Abstract
The HeLa cell line is one of the most popular cell lines in biomedical research, despite its well-known chromosomal instability. We compared the genomic and transcriptomic profiles of 4 different HeLa batches and showed that the gain and loss of genomic material varies widely between batches, drastically affecting basal gene expression. Moreover, different pathways were activated in response to a hypoxic stimulus. Our study emphasizes the large genomic and transcriptomic variability among different batches, to the point that the same experiment performed with different batches can lead to distinct conclusions and irreproducible results. The HeLa cell line is thought to be a unique cell line but it is clear that substantial differences between the primary tumour and the human genome exist and that an indeterminate number of HeLa cell lines may exist, each with a unique genomic profile.
Since HeLa was first established as a human cancer cell line in 19521, it has become probably the most-used human cell line in biological research. The cell line was established from the invasive cervical adenocarcinoma of a young patient, Henrietta Lacks, who eventually died in 1951, and it was the first successful attempt to establish a culture of immortalized human cells. Starting with the pioneering research of Jonas Salk, who developed the first polio vaccine2, the use of HeLa cells has contributed to many fundamental scientific breakthroughs.
HeLa cells have been employed to investigate cancer, AIDS mechanisms3, and the effects of drugs4, toxins5 and radiation6. These cells have also been used in the Nobel-winning experiments that led to the discovery of telomeric activity7,8.
Furthermore, to verify the gene-editing effects involved in specific cellular processes, gene expression profiles and proteome analyses have been applied to HeLa cells9,10,11. The widespread use of this cell line is mainly due to the ease with which they can be handled and manipulated in different conditions.
Starting from the 1950 s, approximately 80,000 studies have reported results obtained using the HeLa cell line as a physiological model. In the majority of these studies, the source of the HeLa cell line is not specified, implying a lab-to-lab origin (Supplementary Table S1).
Several published studies reported that HeLa cells are characterized by extensive chromosome instability (CIN)12,13,14,15,16,17,18,19 (Supplementary Table S1), although one study concluded that they are stable and able to maintain a constant number of chromosomes mitosis after mitosis20.
A few years after the establishment of the line, one of the first studies on the metaphase spreads of clonal cell strains of HeLa was published in 195821, showed a relatively narrow chromosome number distribution between 75 and 82. The large number of duplications and deletions compared with the normal human genome has already been extensively described19 (Supplementary Table S1), and as chromosomes provide genome identity, it might even be argued that the HeLa genome is no longer a human one22. In 1991, it was even proposed that the HeLa cell could be considered a new species (Helacyton gartleri)23.
To our knowledge, all of the reports regarding HeLa CIN have been conducted on a single cell clone or on sub-clones20,24. Recently, the HeLa genome and transcriptome was exhaustively characterized by deep RNA and DNA sequencing performed on a single clone from CLS Cell Lines Service GmbH (the HeLa Kyoto cell line)25. To investigate the reported instability of the HeLa cell lines and to verify the possibility that erroneous conclusions could be generated by the use of HeLa cells obtained from different laboratories, we compared different “batches” obtained from four Italian laboratories (HeLaSR, HeLaV, HeLaP, and HeLaH). HeLaP and HeLaH were derived from the same batch but had been cultured in two different laboratories for approximately 8 years; similarly, HeLaV was derived from the same batch as HeLaSR but had been cultured in two different laboratories for approximately 12 years.
Results
HeLa karyotyping and a-CGH show genomic instability
We evaluated the genomic stability of these “batches” by karyotyping, array-comparative genomic hybridization (a-CGH) and fluorescent in situ hybridization (FISH described in the next section).
Cytogenetic analysis was performed on 20 metaphase spreads stained using the quinacrine-based Q-banding technique (QFQ). The HeLa cells were near-triploid, containing a range of 60–66 chromosomes in HeLaSR, 56–79 in HeLaV, 71–79 in HeLaP, and 63–79 in HeLaH. In addition, approximately 10% of the metaphase spreads of clone HeLaSR exhibited chromosome numbers ranging from 101 to 148, whereas HeLaV (derived from the HeLaSR batch) exhibited a range of 124–198 chromosomes in approximately 30% of its metaphase spreads. We excluded the coexistence of two different sub-clones by monoclonal sub-culturing.
