Abstract
The basic structural and functional unit of a living organism is a single cell. To understand the variability and to improve the biomedical requirement of a single cell, its analysis has become a key technique in biological and biomedical research. With a physical boundary of microchannels and microstructures, single cells are efficiently captured and analyzed, whereas electric forces sort and position single cells. Various microfluidic techniques have been exploited to manipulate single cells through hydrodynamic and electric forces. Digital microfluidics (DMF), the manipulation of individual droplets holding minute reagents and cells of interest by electric forces, has received more attention recently. Because of ease of fabrication, compactness and prospective automation, DMF has become a powerful approach for biological application. We review recent developments of various microfluidic chips for analysis of a single cell and for efficient genetic screening. In addition, perspectives to develop analysis of single cells based on DMF and emerging functionality with high throughput are discussed.
Keywords: microfluidic chips, digital microfluidics, single cell analysis, genetic screening, assisted reproductive technologies
1. Introduction
Multicellular organisms are composed of varied cells grouped into specialized tissues and organs, which typically comprise cells of diverse types present in widely varying abundance. The single cell is the basic structural and functional unit of a living organism. The most critical knowledge for biological and biomedical science is constructed from research on cell biology [1,2]; this information about cell functionality and behavior has been applied in many clinical and biomedical applications [3], such as drug development, disease diagnosis, cancer research and assisted reproductive technology (ART).
Traditional cell assays to study cell differentiation, gene expression and drug response were focused mainly on a population of cells. For an average result of multiple cells, the outcome of all cells is assumed to be homogeneous, but several authors have found cellular heterogeneity or multi-modal distributions in a cell population [4,5,6]. The heterogeneity might arise through genetic or non-genetic processes, which induce distinct cellular decision-making [7]. If an average response of multiple cells is taken to be representative of a typical population, cellular heterogeneity might result in a misleading interpretation [8]. Analysis of a single cell indicated that individual cells that form a cell population might have a complicated distribution. This information is ignored in a traditional cell assay, which emphasizes evaluating the mean of a cell population [9]. To understand the variability from cell to cell and to improve the clinical and biomedical applicability, development of new approaches to isolate, to manipulate, to treat and to analyze single cells are required. Analysis of single cells has become a key technique in biological and biomedical research, as one can thereby analyze individual cells within a cell population [1,7].
Various techniques for analysis of a single cell have been developed, such as flow cytometry and microfluidic chips [10]. Flow cytometry, which has been under development for many years, has become a mature technique for single-cell analysis. In particular, fluorescence-activated cell sorting (FACS), builds upon flow cytometry in order to sort single cells that are tagged with specific fluorescent markers [11,12]. Flow cytometry is, however, a complicated and costly system: it cannot support analysis of cells in real time in their natural environment. Furthermore, integration of an entire assay based on a single cell typically entails processes such as cell manipulation, treatment and final detection. To overcome these problems, integrating various methods of cell manipulation with microfluidic lab-on-a-chip (LOC) platforms, also known as microfluidic chips, has become a major activity in assays of single cells [8,11,13,14,15]. Microfluidic chips could provide efficient genetic screening through a well-controlled microenvironment for analysis and treatment of a single cell. On these platforms, isolation of a single cell, purification of mRNA and subsequent multiplex quantitative polymerase chain reaction (qPCR) in real time must be effected on a chip [16]. Single-cell genomics, transcriptomics, epigenomics and proteomics will allow many enduring questions in biological and biomedical sciences to be answered [17,18,19,20]. Digital microfluidics (DMF), the manipulation of individual droplets by electric forces, is one of the particularly important techniques for single cell manipulation and analysis on a microfluidic chip. In this paper, we review recent developments in DMF chips for analysis of a single cell and efficient genetic screening, which involve manipulation of a single cell, next-generation sequencing (NGS) and assisted-reproductive applications. We conclude with views on the future development of DMF chips for analysis of a single cell and discuss how this emerging efficient tool can advance biological and biomedical research.
2. Microfluidic Chips for Analysis of a Single Cell
In an organism, a single cell is small: its volume is about 1 pL. Cells constituting a tissue or organ are complicated and diverse. An extracellular matrix (ECM) provides structural and biochemical support of the structural connection. To analyze a single cell on a chip, a microfluidic chip typically entails integration of four functions: (1) cell sorting, through microwells [21,22,23,24,25], traps [26,27,28,29], optical tweezers [30,31,32,33] or dielectrophoresis (DEP) [34,35,36,37] to isolate a single cell; (2) cell manipulation, through a traditional syringe pump [38] or an electrowetting-on-dielectric (EWOD) technique [39]; (3) cell lysis, through a mechanical [40], chemical [24] or electrical [23,41,42] process; and (4) analysis of an individual cell, such as with PCR [21,43] or NGS [44].
Microfluidic chips have been applied in biological and biomedical research, including culturing, sorting, patterning and genetic screening of single cells for clinical diagnostics [45,46,47,48,49]. These platforms have become a promising tool for efficient genetic screening through analysis of single cells, because these microfluidic chips can provide rapid, real-time and automated analysis. Gossett et al. [50] demonstrated an automated microfluidic technique capable of probing single cells. A rapid assay of the deformability of native populations of leukocytes and malignant cells in pleural effusions has been enabled on this chip. Guan et al. [51] introduced a new microfluidic chip with real-time feedback control to evaluate single-cell deformability, which was used to discriminate different kinds of cells for cancer diagnosis [30]. Guo et al. [52] produced a microfluidic chip to distinguish red blood cells containing parasitic Plasmodium falciparum from uninfected cells. Several microfluidic chips have been generated to capture single cells and to measure the impedance of the cells, such as human cervical epithelioid carcinoma (HeLa) cells [53,54] or circulating tumor cells (CTCs) from blood [55,56]. Kurz et al. [57] reported a microfluidic chip to trap single cells and to measure the impedance for the monitoring of sub-toxic effects on cell membranes.
