using the iHS test, 35, 17, and 37 candidate selection regions with lengths of 14.78, 7.59, and 15.93 Mb were identified in German Mutton, Dorper, and Sunit, respectively. As shown in Table 1, using the FST test, 10 candidate selection regions with lengths of 4.35 Mb were identified for German Mutton-Dorper-Sunit. Overall, approximately 19.33, 11.82, and 20.10 Mb selection regions were detected in German Mutton, Dorper, and Sunit. The Sunit, a representative Chinese indigenous breed, shared approximately 5.46 Mb candidate selection regions with German Mutton. This was less than the overlapping regions between Sunit and Dorper. In addition, there were approximately 7.29 Mb candidate selection regions shared by German Mutton and Dorper, with an overlapping length longer than that shared by Sunit and German Mutton or by Sunit and Dorper.
2.5. Identification of Candidate Genes in Selection Regions and Functional Analysis
The genes in the selection regions were identified using the Ovis_aries_3.1 database. After a comparison analysis, 102, 76, 102 genes were detected within regions positive for selection signatures in German Mutton, Dorper, and Sunit, respectively. Table 2 summarizes the genes in regions with the top five significant |iHS| values on the X chromosomes of the three breeds.
Table 2.
Selection regions on the X chromosome and corresponding genes for the top five significant iHS values in each sheep breed.
| Breed | Region (Mb) | |iHS| | Gene |
|---|---|---|---|
| German Mutton | 122.16–122.56 | 5.85 | TCEAL4, TCEAL1, MORF4L2, GLRA4, PLP1, RAB9B, TMSB15B, CYSLTR1 |
| 118.98–119.38 | 5.07 | NXT2 | |
| 19.83–20.23 | 4.80 | - | |
| 40.51–40.91 | 4.39 | FAM192A | |
| 3.77–4.17 | 3.66 | - | |
| Dorper | 30.58–30.98 | 6.63 | LAMP1 |
| 41.36–41.76 | 4.12 | EFHC2 | |
| 23.67–24.07 | 3.72 | - | |
| 134.90–135.30 | 3.56 | PABPC5 | |
| 31.27–31.67 | 3.29 | - | |
| Sunit | 103.45–103.85 | 3.63 | TEX13A |
| 31.98–32.38 | 3.44 | TMEM47 | |
| 40.13–40.53 | 3.32 | - | |
| 116.80–117.2 | 3.30 | TRPC5, ALG13 | |
| 94.82–95.22 | 3.25 | - |
The locus with the greatest FST value (0.705) was within the DNA region of the SRSF10 gene. The genes that overlapped with three other FST peaks were TMEM164, ZXDB, and SLITRK2. Twelve other SNPs with significant FST values also existed within selection regions. The genes harbored in the selection regions are shown in Table 3.
Table 3.
Selection and candidate genes detected by FST among the three breeds.
| Breed Pair | Position (Mb) | FST Value | Gene Name |
|---|---|---|---|
| German Mutton-Dorper-Sunit | 51.51–51.91 | 0.71 | SRSF10, DGKK, CCNB3 |
| 117.91–118.31 | 0.64 | TMEM164 | |
| 42.80–43.21 | 0.64 | ZXDB | |
| 86.91–87.31 | 0.63 | SLITRK2 | |
| 45.82–46.22 | 0.59 | MSN, MMD, VSIG4, HEPH | |
| 134.78–135.18 | 0.59 | PCDH11X, ZGC:112234, PABPC5 | |
| 125.43–125.83 | 0.58 | TIMM8A, BTK, RPL36A, GLA, HNRNPH2, ARMCX4, CSNK1A1, ARMCX1, ARMCX6, ARMCX3, ARMCX2 | |
| 117.84–118.24 | 0.58 | RGAG1, AMMECR1 | |
| 73.69–74.09 | 0.57 | CHM, DACH2 | |
| 77.77–78.17 | 0.57 | FAM58A, ATP2B3, BGN, HAUS7, TREX2, ZNF275, KIR3DL1 | |
| 125.15–125.55 | 0.57 | XKRX, ARL13A, TRMT2B, TMEM35, CENPI, DRP2, TAF7L | |
| 84.41–84.81 | 0.56 | SLITRK2, SLITRK4 | |
| German Mutton-Dorper | 86.65–87.30 | 0.71 | SLITRK4 |
| 45.82–46.22 | 0.67 | MSN, MMD, VSIG4, HEPH | |
| 125.15–125.55 | 0.64 | XKRX, ARL13A, TRMT2B, TMEM35, CENPI, DRP2, TAF7L | |
| 9.35–9.75 | 0.63 | - | |