Ten HeLaV clones, derived from a monoclonal sub-culture, showed the presence of both metaphase populations (with a range of 49–81 and a range of 92–240 chromosomes), suggesting that a single cell could carry the inherent defect, possibly generated by new mutation events. For our FISH and transcriptomic analysis, we used the batch HeLaSR because it showed fewer mitoses with double the modal chromosome number compared to HeLaV.
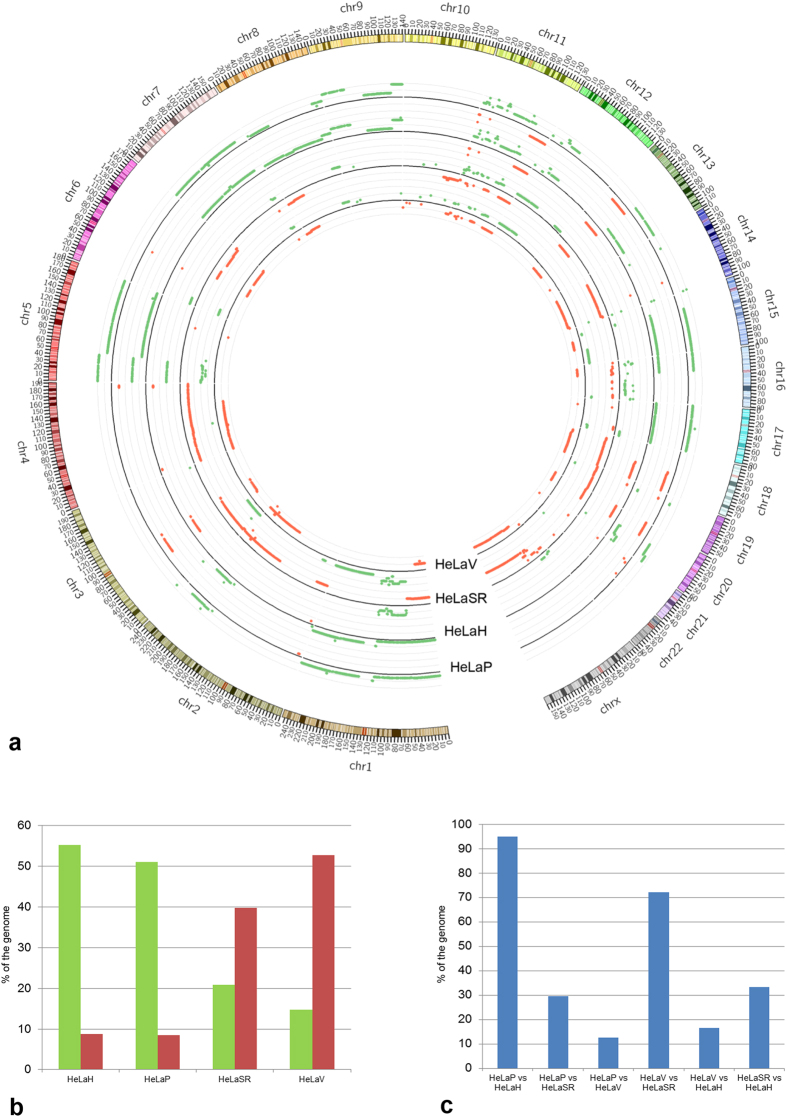
To characterize all of the genomic imbalances of the four “batches”, we performed a-CGH. As showed in the Circos plot26 (Fig. 1A), a comparison between the HeLa DNA content and the diploid human genome evinced a substantial difference among the four lines analysed. The comparison of the 4 HeLa clones underscored that although the amount of gain and loss of genomic material compared to the human diploid genome is highly variant among the 4 clones, there is similarity within the pairs with a common origin, i.e., HeLaH/HeLaP vs HeLaSR/HeLaV (Fig. 1B).
Figure 1. The genomic landscape of the 4 HeLa cell lines.
(a) Circos plot of the four HeLa genomes compared with the diploid human genome, with tracks representing the gain (green) and loss (red) of genomic material. (b) The histogram summarizes the percentage of the gain (green) and loss (red) of genomic material in the 4 cell lines compared to the diploid human genome, highlighting the separate evolution of the two pairs of HeLa clones (HeLaH and HeLP vs HeLaSR and HeLaV). (c) The histogram shows the percentage of similarity in genomic content in each HeLa clone compared with the other lines and emphasizes that the two more similar lines, based on their common origin (HeLaH and HeLaP; HeLaSR and HeLaV), bear differences in their genomic content.