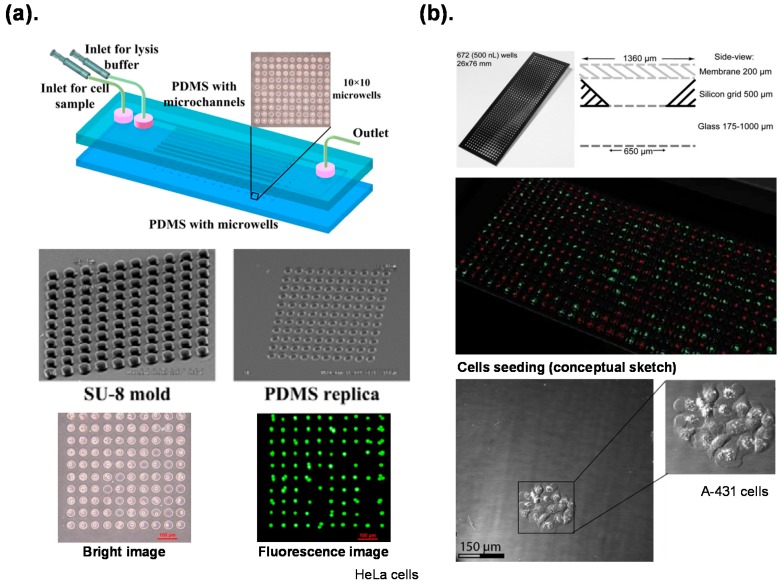
The method most frequently used to isolate a single cell is physical separation. At designed physical boundaries, an individual cell is isolated, captured and sorted with mechanical structures on a chip. Capturing an individual cell with microwells is an attractive strategy, because it is simple and easily operated. Jen et al. [23,24] reported microfluidic chips with arrays of microwells that isolated individual cells and provided chemical and electric lysis of a single cell with high throughput (Figure 1a). Lindstrom et al. [21,22,58,59] developed a novel microplate with microwells for efficient analyses of single cells. This platform allowed each single cell to be cultivated and analyzed individually for reprogramming factor evaluation on stem cells [22], PCR amplification and genetic analysis [21] (Figure 1b).
Figure 1.
Individual cells isolated on a chip with microwells described in: (a) Jen et al., 2012 [24]; (b) Lindstrom et al., 2009 [21]. Reproduction of the figures has been made with permission from Multidisciplinary Digital Publishing Institute and The Royal Society of Chemistry.
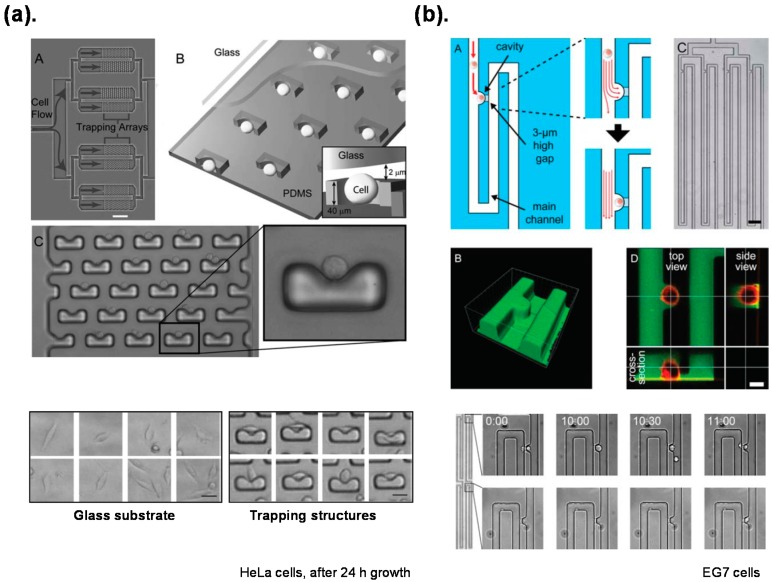
Microfluidic hydrodynamic traps combine dynamic cell isolation with prospective high throughput on a chip [60,61]. Di Carlo et al. [26,62] produced a dynamic platform that allows culture of a single cell with a consistent environment and dynamic control of individual cells (Figure 2a). Kobel et al. [60] reported a microfluidic chip with efficiency of trapping a single cell enhanced up to 97% (Figure 2b).
Figure 2.
Individual cell isolated on a chip with microfluidic hydrodynamic traps described in: (a) Di Carlo et al., 2006 [26]: (A) A photograph of the cell trapping device; (B) A diagram of the device and mechanism of trapping; (C) A high resolution micrograph of the trapping device; (b) Kobel et al., 2010 [60]: (A) Schematic illustration of the single cell trap; (B) A three-dimensional reconstruction image of the cell trap; (C) An array of the single cell traps; (D) An orthogonal view of a fluorescently labeled single cell. Reproduction of the figures has been made with permission from The Royal Society of Chemistry.
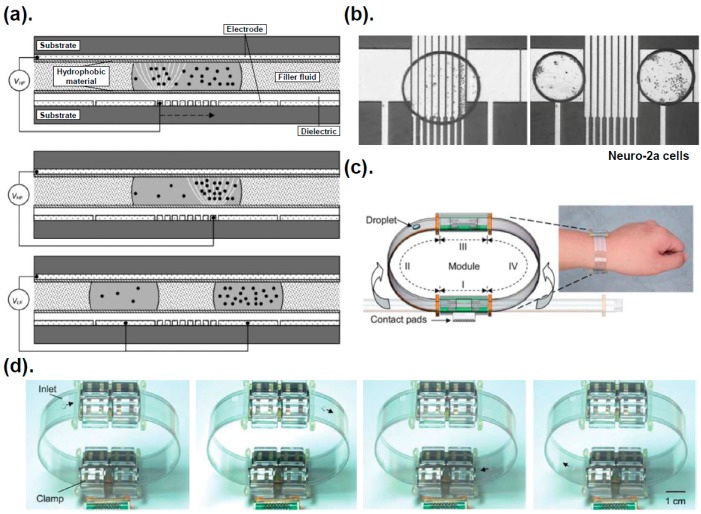
Compared with use of mechanical structures, isolating a cell with an optical or electric force is a contact-free method that eliminates deformation and damage of cells [63,64]. Here we focus more on the electric forces, including DEP and EWOD. The DEP force generated with a non-uniform electric field interacting with polarized, suspended particles [65,66,67] has been widely used to manipulate cells [39,63,68,69]. The DEP force is classified as positive DEP (pDEP) and negative DEP (nDEP) based on the polarizability of the particles or cells and the liquid. Fan et al. [39] used DEP forces to concentrate suspended particles in a liquid droplet with dielectric-coated electrodes patterned on a plate (Figure 3a). Creating two droplets with mammalian cells and polystyrene beads at distinct concentrations was achieved with DEP and EWOD (Figure 3b).