| 4.85–5.25 | 0.61 | PNPLA4 | |
| 116.23–116.63 | 0.61 | - | |
| 37.26–37.66 | 0.61 | BCOR, DUT | |
| 17.56–17.96 | 0.60 | RPS6KA3 | |
| 117.84–118.24 | 0.59 | RGAG1, AMMECR1 | |
| Sunit-German Mutton | 117.91–118.31 | 0.97 | RGAG1, AMMECR1, TMEM164 |
| 134.78–135.18 | 0.95 | PCDH11X, ZGC:112234, PABPC5 | |
| 42.80–43.20 | 0.94 | ZXDB | |
| 51.17–51.61 | 0.93 | SHROOM4, SRSF10 | |
| 10.63–11.03 | 0.91 | EGFL6, RAD51L3 | |
| 52.36–52.76 | 0.91 | USP27X, SUMO2, PPP1R3F, FOXP3, CCDCC22, CACNA1F, SYP, PRICKLE3, PLP2, MAGIX, GPKOW, RPL36, WDR45, PRAF2 | |
| 79.89–80.29 | 0.90 | CD99L2, MTMR1, MTM1, MAMLD1 | |
| 20.82–21.22 | 0.89 | PRDX4, ACOT9, SAT1 | |
| 73.69–74.09 | 0.88 | CHM, DACH2 | |
| 117.91–118.31 | 0.88 | RGAG1, AMMECR1, TMEM164 | |
| Sunit-Dorper | 60.80–61.20 | 0.87 | OGT, ACRC, CXCR3, DMRTC2 |
| 74.29–74.69 | 0.87 | DACH2 | |
| 118.09–118.49 | 0.82 | AMMECR1, TMEM164 | |
| 80.30–80.76 | 0.82 | MAMLD1 | |
| 117.72–118.12 | 0.81 | CHRDL1, RGAG1 | |
| 81.49–81.89 | 0.81 | AFF2 | |
| 51.51–51.91 | 0.80 | SRSF10, DGKK, CCNB3 | |
| 112.66–113.06 | 0.79 | PLS3 | |
| 92.61–93.01 | 0.79 | - | |
| 17.08–17.48 | 0.77 | MAP3K15, RPL17, SH3KBP1, CXORF23 |
DAVID v2.1 was used to conduct the GO and KEGG pathway enrichment analyses to investigate the functions of the candidate genes. After this enrichment analysis, followed by the Benjamini correction procedure, there were almost no significant functional terms.
The genes were found to be involved in metabolism, muscle development, and reproduction based on information in the NCBI gene database, although some genes were not entirely harbored within the potential selection regions. Among them, BMP15, identified by iHS, overlapped with the potential selection region of 51.07–51.91 Mb and functions in reproduction [21].
3. Discussion
In recent years, extensive research using SNP chip array data to detect positive selection signatures has made considerable progress [9,22,23,24,25]. During this same time, a number of statistical methods have been developed to identify selection regions in the genome. To increase the accuracy of detection, we used two methods in the present study to detect selection regions on the X chromosome: iHS and FST approaches. The iHS relies on the EHH statistic, providing a more powerful approach to identify selection footprints at loci that are fixed or probably fixed [26]. The iHS test is based on linkage disequilibrium and is dependent on SNP spacing and frequency because it is a multi-marker test [16]. On the X chromosome, 1226 SNPs may not affect the accuracy of the iHS. Additionally, the power of the iHS method also relies on ancestral allele information, which is available for only a portion of the SNPs on the ovine chip [16]. However, Zhao et al. [27] used iHS and FST to successfully identify selection signatures in dairy and beef cattle .The FST measures population differentiation by detecting allele frequencies at a locus using a between-population method [28]. Detecting recent positive selection with FST is complicated because the distribution of the genetic variation due to selection can be difficult to distinguish from that which arises after certain demographic events [15]. However, ascertainment bias and demographic events would be expected to change patterns of FST in the same way genome-wide, whereas FST values only in selected and nearby loci would be altered by selection events [29,30,31,32].