To express the similarities among the HeLa clones, we calculated the percentage of the genomes with the same annotation (diploid, deleted or amplified). The results, depicted in Fig. 1C, show high similarity within the two pairs HeLaSR/HeLaV and HeLaH/HeLaP. We emphasize that even recently split-out batches (HeLaSR vs HeLaV and HeLaH vs HeLaP) present specific gains or losses and that these new events are acquired and stably maintained. A possible hypothesis to explain the different chromosome imbalances between the two pairs could be ancestral mutational events that conferred an evolutionary advantage in DNA losses or gains.
FISH analysis highlights new specific markers
To identify chromosomal rearrangements and to verify the presence of HeLa-specific markers observed by karyotyping17,20, we performed a FISH analysis to paint chromosomes 1, 3, 5, 9, 13 and 19 in HeLaSR and HeLaP because they appeared to be the two most different clones by a-CGH. As shown in Table 1, HeLaP displays almost all of the HeLa-specific markers previously identified12,20,25, whereas HeLaSR has lost most of them. In addition, we found in each clone the presence of new specific markers (Supplementary Fig. S1), confirming an independent evolution of each batches.
Table 1. HeLa cell line’s specific markers of HeLaSR and HeLaP lines.
| Chromosome markers previously reported17,18,19,20 | HeLaSR | HeLaP |
|---|---|---|
| M1 der(1)t(1;3)(q11;q11) | 10/10 | 9/10 |
| M2 der(1;9)(p11;q11) | 0/10 | 4/10 |
| M4 der(3;5)(p11;q11) | 10/10 | 0/10 |
| M5 der(3;20)(q10;q?10) | 8/10 | 8/10 |
| M7 i(5)(p10) | 10/10 | 10/10 |
| M8 der(7)t(7;19)(q35;?) | 0/10 | 9/10 |
| M10 der(9)t(3;9)(p21;p11) | 0/10 | 8/10 |
| M11 der(11)t(9;11;9)(?;p14?q33?;?dup(11)(p?)dup(11)(q?) | 0/10 | 10/10 |
| M14 der(19)t(13;19)(q21;p13) | 10/10 | 10/10 |
| M22 der (5;9)(p10;p10) | 0/10 | 10/10 |
| Loss or acquired chromosomes | ||
| +chr.1 | 9/10 | 5/10 |
| − chr. 3 | 10/10 | 10/10 |
| − 2 chrs. 5 | 0/10 | 10/10 |
| +chr.9 | 2/10 | 1/10 |
| − chr. 9 | 0/10 | 3/10 |
| − chr. 13 | 10/10 | 1/10 |
| − chr. 19 | 2/10 | 10/10 |
| Specific new markers | ||
| i(1p) | 1/10 | 0/10 |
| i(5q) | 0/10 | 10/10 |
| t(3p;13q) | 10/10 | 1/10 |
| t(6;19) | 10/10 | 1/10 |
Marker identification by FISH using WCP (Whole Chromosome Painting) for chr.1 vs chr. 3; chr. 5 vs chr. 9; chr.13 vs chr.19 (see Supplementary Fig. S1). Ten metaphases for each HeLa clone were analysed.
Transcriptomic analysis reveals that different batches exposed to hypoxic stimulus display differences in gene expression
Our genomic analysis uncovered deep variability between thebatches , and because genomic gains or losses or whole-chromosome aneuploidy can have drastic consequences on gene expression27, we performed a whole-transcriptome analysis to verify the transcriptional effects of this differential chromosome instability. The microarray expression analysis was performed on HeLaSR and HeLaP, the batches most different in terms of genomic content. These cell lines were exposed to hypoxic conditions, chosen because the induced hypoxic pathway is well characterized28,29.