Figure 3.
Dielectrophoresis (DEP) forces exerting on the suspended particles described by Fan et al., 2008 [39]. (a) A parallel-plate device with square and strip electrodes to manipulate droplets and a particle; (b) DEP forces exerted on suspended particles including mammalian cells (Neuro-2a); (c,d) Prototype of EWOD-based, continuously microfluidic module for a portable system reported by Fan et al., 2011 [70]. Reproduction of the figures has been made with permission from The Royal Society of Chemistry.
3. Digital Microfluidic Chips and Biological Application
Digital microfluidics (DMF), according to which tiny droplets are manipulated with electric forces, including EWOD and DEP, have been confirmed to be a powerful platform for reagent addition, droplet transmission, solution mixing, splitting and dispensing for biological application [70,71,72,73,74,75,76,77,78,79,80]. For a simple configuration of device and easy modular interfaces, portable and wearable DMF systems with assembled modules for continuous actuation of droplets were demonstrated [70] (Figure 3c).
A DMF chip with arrays of electrodes is typically fabricated on indium tin oxide (ITO) glass plates using photolithography and wet etching. The patterned ITO electrodes are then covered with a dielectric and a hydrophobic layer [16,39,73,81]. The advantages of DMF include ease of fabrication, simple device structure, small consumption of reagents, easy integration with analytical instruments and prospective automation. Thus, DMF has become highly suitable for biological application. Barbulovic-Nad et al. [80] introduced a DMF chip to implement cell-based assays; the platform was demonstrated to be advantageous for cell-based assays because of potential for automated manipulation of multiple reagents. Vergauwe et al. [78] reported a DMF chip for homogeneous and heterogeneous bio-assays with great analytical performance capable of medical applications. Kumar et al. [75] demonstrated the first use of a DMF technique for individual protoplasts from Arabidopsis thaliana plants. Shih et al. developed the first DMF chip capable of cell impedance sensing [76]; they also integrated droplet-in-channel microfluidics with DMF to develop a novel chip to perform complicated assays [81].
This work demonstrates that DMF chips would be a generic and powerful platform for the biological assays, including drug screening, immunoassays, analysis of single cells and digital PCR. This promising new technique might allow the efficient genetic screening based on a single cell to become a reality.
4. Digital Microfluidic Chips for Genetic Screening
Investigating gene expression and developing genetic screening at a level of a single cell provides an important capability to resolve the problem of disease etiology, cancer pathology and other biomedical applications [82]. Traditional methods of genetic screening require a large amount of sample for an analysis, which typically decreases the sensitivity and accuracy on analysis of only a single cell [83,84]. Various microfluidic techniques have been developed to address this problem. Digital polymerase chain reaction (digital PCR) platforms have measured DNA or cDNA of a single cell [85,86], but challenges persist in treating the integration of varied programs for genetic analyses of a single cell on a device, including cell sorting, manipulation, lysis, PCR and genetic screening. Within the past decade, microfluidic chips have become one of the most powerful platforms to achieve efficient genetic screening at the level of a single cell [16,87]. Toriello et al. [88] and Bontoux et al. [89] reported a polydimethylsiloxane (PDMS) device to analyze the gene expression of single cells, and Marcus et al. [90] developed a microfluidic chip with integrated flow that conducted cell capture, lysis, mRNA purification, cDNA synthesis and purification but with a complicated auxiliary system for control.
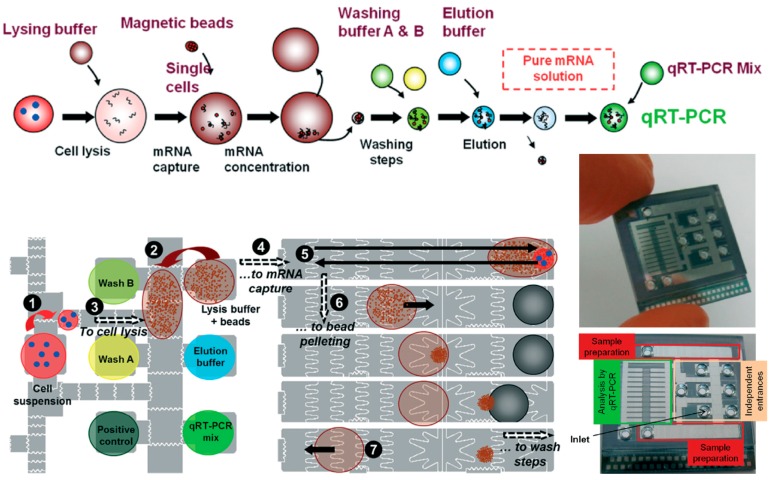
As mentioned above, the DMF chip has advantages of ease of fabrication, simple supporting instrumentation and prospective automation. Rival et al. [16] described an integrated and automated EWOD system to perform a complete workflow from the isolation of a single cell to a genetic analysis (Figure 4). DMF is becoming a powerful approach for biological applications, even enabling sample preparation for PCR to develop an efficient genetic screening platform based on a single cell [16,91,92].
Figure 4.
An integrated EWOD system for genetic analysis based on a single cell described by Rival et al., 2014 [16]. The lysis and mRNA capture steps: (1) A droplet containing a few cells is dispensed; (2) A droplet of lysis buffer and magnetic beads is dispensed; (3) A droplet with the cells and a droplet with the beads are merged for cell lysis; (4) This merged droplet is moved to the “sample preparation” electrodes for operating magnetic beads; (5) The droplet is moved back and forth to enable bead mixing and mRNA capture; (6) The droplet is moved towards the magnet, the beads are concentrated to result in bead extraction; (7) An empty droplet is moved towards the waist. Reproduction of the figures has been made with permission from The Royal Society of Chemistry.