In this study, the results obtained using the iHS method followed strict distributions. By contrast, the results obtained using the FST approach did not follow strict distributions, but followed an approximately normal distribution. This difference indicates that the risk of false positives when using the traditional significance test remains high because of the uncertainty in the null distribution for the test statistic [6]. For this reason, a boxplot strategy was used to determine the upper and lower threshold values, confirming the outliers for the FST values at each SNP locus. Thus, the results of the methods used to detect selection signatures on the X chromosome were subjected to strict criteria to prevent the occurrence of false positives. As stated, for the FST statistic, the boxplot strategy [29] was adopted to define upper and lower threshold FST values for each SNP locus to determine outliers. We first calculated the distribution of the X chromosome FST interquartile range; then, FST values greater than the upper threshold or less than the lower threshold values were defined as outliers.
In this study, 49, 34, and 55 selection regions were detected in German Mutton, Dorper, and Sunit breeds, respectively. German Mutton sheep were imported into China from Germany at the end of 20th century, and Dorper sheep from Australia in 2001 [30]. As these livestock populations migrated across the globe, they encountered numerous environments, each with unique ecological conditions. The populations were exposed to artificial selection through breeding programs. Hence, their genomes would be expected to be marked with many signals of positive selection. Sunit sheep are indigenous to northern China and are used for both their meat and fat in the Inner Mongolia Autonomous Region of China. Sunit sheep have adapted to the natural conditions of the Gobi Desert after approximately 800 years of natural and artificial selection. German Mutton shared approximately 7.29 Mb candidate selection regions with Dorper. This is a longer overlapping length than that shared by Sunit and German Mutton, which shared 5.46 Mb, or Dorper and Sunit, which shared approximately 5.90 Mb selection regions. This result suggests that selection intensity may be greater for German Mutton and Dorper than for Sunit sheep.
Following the Benjamini correction for the enrichment analysis of the genes located in candidate selection regions, no significant functional terms were detected. However, some GO terms with p values less than 0.05 were related to biological process, molecular functions, and cellular components, indicating that some traits may have undergone selection during the domestication of these sheep. We also detected some genes associated with the immune system on the X chromosome in sheep, consistent with the identification of selection footprints on the X chromosome in pigs [6].
Among our candidate genes, VSIG4 was identified and characterized by Helmy et al. [31] to be a complement receptor in the immunoglobulin superfamily, binding complement fragments C3b and iC3b to remove pathogenic microorganisms. The PCDH19 gene was another candidate gene for selection detected in our study, and it is expressed in the nervous system and skin and its derivative tissues [32]. The BTK gene is known to function in adaptive immunity, mainly in B cell signaling pathways, playing a key role in B cell proliferation, development, differentiation, survival, and apoptosis [33,34]. Demars et al. [35] identified two novel BMP15 mutations responsible for an atypical hyperprolificacy phenotype in sheep.
To date, a number of studies have identified selection signatures in sheep [36,37], but these studies focused on autosomal genes. Amaral et al. [38] used sequencing of pooled DNA to detect selection signatures genome-wide, but found it was difficult to analyze the isolated X chromosome. Rubin et al. [13] proposed that the X chromosome should be separately analyzed to detect selection signatures, as we did in this study. Thus, using iHS and FST, we detected 222 genes on the X chromosome in sheep with signatures indicating that they had undergone selection during domestication. These genes can be used as molecular markers in future sheep breeding.
4. Experimental Section
4.1. Experimental Animals and DNA Samples
The sheep population initially consisted of 161 (71 males and 90 females) German Mutton, 99 (49 males and 50 females) Dorper, and 69 (57 males and 12 females) Sunit. After a principal component analysis (PCA) was performed to identify population structure and the relatedness of animals, 148 females, including 89 German Mutton, 47 Dorper, and 12 Sunit, were finally chosen for selection signature identification on the X chromosome.
Blood samples were collected from six-month-old lambs using standard methods. Whole genomic DNA was extracted from blood samples using a TIANamp Blood DNA kit (Tiangen Biotech Co., Ltd., Beijing, China).
4.2. Genotyping and Quality Control
Genomic DNA was genotyped using the Illumina OvineSNP50 BeadChip containing 54,241 SNPs with an average gap spacing distance of 50.9 kb. The genotyping platform used was the Infinium II Multi-Sample Assay (Illumina, Inc., San Diego, CA, USA). The SNP chips were scanned using iScan, and the data were analyzed using GenomeStudio software (Illumina).
PLINK software (v1.07; http://pngu.mgh.harvard.edu/purcell/plink) was used to control the quality of the X chromosome genotype data. An individual was removed if the call rate was less than 90% or the sample was a duplicate. A SNP locus was excluded if (1) the SNP call rate was less than 90%; (2) its minor allele frequency was less than 0.05; or (3) it did not obey Hardy-Weinberg equilibrium (p value < 10−6). After quality control, BEAGLE software [39] was used to impute the missing genotypes and infer haplotypes.