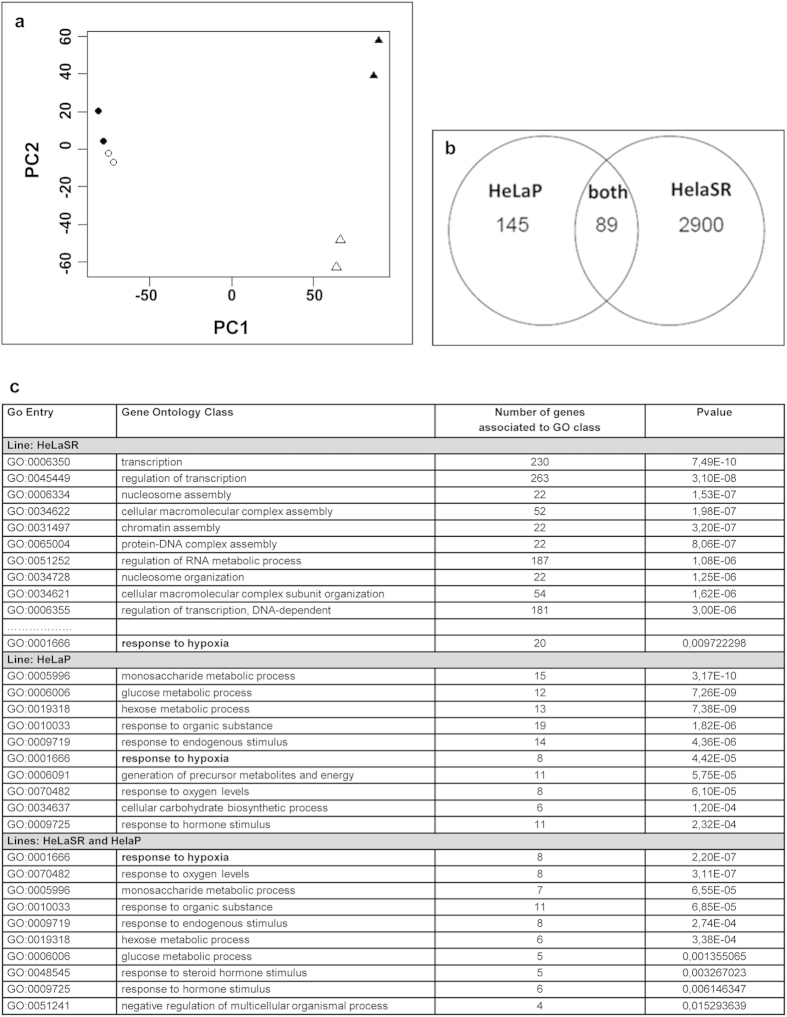
A principal-component analysis (PCA) was performed to determine the expression trends within the dataset. PCA is a useful technique to reduce the dimensionality of large data sets and to visually assess similarities and differences between samples30. PCA was used to identify trends in the regulation of genes induced by hypoxic exposure and to map the entire dataset on a two-axis graph (principal components, PC1 and PC2) where the distances account for similarity (Fig. 2A); the closer the distance between samples, the more they are similar. The main divergence was due to two different clones, HeLaSR vs HeLaP (triangles and circles, respectively), as demonstrated by the first principal component (PC1). The second component (PC2), summarizing the effects of hypoxia, highlighted strong changes in HeLaSR and mild changes in HeLaP. Indeed, untreated HeLaP and HeLaP after hypoxia are closer to each other than untreated HeLaSR is to HeLaSR after hypoxia, meaning that hypoxia has a stronger effect in HeLaSR than in HeLaP (Fig. 2A). Furthermore, we identified the genes regulated by hypoxic treatment in these two clones: 2,900 (1,666 genes upregulated and 1,234 downregulated) genes were mis-regulated only in HeLaSR, whereas 145 genes were mis-regulated only in HeLaP (104 up and 41 down). A total of 89 genes (88 up and 1 down) were mis-regulated in both cell lines (Fig. 2B).
Figure 2. Transcriptional landscape.
(a) Principal component analysis (PCA) of the transcriptome. The PCA maps the entire dataset on a two-axis graph (principal components, PC1 and PC2), where the distances account for similarity. Untreated HeLaP cells (white circles) and HeLaP cells after hypoxia (black circles) are closer to each other than untreated HeLaSR cells (white triangles) are to hypoxic HeLaSR cells (black triangles), suggesting that hypoxia exerts a stronger effect on HeLaSR cells than on HeLaP cells. (b) Venn diagram showing the number of genes either upregulated or downregulated due to hypoxia in the HeLaP and HeLaSR clones compared to their respective controls. (c) Gene ontology of genes either upregulated or downregulated by hypoxia in HeLaSR and HeLaP cells.
These three gene lists were analysed for gene ontology (GO) enrichment (Fig. 2C). The more significant GO classes those that are differentially expressed between HeLaSR and HeLaP, demonstrating the large differences in the expression profiles of the two cell lines in response to hypoxia.