5. Efficient Genetic Screening for Assisted Reproductive Techniques
The early development of a mammalian embryo is a complicated process involving an upheaval of a transcriptional architecture [93,94,95]. In applications, human in vitro fertilization (IVF) is an important scientific achievement in the twentieth century, but until recently there has been little knowledge of regulatory mechanisms in genes of early mammalian embryos. The early embryo undergoes cleavage divisions in a series from two cell, four cells, eight cells, morula, even to blastocyst. A platform for genetic screening based on a single cell could provide critical knowledge to clarify regulatory mechanisms of genes in early mammalian embryos [93]. This emerging efficient technique would benefit a biomedical approach, such as assisted reproductive technology (ART).
The microfluidic ART platforms under development are focused on simulation of the Fallopian tube to optimize the IVF, especially the early embryo culture in vitro [96,97,98]. Pre-implantation genetic diagnosis (PGD) has been recently developed to detect genetic diseases [99,100]. The current methods of sorting single cells include taking trophectoderm cells from blastocyst, blastomeres from embryos at a cleavage stage and polar bodies from the oocyte or zygote [101]. The advantages of a DMF chip include simple accompanying instrumentation and prospective automation for sorting single cells from an early embryo. Huang et al. [98] demonstrated a EWOD-based microfluidic device for culture of early mammalian embryos in vitro, presaging future clinical application. Although a DMF chip applied in efficient genetic diagnosis is still at an initial stage, we believe that this powerful platform will have a major impact on the ART field.
6. Conclusions
Recent developments of microfluidic chips for single cell analysis were reviewed; microfluidic techniques provide numerous advantages for biological and biomedical research, including ease for modularity, small sample requirement, potential of automation, and high-throughput. Perspectives on DMF in analysis of a single cell and efficient genetic screening were particularly focused on and described. We expect that these novel single-cell techniques on microfluidic chips will be important in biomedical areas.
Acknowledgments
Jie-Long He thanks the Ministry of Science and Technology, Taiwan for providing a post-doctoral fellowship.
Author Contributions
Jie-Long He and Shih-Kang Fan wrote the manuscript. An-Te Chen and Jyong-Huei Lee assisted in collecting and integrating the references.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Svahn H.A., van den Berg A. Single cells or large populations? Lab Chip. 2007;7:544–546. doi: 10.1039/b704632b. [DOI] [PubMed] [Google Scholar]
- 2.Bahcall O.G. Single cell resolution in regulation of gene expression. Mol. Syst. Biol. 2005;1:2005.0015. doi: 10.1038/msb4100020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dittrich P.S., Tachikawa K., Manz A. Micro total analysis systems. Latest advancements and trends. Anal. Chem. 2006;78:3887–3908. doi: 10.1021/ac0605602. [DOI] [PubMed] [Google Scholar]
- 4.Graf T., Stadtfeld M. Heterogeneity of embryonic and adult stem cells. Cell Stem Cell. 2008;3:480–483. doi: 10.1016/j.stem.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Zhou L., Shen Y., Jiang L., Yin D., Guo J., Zheng H., Sun H., Wu R., Guo Y. Systems mapping for hematopoietic progenitor cell heterogeneity. PLoS ONE. 2015;10:e0126937. doi: 10.1371/journal.pone.0126937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donati G., Watt F.M. Stem cell heterogeneity and plasticity in epithelia. Cell Stem Cell. 2015;16:465–476. doi: 10.1016/j.stem.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 7.Perkins T.J., Swain P.S. Strategies for cellular decision-making. Mol. Syst. Biol. 2009;5:326. doi: 10.1038/msb.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin H., Marshall D. Microfluidics for single cell analysis. Curr. Opin. Biotechnol. 2012;23:110–119. doi: 10.1016/j.copbio.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Wang D., Bodovitz S. Single cell analysis: The new frontier in “omics”. Trends Biotechnol. 2010;28:281–290. doi: 10.1016/j.tibtech.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah P., Zhu X., Chen C., Hu Y., Li C.Z. Lab-on-chip device for single cell trapping and analysis. Biomed. Microdevices. 2014;16:35–41. doi: 10.1007/s10544-013-9803-7. [DOI] [PubMed] [Google Scholar]
- 11.Cheung K.C., di Berardino M., Schade-Kampmann G., Hebeisen M., Pierzchalski A., Bocsi J., Mittag A., Tarnok A. Microfluidic impedance-based flow cytometry. Cytom. Part A J. Int. Soc. Anal. Cytol. 2010;77:648–666. doi: 10.1002/cyto.a.20910. [DOI] [PubMed] [Google Scholar]
- 12.Ning B., Yihong Z., Chang L. Microfluidic electroporative flow cytometry for studying single-cell biomechanics. Anal. Chem. 2008;80:7714–7719. doi: 10.1021/ac801060t. [DOI] [PubMed] [Google Scholar]