4.3. Analyses Integrated Haplotype Score
The iHS, calculated as described by Voight et al. [9], was defined as the log of the ratio of the integrated extended haplotype homozygosity (EHH) score for haplotypes centering the ancestral allele to the integrated EHH score for haplotypes centering the derived allele. The iHS score was computed at X chromosome SNPs for each breed using the R package “rehh” [40]. The formula for the standardized iHS is as follows:
| (1) |
where iHHA and iHHD represent the integrated EHH score for ancestral and derived core alleles, respectively.
4.4. Population Differentiation Index
The classic measure of population genetic differentiation, FST, was used to detect signatures of diversifying selection, based on genetic polymorphism data. Here, FST was calculated as described by MacEachern et al. [41]:
| (2) |
where HT represents the expected heterozygosity for the overall total population such that
| (3) |
where p and q denote the frequency of alleles A1 and A2 over the total population.
In Equation (2), HS represents the expected heterozygosities in subpopulations and is calculated as follows:
| (4) |
with Hexpi denoting expected heterozygosity and ni denoting the sample size in subpopulation i.
4.5. Identifying the Region in the X Chromosome under Selection
For FST, A boxplot strategy was used to determine the upper and lower threshold values to confirm outliers of the FST values for each SNP locus.
First, the interquartile range (Q) of the FST empirical distribution on X chromosome was calculated as
| (5) |
with FU and FL representing the upper and lower interquartile ranges, respectively. The upper (UL) and lower (LL) threshold values were then calculated as follows:
| (6) |
| (7) |
All values greater than the upper threshold or less than the lower threshold values were defined as outliers.
For iHS, the thresholds of empirical cutoffs for the X chromosome were based on the autosomal cutoffs. The threshold of |iHS| on the X chromosome was |iHS| > 2.
4.6. Enrichment Analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway mapping, molecular function, cellular components, and biological process were determined for the candidate selection regions. An abundant database of human genomic information was referred to identify genes on the sheep genome using the many available annotations on the sheep genome. The program Database for Annotation, Visualization and Integrated Discovery (DAVID) 6.7 (http://david.abcc.ncifcrf.gov/) [42] was used to generate the homology gene set and gene enrichment analysis.
4.7. Gene Annotation
In the selection region, the outlier or selection footprint was extended approximately 100 kb in the upstream and downstream directions. The National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/gene/) database and the most recent sheep genome Ovis_aries_v3.1 [43] (http://www.livestockgenomics.csiro.au/sheep/oar3.1.php) were used to identify the biological function of genes in the selection regions. In addition, the genomic information of other species, including human, mouse, and bovine, were used to predict gene function.
5. Conclusions
In this study, we were able to identify many novel regions and genes on X chromosome by different methods, we demonstrated that X chromosome has undergo selection in the process of sheep domesticated.
Acknowledgments
This work was supported by the Agricultural Science and Technology Innovation Program (No. ASTIP-IAS-TS-6), National Natural Science Foundations of China (No. 31200927), and the National Modern Agricultural Industry Technology Fund for Scientists in Sheep Industry System (No. CARS-39-04B).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/16/09/20360/s1.