The classes related to hypoxia were present in both, but they were not the most significant (Fig. 2C). On the contrary, when we analysed the 89 mis-regulated genes common to both cell lines, the most enriched classes are related to hypoxia (Fig. 2B,C). These results suggest that the different genomic contents of the two clones have a remarkable influence on basal gene expression and, consequently, on the response to a hypoxic stimulus. The cells activate different pathways, but interestingly, the regulation of hypoxia-related genes is preserved.
Real-time RT-PCR confirmed the microarray data
A subset of regulated genes identified for their relevance in the hypoxic response were validated by real-time RT-PCR (qPCR) (Supplementary Table S2). The trend of the array data was confirmed by qRT-PCR (Supplementary Table S3).
Copy number variation influences gene expression
Because significant chromosome copy number alterations are frequently associated with gene expression changes in the affected regions31, we evaluated the possible effects of copy number variation on gene expression32 in our samples. Based on the data obtained with a-CGH and the expression array performed on HeLaP and HeLaSR samples as previously described, a general lack of dosage compensation was observed in HeLaP (Fig. 3A) and in HeLaSR (Fig. 3B), meaning that gene dosage is correlated to gene expression.
Figure 3. Gene expression by copy number in HeLaP and HeLaSR cells.
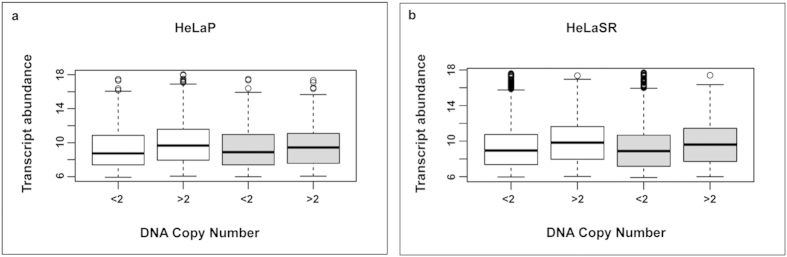
Transcript abundance (arbitrary unit) is positively correlated with gene copy number. (a) HeLaP gene expression in untreated (white) and hypoxic (grey) cells. (b) HeLaSR gene expression in untreated (white) and hypoxic (grey) cells. A general lack of dosage compensation was observed in both cell lines.
Discussion
Genetic aberrations, such as gene amplification, deletions, and loss of heterozygosity, are hallmarks of cancer and are major contributors to the neoplastic process through the accumulation of mutations in specific genes. The mechanisms through which these mutations are generated are the subject of continued debate33,34,35,36,37, and several cancer-predisposing mutations affect genes that are responsible for maintaining the integrity and number of chromosomes during cell division.
The main factors that control chromosome stability are telomere maintenance, cell division mechanisms, and the mitotic checkpoint that ensures correct chromosome segregation38,39,40.
Consequently, the archetypical transformation of tumour cells results in CIN41,42. Established cell lines have been traditionally used to characterize the biological significance of specific genomic aberrations identified in primary tumours43 and to test the therapeutic efficacy of anticancer agents44.
HeLa was the first cultured cancer line45. Although its CIN has already been extensively investigated19 (see Supplementary Table S1), demonstrating the low degree of similarity with the initial tumour and with the human diploid genome, this cell line is largely used in biomedical research. The HeLa cell line is among the most frequently utilized models in research to validate drugs for cancer therapy46 and for genetic/epigenetic manipulation using demethylation agents47, siRNA and expression vectors to study gene function48,49,50.
Here, we report the high level of CIN in HeLa clones obtained from different laboratories. We demonstrated that each clone accumulates genomic variability in a time-dependent manner. We speculate that some chromosomal rearrangements or point mutations that randomly arose in each clone increased survival, leading to a kind of “genetic drift” in clonal variants.
This conclusion is evident in the differing DNA content of the clones HeLaSR and HeLaV, which showed a loss of genomic DNA, whereas HeLaH and HeLaP exhibited a gain of genomic DNA in comparison with the diploid human genome. We speculate that even slightly different culture conditions may produce this divergence. The number of passages that a cell line undergoes can certainly lead to extensive modifications in growth rate, morphology, aneuploidy, chromosome alterations, gene expression and drug sensitivity, depending on the culture environment51,52,53,54,55. In addition, we demonstrated that the genomic gains or losses or whole-chromosome aneuploidy is specific to each analysed clone (Fig. 1A) and that these differences in genomic content have a remarkable influence on basal gene expression (Fig. 2A). We showed that in response to a hypoxic stimulus, each cell line activates different pathways, although the regulation of genes related to hypoxia is conserved (Fig. 2A,C).