- 13.Shah P., Kaushik A., Zhu X., Zhang C., Li C.Z. Chip based single cell analysis for nanotoxicity assessment. Analyst. 2014;139:2088–2098. doi: 10.1039/c3an02280c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindstrom S., Andersson-Svahn H. Miniaturization of biological assays—Overview on microwell devices for single-cell analyses. Biochim. Biophys. Acta. 2011;1810:308–316. doi: 10.1016/j.bbagen.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 15.Le Gac S., van den Berg A. Single cells as experimentation units in lab-on-a-chip devices. Trends Biotechnol. 2010;28:55–62. doi: 10.1016/j.tibtech.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Rival A., Jary D., Delattre C., Fouillet Y., Castellan G., Bellemin-Comte A., Gidrol X. An EWOD-based microfluidic chip for single-cell isolation, mRNA purification and subsequent multiplex qPCR. Lab Chip. 2014;14:3739–3749. doi: 10.1039/C4LC00592A. [DOI] [PubMed] [Google Scholar]
- 17.Shapiro E., Biezuner T., Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 18.Ullal A.V., Peterson V., Agasti S.S., Tuang S., Juric D., Castro C.M., Weissleder R. Cancer cell profiling by barcoding allows multiplexed protein analysis in fine-needle aspirates. Sci. Transl. Med. 2014;6:219ra219. doi: 10.1126/scitranslmed.3007361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salehi-Reyhani A., Burgin E., Ces O., Willison K.R., Klug D.R. Addressable droplet microarrays for single cell protein analysis. Analyst. 2014;139:5367–5374. doi: 10.1039/C4AN01208A. [DOI] [PubMed] [Google Scholar]
- 20.Kalisky T., Quake S.R. Single-cell genomics. Nat. Methods. 2011;8:311–314. doi: 10.1038/nmeth0411-311. [DOI] [PubMed] [Google Scholar]
- 21.Lindstrom S., Hammond M., Brismar H., Andersson-Svahn H., Ahmadian A. PCR amplification and genetic analysis in a microwell cell culturing chip. Lab Chip. 2009;9:3465–3471. doi: 10.1039/b912596e. [DOI] [PubMed] [Google Scholar]
- 22.Lindstrom S., Eriksson M., Vazin T., Sandberg J., Lundeberg J., Frisen J., Andersson-Svahn H. High-density microwell chip for culture and analysis of stem cells. PLoS ONE. 2009;4:e6997. doi: 10.1371/journal.pone.0006997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jen C.P., Amstislavskaya T.G., Liu Y.H., Hsiao J.H., Chen Y.H. Single-cell electric lysis on an electroosmotic-driven microfluidic chip with arrays of microwells. Sensors (Basel) 2012;12:6967–6977. doi: 10.3390/s120606967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jen C.P., Hsiao J.H., Maslov N.A. Single-cell chemical lysis on microfluidic chips with arrays of microwells. Sensors (Basel) 2012;12:347–358. doi: 10.3390/s120100347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osada K., Hosokawa M., Yoshino T., Tanaka T. Monitoring of cellular behaviors by microcavity array-based single-cell patterning. Analyst. 2014;139:425–430. doi: 10.1039/C3AN01698F. [DOI] [PubMed] [Google Scholar]
- 26.Di Carlo D., Wu L.Y., Lee L.P. Dynamic single cell culture array. Lab Chip. 2006;6:1445–1449. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- 27.Skelley A.M., Kirak O., Suh H., Jaenisch R., Voldman J. Microfluidic control of cell pairing and fusion. Nat. Methods. 2009;6:147–152. doi: 10.1038/nmeth.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee S.-W., Kang J.Y., Lee I.-H., Ryu S.-S., Kwak S.-M., Shin K.-S., Kim C., Jung H.-I., Kim T.-S. Single-cell assay on CD-like lab chip using centrifugal massive single-cell trap. Sens. Actuators A Phys. 2008;143:64–69. doi: 10.1016/j.sna.2007.06.043. [DOI] [Google Scholar]
- 29.Liu W., Dechev N., Foulds I.G., Burke R., Parameswaran A., Park E.J. A novel permalloy based magnetic single cell micro array. Lab Chip. 2009;9:2381–2390. doi: 10.1039/b821044f. [DOI] [PubMed] [Google Scholar]
- 30.Remmerbach T.W., Wottawah F., Dietrich J., Lincoln B., Wittekind C., Guck J. Oral cancer diagnosis by mechanical phenotyping. Cancer Res. 2009;69:1728–1732. doi: 10.1158/0008-5472.CAN-08-4073. [DOI] [PubMed] [Google Scholar]
- 31.Guck J., Schinkinger S., Lincoln B., Wottawah F., Ebert S., Romeyke M., Lenz D., Erickson H.M., Ananthakrishnan R., Mitchell D., et al. Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys. J. 2005;88:3689–3698. doi: 10.1529/biophysj.104.045476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang M.M., Tu E., Raymond D.E., Yang J.M., Zhang H., Hagen N., Dees B., Mercer E.M., Forster A.H., Kariv I., et al. Microfluidic sorting of mammalian cells by optical force switching. Nat. Biotechnol. 2005;23:83–87. doi: 10.1038/nbt1050. [DOI] [PubMed] [Google Scholar]
- 33.Yang T., Paie P., Nava G., Bragheri F., Martinez Vazquez R., Minzioni P., Veglione M., di Tano M., Mondello C., Osellame R., et al. An integrated optofluidic device for single-cell sorting driven by mechanical properties. Lab Chip. 2015;15:1262–1266. doi: 10.1039/C4LC01496K. [DOI] [PubMed] [Google Scholar]
- 34.Taff B.M., Voldman J. A scalable addressable positive-dielectrophoretic cell-sorting array. Anal. Chem. 2005;77:7976–7983. doi: 10.1021/ac0513616. [DOI] [PubMed] [Google Scholar]
- 35.Hu X., Bessette P.H., Qian J., Meinhart C.D., Daugherty P.S., Soh H.T. Marker-specific sorting of rare cells using dielectrophoresis. Proc. Natl. Acad. Sci. USA. 2005;102:15757–15761. doi: 10.1073/pnas.0507719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah G.J., Ohta A.T., Chiou E.P., Wu M.C., Kim C.J. EWOD-driven droplet microfluidic device integrated with optoelectronic tweezers as an automated platform for cellular isolation and analysis. Lab Chip. 2009;9:1732–1739. doi: 10.1039/b821508a. [DOI] [PubMed] [Google Scholar]