Author Contributions
Fuping Zhao and Lixin Du conceived and designed the experiments; Caiye Zhu performed the experiments; Caiye Zhu and Zehu Yuan analyzed the data; Hongying Fan, Shijin Hu, Li Zhang, Caihong Wei, and Qin Zhang contributed reagents/materials/analysis tools; Caiye Zhu wrote the paper. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Zeder M.A. Domestication and early agriculture in the Mediterranean Basin: Origins, diffusion, and impact. Proc. Natl. Acad. Sci. USA. 2008;105:11597–11604. doi: 10.1073/pnas.0801317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chessa B., Pereira F., Arnaud F., Amorim A., Goyache F., Mainland I., Kao R.R., Pemberton J.M., Beraldi D., Stear M.J. Revealing the history of sheep domestication using retrovirus integrations. Science. 2009;324:532–536. doi: 10.1126/science.1170587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kijas J.W., Lenstra J.A., Hayes B., Boitard S., Neto L.P., Cristobal M.S., Servin B., McCulloch R., Whan V., Gietzen K., et al. Genome-wide analysis of the world’s sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao F., Wei C., Zhang L., Liu J., Wang G., Zeng T., Du L. A genome scan of recent positive selection signatures in three sheep populations. J. Integr. Agric. 2015;15 doi: 10.1016/S2095-3119(15)61080-2. [DOI] [Google Scholar]
- 5.Stainton J.J., Haley C.S., Charlesworth B., Kranis A., Watson K., Wiener P. Detecting signatures of selection in nine distinct lines of broiler chickens. Anim. Genet. 2014;46:37–49. doi: 10.1111/age.12252. [DOI] [PubMed] [Google Scholar]
- 6.Ma Y., Zhang H., Zhang Q., Ding X. Identification of Selection Footprints on the X Chromosome in Pig. PLoS ONE. 2014;9:e94911. doi: 10.1371/journal.pone.0094911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkinson S., Lu Z.H., Megens H.J., Archibald A.L., Haley C., Jackson I.J., Groenen M.A., Crooijmans R.P., Ogden R., Wiener P. Signatures of diversifying selection in European pig breeds. PLoS Genet. 2013;9:e1003453. doi: 10.1371/journal.pgen.1003453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki Y. Statistical methods for detecting natural selection from genomic data. Genes Genet. Syst. 2010;85:359–376. doi: 10.1266/ggs.85.359. [DOI] [PubMed] [Google Scholar]
- 9.Voight B.F., Kudaravalli S., Wen X., Pritchard J.K. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weir B.S., Cockerham C.C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.2307/2408641. [DOI] [PubMed] [Google Scholar]
- 11.Riebler A., Held L., Stephan W. Bayesian variable selection for detecting adaptive genomic differences among populations. Genetics. 2008;178:1817–1829. doi: 10.1534/genetics.107.081281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mcrae K.M., Mcewan J.C., Dodds K.G., Gemmell N.J. Signatures of selection in sheep bred for resistance or susceptibility to gastrointestinal nematodes. BMC Geno. 2014;15:637–637. doi: 10.1186/1471-2164-15-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubina C.J., Megensb H.-J., Barrioa A.M., Maqboolc K., Sayyabc S., Schwochowc D., Wang C., Örjan C., Jerna P., Jørgensen C.B., et al. Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. USA. 2012;109 doi: 10.1073/pnas.1217149109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heyer E., Segurel L. Looking for signatures of sex-specific demography and local adaptation on the X chromosome. Genome Biol. 2010;11 doi: 10.1186/gb-2010-11-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McVicker G., Gordon D., Davis C., Green P. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 2009;5:e1000471. doi: 10.1371/journal.pgen.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moradi M.H., Nejati-Javaremi A., Moradi-Shahrbabak M., Dodds K.G., Mcewan J.C. Genomic scan of selective sweeps in thin and fat tail sheep breeds for identifying of candidate regions associated with fat deposition. BMC Genet. 2012;13 doi: 10.1186/1471-2156-13-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Disteche C.M. Dosage compensation of the active X chromosome in mammals. Nat. Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- 18.Graves J.A.M. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 19.Zhang W., Shen M., Li H., Gao L., Liang Y., Yang J., Liu S., Wang X., Gan S. Detection and analysis of polymorphisms of 59571364 and 59912586 loci on X chromosome in fat-tail and thin-tail sheep flocks (In Chinses) Yi Chuan = Hereditas/Zhongguo Yi Chuan Xue Hui Bian Ji. 2013;35:1384–1390. doi: 10.3724/SP.J.1005.2013.01384. [DOI] [PubMed] [Google Scholar]