- 37.Park S., Wijethunga P.A., Moon H., Han B. On-chip characterization of cryoprotective agent mixtures using an EWOD-based digital microfluidic device. Lab Chip. 2011;11:2212–2221. doi: 10.1039/c1lc20111e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenbluth M.J., Lam W.A., Fletcher D.A. Analyzing cell mechanics in hematologic diseases with microfluidic biophysical flow cytometry. Lab Chip. 2008;8:1062–1070. doi: 10.1039/b802931h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan S.K., Huang P.W., Wang T.T., Peng Y.H. Cross-scale electric manipulations of cells and droplets by frequency-modulated dielectrophoresis and electrowetting. Lab Chip. 2008;8:1325–1331. doi: 10.1039/b803204a. [DOI] [PubMed] [Google Scholar]
- 40.Di Carlo D., Jeong K.H., Lee L.P. Reagentless mechanical cell lysis by nanoscale barbs in microchannels for sample preparation. Lab Chip. 2003;3:287–291. doi: 10.1039/b305162e. [DOI] [PubMed] [Google Scholar]
- 41.Zheng S., Lin H., Liu J.Q., Balic M., Datar R., Cote R.J., Tai Y.C. Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells. J. Chromatogr. A. 2007;1162:154–161. doi: 10.1016/j.chroma.2007.05.064. [DOI] [PubMed] [Google Scholar]
- 42.Bao N., Kodippili G.C., Giger K.M., Fowler V.M., Low P.S., Lu C. Single-cell electrical lysis of erythrocytes detects deficiencies in the cytoskeletal protein network. Lab Chip. 2011;11:3053–3056. doi: 10.1039/c1lc20365g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ginsberg S.D. RNA amplification strategies for small sample populations. Methods. 2005;37:229–237. doi: 10.1016/j.ymeth.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 44.Feng X., Du W., Luo Q., Liu B.F. Microfluidic chip: Next-generation platform for systems biology. Anal. Chim. Acta. 2009;650:83–97. doi: 10.1016/j.aca.2009.04.051. [DOI] [PubMed] [Google Scholar]
- 45.Koster S., Angile F.E., Duan H., Agresti J.J., Wintner A., Schmitz C., Rowat A.C., Merten C.A., Pisignano D., Griffiths A.D., et al. Drop-based microfluidic devices for encapsulation of single cells. Lab Chip. 2008;8:1110–1115. doi: 10.1039/b802941e. [DOI] [PubMed] [Google Scholar]
- 46.Clausell-Tormos J., Lieber D., Baret J.C., El-Harrak A., Miller O.J., Frenz L., Blouwolff J., Humphry K.J., Koster S., Duan H., et al. Droplet-based microfluidic platforms for the encapsulation and screening of mammalian cells and multicellular organisms. Chem. Biol. 2008;15:427–437. doi: 10.1016/j.chembiol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 47.Brouzes E., Medkova M., Savenelli N., Marran D., Twardowski M., Hutchison J.B., Rothberg J.M., Link D.R., Perrimon N., Samuels M.L. Droplet microfluidic technology for single-cell high-throughput screening. Proc. Natl. Acad. Sci. USA. 2009;106:14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu L., Chen M.C., Cheung K.C. Droplet-based microfluidic system for multicellular tumor spheroid formation and anticancer drug testing. Lab Chip. 2010;10:2424–2432. doi: 10.1039/c004590j. [DOI] [PubMed] [Google Scholar]
- 49.Pekin D., Skhiri Y., Baret J.C., le Corre D., Mazutis L., Salem C.B., Millot F., El Harrak A., Hutchison J.B., Larson J.W., et al. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip. 2011;11:2156–2166. doi: 10.1039/c1lc20128j. [DOI] [PubMed] [Google Scholar]
- 50.Gossett D.R., Tse H.T., Lee S.A., Ying Y., Lindgren A.G., Yang O.O., Rao J., Clark A.T., di Carlo D. Hydrodynamic stretching of single cells for large population mechanical phenotyping. Proc. Natl. Acad. Sci. USA. 2012;109:7630–7635. doi: 10.1073/pnas.1200107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guan G., Chen P.C.Y., Peng W.K., Bhagat A.A., Ong C.J., Han J. Real-time control of a microfluidic channel for size-independent deformability cytometry. J. Micromech. Microeng. 2012;22:105037. doi: 10.1088/0960-1317/22/10/105037. [DOI] [Google Scholar]
- 52.Guo Q., Reiling S.J., Rohrbach P., Ma H. Microfluidic biomechanical assay for red blood cells parasitized by Plasmodium falciparum. Lab Chip. 2012;12:1143–1150. doi: 10.1039/c2lc20857a. [DOI] [PubMed] [Google Scholar]
- 53.Jang L.S., Wang M.H. Microfluidic device for cell capture and impedance measurement. Biomed. Microdevices. 2007;9:737–743. doi: 10.1007/s10544-007-9084-0. [DOI] [PubMed] [Google Scholar]
- 54.Malleo D., Nevill J.T., Lee L.P., Morgan H. Continuous differential impedance spectroscopy of single cells. Microfluid. Nanofluid. 2010;9:191–198. doi: 10.1007/s10404-009-0534-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han K.H., Han A., Frazier A.B. Microsystems for isolation and electrophysiological analysis of breast cancer cells from blood. Biosens. Bioelectron. 2006;21:1907–1914. doi: 10.1016/j.bios.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 56.Chung J., Issadore D., Ullal A., Lee K., Weissleder R., Lee H. Rare cell isolation and profiling on a hybrid magnetic/size-sorting chip. Biomicrofluidics. 2013;7:54107. doi: 10.1063/1.4821923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kurz C.M., Buth H., Sossalla A., Vermeersch V., Toncheva V., Dubruel P., Schacht E., Thielecke H. Chip-based impedance measurement on single cells for monitoring sub-toxic effects on cell membranes. Biosens. Bioelectron. 2011;26:3405–3412. doi: 10.1016/j.bios.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 58.Lindstrom S., Mori K., Ohashi T., Andersson-Svahn H. A microwell array device with integrated microfluidic components for enhanced single-cell analysis. Electrophoresis. 2009;30:4166–4171. doi: 10.1002/elps.200900572. [DOI] [PubMed] [Google Scholar]
- 59.Lindstrom S., Larsson R., Svahn H.A. Towards high-throughput single cell/clone cultivation and analysis. Electrophoresis. 2008;29:1219–1227. doi: 10.1002/elps.200700536. [DOI] [PubMed] [Google Scholar]
- 60.Kobel S., Valero A., Latt J., Renaud P., Lutolf M. Optimization of microfluidic single cell trapping for long-term on-chip culture. Lab Chip. 2010;10:857–863. doi: 10.1039/b918055a. [DOI] [PubMed] [Google Scholar]
- 61.Bow H., Pivkin I.V., Diez-Silva M., Goldfless S.J., Dao M., Niles J.C., Suresh S., Han J. A microfabricated deformability-based flow cytometer with application to malaria. Lab Chip. 2011;11:1065–1073. doi: 10.1039/c0lc00472c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Di Carlo D., Lee L.P. Dynamic single-cell analysis for quantitative biology. Anal. Chem. 2006;78:7918–7925. doi: 10.1021/ac069490p. [DOI] [PubMed] [Google Scholar]
- 63.Lan K.C., Jang L.S. Integration of single-cell trapping and impedance measurement utilizing microwell electrodes. Biosens. Bioelectron. 2011;26:2025–2031. doi: 10.1016/j.bios.2010.08.080. [DOI] [PubMed] [Google Scholar]
- 64.Huang N.T., Zhang H.L., Chung M.T., Seo J.H., Kurabayashi K. Recent advancements in optofluidics-based single-cell analysis: Optical on-chip cellular manipulation, treatment, and property detection. Lab Chip. 2014;14:1230–1245. doi: 10.1039/c3lc51211h. [DOI] [PubMed] [Google Scholar]
- 65.Rahman A.R.A., Price D.T., Bhansali S. Effect of electrode geometry on the impedance evaluation of tissue and cell culture. Sens. Actuators B Chem. 2007;127:89–96. [Google Scholar]
- 66.Fan S.K., Chiu C.P., Hsu C.H., Chen S.C., Huang L.L., Lin Y.H., Fang W.F., Chen J.K., Yang J.T. Particle chain display—An optofluidic electronic paper. Lab Chip. 2012;12:4870–4876. doi: 10.1039/c2lc40480j. [DOI] [PubMed] [Google Scholar]
- 67.Fan S.K., Chiu C.P., Huang P.W. Transmittance tuning by particle chain polarization in electrowetting-driven droplets. Biomicrofluidics. 2010;4:43011. doi: 10.1063/1.3516656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Choi H., Kim K.B., Jeon C.S., Hwang I., Lee S., Kim H.K., Kim H.C., Chung T.D. A label-free DC impedance-based microcytometer for circulating rare cancer cell counting. Lab Chip. 2013;13:970–977. doi: 10.1039/c2lc41376k. [DOI] [PubMed] [Google Scholar]
- 69.Mernier G., Duqi E., Renaud P. Characterization of a novel impedance cytometer design and its integration with lateral focusing by dielectrophoresis. Lab Chip. 2012;12:4344–4349. doi: 10.1039/c2lc40551b. [DOI] [PubMed] [Google Scholar]
- 70.Fan S.K., Yang H., Hsu W. Droplet-on-a-wristband: Chip-to-chip digital microfluidic interfaces between replaceable and flexible electrowetting modules. Lab Chip. 2011;11:343–347. doi: 10.1039/C0LC00178C. [DOI] [PubMed] [Google Scholar]
- 71.Fan S.K., Hsieh T.H., Lin D.Y. General digital microfluidic platform manipulating dielectric and conductive droplets by dielectrophoresis and electrowetting. Lab Chip. 2009;9:1236–1242. doi: 10.1039/b816535a. [DOI] [PubMed] [Google Scholar]
- 72.Fan S.K., Chen W.J., Lin T.H., Wang T.T., Lin Y.C. Reconfigurable liquid pumping in electric-field-defined virtual microchannels by dielectrophoresis. Lab Chip. 2009;9:1590–1595. doi: 10.1039/b900790c. [DOI] [PubMed] [Google Scholar]
- 73.Fan S.K., Yang H., Wang T.T., Hsu W. Asymmetric electrowetting—Moving droplets by a square wave. Lab Chip. 2007;7:1330–1335. doi: 10.1039/b704084a. [DOI] [PubMed] [Google Scholar]
- 74.Choi K., Ng A.H., Fobel R., Wheeler A.R. Digital microfluidics. Annu. Rev. Anal. Chem. 2012;5:413–440. doi: 10.1146/annurev-anchem-062011-143028. [DOI] [PubMed] [Google Scholar]
- 75.Kumar P.T., Toffalini F., Witters D., Vermeir S., Rolland F., Hertog M.L.A.T.M., Nicolai B.M., Puers R., Geeraerd A., Lammertyn J. Digital microfluidic chip technology for water permeability measurements on single isolated plant protoplasts. Sens. Actuators B Chem. 2014;199:479–487. doi: 10.1016/j.snb.2014.04.018. [DOI] [Google Scholar]
- 76.Shih S.C., Barbulovic-Nad I., Yang X., Fobel R., Wheeler A.R. Digital microfluidics with impedance sensing for integrated cell culture and analysis. Biosens. Bioelectron. 2013;42:314–320. doi: 10.1016/j.bios.2012.10.035. [DOI] [PubMed] [Google Scholar]
- 77.Witters D., Vergauwe N., Vermeir S., Ceyssens F., Liekens S., Puers R., Lammertyn J. Biofunctionalization of electrowetting-on-dielectric digital microfluidic chips for miniaturized cell-based applications. Lab Chip. 2011;11:2790–2794. doi: 10.1039/c1lc20340a. [DOI] [PubMed] [Google Scholar]
- 78.Vergauwe N., Witters D., Ceyssens F., Vermeir S., Verbruggen B., Puers R., Lammertyn J. A versatile electrowetting-based digital microfluidic platform for quantitative homogeneous and heterogeneous bio-assays. J. Micromech. Microeng. 2011;21:054026. doi: 10.1088/0960-1317/21/5/054026. [DOI] [Google Scholar]
- 79.Vergauwe N., Witters D., Atalay Y.T., Verbruggen B., Vermeir S., Ceyssens F., Puers R., Lammertyn J. Controlling droplet size variability of a digital lab-on-a-chip for improved bio-assay performance. Microfluid. Nanofluid. 2011;11:25–34. doi: 10.1007/s10404-011-0769-6. [DOI] [Google Scholar]