- 20.Sabeti P.C., Patrick V., Ben F., Jason L., Elizabeth H., Chris C., Xiaohui X., Byrne E.H., Mccarroll S.A., Rachelle G. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W., Liu S., Li F., Pan X., Li C., Zhang X., Ma Y., La Y., Xi R., Li T. Polymorphisms of the Ovine BMPR-IB, BMP-15 and FSHR and Their Associations with Litter Size in Two Chinese Indigenous Sheep Breeds. Int. J. Mol. Sci. 2015;16:11385–11397. doi: 10.3390/ijms160511385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doerks T., Copley R.R., Schultz J., Ponting C.P., Bork P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002;12:47–56. doi: 10.1101/gr.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelley J.L., Madeoy J., Calhoun J.C., Swanson W., Akey J.M. Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Res. 2006;16:980–989. doi: 10.1101/gr.5157306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang E.T., Kodama G., Baldi P., Moyzis R.K. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc. Natl. Acad. Sci. USA. 2006;103:135–140. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang C., Bailey D.K., Awad T., Liu G., Xing G., Cao M., Valmeekam V., Retief J., Matsuzaki H., Taub M., et al. A whole genome long-range haplotype (WGLRH) test for detecting imprints of positive selection in human populations. Bioinformatics. 2006;22:2122–2128. doi: 10.1093/bioinformatics/btl365. [DOI] [PubMed] [Google Scholar]
- 26.Tang K., Thornton K.R., Stoneking M. A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol. 2007;5:e171. doi: 10.1371/journal.pbio.0050171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao F., McParland S., Kearney F., Du L., Berry D.P. Detection of selection signatures in dairy and beef cattle using high-density genomic information. Genet. Sel. Evol. 2015;47:1–12. doi: 10.1186/s12711-015-0127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright S. The genetical structure of populations. Ann. Eugen. 1949;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 29.McGill R., Tukey J.W., Larsen W.A. Variations of box plots. Am. Stat. 1978;32:12–16. [Google Scholar]
- 30.China National Commission of Animal Genetic Resources: 2012 . Animal genetic resource in China-Sheep and Goats. China Agriculture Press; Beijing, China: 2012. [Google Scholar]
- 31.Helmy K.Y., Katschke K.J., Jr., Gorgani N.N., Kljavin N.M., Elliott J.M., Diehl L., Scales S.J., Ghilardi N., van Lookeren Campagne M. CRIg: A macrophage complement receptor required for phagocytosis of circulating pathogens. Cell. 2006;124:915–927. doi: 10.1016/j.cell.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 32.Liu Q., Chen Y., Kubota F., Pan J.J., Murakami T. Expression of protocadherin-19 in the nervous system of the embryonic zebrafish. Int. J. Dev. Biol. 2010:54. doi: 10.1387/ijdb.092882ql. [DOI] [PubMed] [Google Scholar]
- 33.Satterthwaite A.B., Li Z., Witte O.N. Btk function in B cell development and response. Semi Immunol. 1998;10:309–316. doi: 10.1006/smim.1998.0123. [DOI] [PubMed] [Google Scholar]
- 34.Gauld S.B., Porto J.M.D., Cambier J.C.B. Cell Antigen Receptor Signaling: Roles in Cell Development and Disease. Science. 2002;296:1641–1642. doi: 10.1126/science.1071546. [DOI] [PubMed] [Google Scholar]
- 35.Demars J., Fabre S., Sarry J., Rossetti R., Gilbert H., Persani L., Tosser-Klopp G., Mulsant P., Nowak Z., Drobik W. Genome-Wide Association Studies Identify Two Novel BMP15 Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep. PLoS Genet. 2013;9:269–271. doi: 10.1371/journal.pgen.1003482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Worley K., Carey J., Veitch A., Coltman D.W. Detecting the signature of selection on immune genes in highly structured populations of wild sheep (Ovis dalli) Mol. Ecol. 2006;15:623–637. doi: 10.1111/j.1365-294X.2006.02829.x. [DOI] [PubMed] [Google Scholar]
- 37.Kijas J.W. Haplotype-based analysis of selective sweeps in sheep. Genome. 2014;57:433–437. doi: 10.1139/gen-2014-0049. [DOI] [PubMed] [Google Scholar]
- 38.Amaral A.J., Ferretti L., Megens H.-J., Crooijmans R.P., Nie H., Ramos-Onsins S.E., Perez-Enciso M., Schook L.B., Groenen M.A. Genome-wide footprints of pig domestication and selection revealed through massive parallel sequencing of pooled DNA. PLoS ONE. 2011;6:e14782. doi: 10.1371/journal.pone.0014782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Browning B.L., Browning S.R. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gautier M., Vitalis R. Rehh: An R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics. 2012;28:1176–1177. doi: 10.1093/bioinformatics/bts115. [DOI] [PubMed] [Google Scholar]
- 41.MacEachern S., Hayes B., McEwan J., Goddard M. An examination of positive selection and changing effective population size in Angus and Holstein cattle populations (Bos taurus) using a high density SNP genotyping platform and the contribution of ancient polymorphism to genomic diversity in Domestic cattle. BMC Genom. 2009;10 doi: 10.1186/1471-2164-10-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 43.Jiang Y., Xie M., Chen W., Talbot R., Maddox J.F., Faraut T., Wu C., Muzny D.M., Li Y., Zhang W. The sheep genome illuminates biology of the rumen and lipid metabolism. Science. 2014;344:1168–1173. doi: 10.1126/science.1252806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.