- 80.Barbulovic-Nad I., Yang H., Park P.S., Wheeler A.R. Digital microfluidics for cell-based assays. Lab Chip. 2008;8:519–526. doi: 10.1039/b717759c. [DOI] [PubMed] [Google Scholar]
- 81.Shih S.C., Gach P.C., Sustarich J., Simmons B.A., Adams P.D., Singh S., Singh A.K. A droplet-to-digital (D2D) microfluidic device for single cell assays. Lab Chip. 2015;15:225–236. doi: 10.1039/C4LC00794H. [DOI] [PubMed] [Google Scholar]
- 82.Thompson A.M., Gansen A., Paguirigan A.L., Kreutz J.E., Radich J.P., Chiu D.T. Self-digitization microfluidic chip for absolute quantification of mRNA in single cells. Anal. Chem. 2014;86:12308–12314. doi: 10.1021/ac5035924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thompson A.M., Paguirigan A.L., Kreutz J.E., Radich J.P., Chiu D.T. Microfluidics for single-cell genetic analysis. Lab Chip. 2014;14:3135–3142. doi: 10.1039/C4LC00175C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee T.M., Hsing I.M. DNA-based bioanalytical microsystems for handheld device applications. Anal. Chim. Acta. 2006;556:26–37. doi: 10.1016/j.aca.2005.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sanders R., Huggett J.F., Bushell C.A., Cowen S., Scott D.J., Foy C.A. Evaluation of digital PCR for absolute DNA quantification. Anal. Chem. 2011;83:6474–6484. doi: 10.1021/ac103230c. [DOI] [PubMed] [Google Scholar]
- 86.White R.A., 3rd, Quake S.R., Curr K. Digital PCR provides absolute quantitation of viral load for an occult RNA virus. J. Virol. Methods. 2012;179:45–50. doi: 10.1016/j.jviromet.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 87.Bennett M.R., Hasty J. Microfluidic devices for measuring gene network dynamics in single cells. Nat. Rev. Genet. 2009;10:628–638. doi: 10.1038/nrg2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Toriello N.M., Douglas E.S., Thaitrong N., Hsiao S.C., Francis M.B., Bertozzi C.R., Mathies R.A. Integrated microfluidic bioprocessor for single-cell gene expression analysis. Proc. Natl. Acad. Sci. USA. 2008;105:20173–20178. doi: 10.1073/pnas.0806355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bontoux N., Dauphinot L., Vitalis T., Studer V., Chen Y., Rossier J., Potier M.C. Integrating whole transcriptome assays on a lab-on-a-chip for single cell gene profiling. Lab Chip. 2008;8:443–450. doi: 10.1039/b716543a. [DOI] [PubMed] [Google Scholar]
- 90.Marcus J.S., Anderson W.F., Quake S.R. Microfluidic single-cell mRNA isolation and analysis. Anal. Chem. 2006;78:3084–3089. doi: 10.1021/ac0519460. [DOI] [PubMed] [Google Scholar]
- 91.Schell W.A., Benton J.L., Smith P.B., Poore M., Rouse J.L., Boles D.J., Johnson M.D., Alexander B.D., Pamula V.K., Eckhardt A.E., et al. Evaluation of a digital microfluidic real-time PCR platform to detect DNA of Candida albicans in blood. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:2237–2245. doi: 10.1007/s10096-012-1561-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hua Z., Rouse J.L., Eckhardt A.E., Srinivasan V., Pamula V.K., Schell W.A., Benton J.L., Mitchell T.G., Pollack M.G. Multiplexed real-time polymerase chain reaction on a digital microfluidic platform. Anal. Chem. 2010;82:2310–2316. doi: 10.1021/ac902510u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xue Z., Huang K., Cai C., Cai L., Jiang C.Y., Feng Y., Liu Z., Zeng Q., Cheng L., Sun Y.E., et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500:593–597. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Niakan K.K., Han J., Pedersen R.A., Simon C., Pera R.A. Human pre-implantation embryo development. Development. 2012;139:829–841. doi: 10.1242/dev.060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zeng F., Baldwin D.A., Schultz R.M. Transcript profiling during preimplantation mouse development. Dev. Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 96.Swain J.E., Smith G.D. Advances in embryo culture platforms: Novel approaches to improve preimplantation embryo development through modifications of the microenvironment. Hum. Reprod. Update. 2011;17:541–557. doi: 10.1093/humupd/dmr006. [DOI] [PubMed] [Google Scholar]
- 97.Smith G.D., Swain J.E., Bormann C.L. Microfluidics for gametes, embryos, and embryonic stem cells. Semin. Reprod. Med. 2011;29:5–14. doi: 10.1055/s-0030-1268699. [DOI] [PubMed] [Google Scholar]
- 98.Huang H.Y., Shen H.H., Tien C.H., Li C.J., Fan S.K., Liu C.H., Hsu W.S., Yao D.J. Digital microfluidic dynamic culture of mammalian embryos on an electrowetting on dielectric (EWOD) chip. PLoS ONE. 2015;10:e0124196. doi: 10.1371/journal.pone.0124196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Egozcue J., Santalo J., Gimenez C., Durban M., Benet J., Navarro J., Vidal F. Preimplantation genetic screening and human implantation. J. Reprod. Immunol. 2002;55:65–72. doi: 10.1016/S0165-0378(01)00137-1. [DOI] [PubMed] [Google Scholar]
- 100.Yan L., Wei Y., Huang J., Zhu X., Shi X., Xia X., Yan J., Lu C., Lian Y., Li R., et al. Advances in preimplantation genetic diagnosis/screening. Sci. China Life Sci. 2014;57:665–671. doi: 10.1007/s11427-014-4683-5. [DOI] [PubMed] [Google Scholar]
- 101.Harper J.C., Harton G. The use of arrays in preimplantation genetic diagnosis and screening. Fertil. Steril. 2010;94:1173–1177. doi: 10.1016/j.fertnstert.2010.04.064. [DOI] [PubMed] [Google Scholar]