These results suggest that different HeLa batches used for the same experiment may yield different results. The large differences in the basal gene expression profiles suggest that the use of uncharacterized clones may lead to faulty conclusions and to irreproducible results in studies of gene function and pathway analysis. With respect to genomic content, the use of uncharacterized batches of HeLa cells56 may result in different behaviours between clones in response to specific stimuli, such as cancer drugs, used in biomedical research.
Our results suggest that not a single HeLa cell line, or even a set of similar HeLa cell lines, exists. Rather, an indeterminate number of clones exist, each carrying large genomic differences that lead to different expression profiles. The HeLa cell recalls the main character of the novel by Pirandello (the Italian poet awarded the 1934 Nobel Prize in Literature): “One, no one, one hundred thousand”57: the HeLa cell line is thought to be a unique cell line (“one”), but it is clear that large differences exist between the tumour from which it was derived and the human genome (“no one”) and that an indeterminate number of HeLa cell lines are scattered in laboratories worldwide (“one hundred thousand” or more), each with a unique genomic profile.
Methods
Cell lines
The cell lines HeLaP, HeLaH, HeLaSR and HeLaV were obtained from four Italian laboratories. HeLaP and HeLaH were derived from the same cell-line batch but were cultured in two different laboratories for approximately 8 years. Similarly, HeLaSR and HeLaV were derived from the same cell-line batch but were cultured in two different laboratories for approximately 12 years. In our lab, the mycoplasma-free cell lines were cultured in DMEM-F12 (Euroclone SpA, Milan, Italy) with 10% heat-inactivated foetal bovine serum (Euroclone), 1 mM glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin and were incubated at 37 °C and 5% CO2. The cells were cultured to reach a confluence of 80–90%.
Cytogenetic analysis
Metaphase spreads were obtained from the four cell lines. After culturing, the cells were treated with 0.04 μg/ml colcemid for 2 hours. The cells were harvested by treatment with a 1× trypsin/EDTA solution for approximately 5 min. A hypotonic treatment was performed with 0.96% Na citrate for 15 min at 37 °C, and the cells were subsequently fixed in a fresh methanol/acetic acid (3/1, v/v) solution. Metaphase spreads were stained overnight with 0.005% quinacrine mustard solution in McIlvaine buffer (QFQ-banding), and standard cytogenetic analyses were performed with a Leica D5000B fluorescent microscope.
Monoclonal subculturing by limiting dilution
To exclude the possibility that the HeLaV and HeLaSR cell lines arose from 2 different HeLa subclones, we obtained a cell culture derived from a single cell by limiting-dilution culture. We prepared serial dilutions to obtain a suspension containing 1 cell/μl. A single μl of the final cell suspension was seeded into each well of a 96-well plate containing growth medium. The presence of a single cell was evaluated by microscopy, and the cells were incubated at 37 °C and 5% CO2 overnight. After one day, we used microscopy to confirm the presence of a single cluster of cells (2 or more) per well, implying a single-cell origin, and we excluded any wells with double or multiple clusters of cells. The monoclonal cell lines were trypsinised and seeded in T25 flasks to obtain sufficient cells for the metaphase spread analysis.
Fluorescence in situ hybridization experiments
Metaphase spreads were hybridized with Cytocell Aquarius Whole Chromosome Painting probes (WCP Probes) using the codenaturation protocol. Briefly, 20 μl of ready-to-use probes were deposited on the slide and sealed with a coverslip using rubber cement. Codenaturation was performed on a HyChrome hybridization machine at 75 °C for 2 min, and hybridization was conducted overnight. The slides were then washed for 2 min in 0.4× SSC buffer at 72 °C and for 30 min in 2× SSC/0.05% Tween 20 at room temperature. Metaphases spreads were then counterstained in 2× SSC buffer with 200 ng/ml of 4,6-diamidino-2-phenylindole (DAPI) and then mounted with an antifade solution (Vector Laboratories INC., Burlingame, CA, USA). The samples were analysed with a Leica DM5000B fluorescence microscope, and images were captured with the Leica QFISH software system.
DNA extraction
DNA was extracted from approximately 10 × 107 cells using the DNeasy Blood and Tissue kit (Qiagen S.r.l. Milan, Italy) according to the manufacturer’s protocol. Genomic DNA was quantified using an ND-1000 UV-Vis spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
a-CGH analysis
a-CGH was performed on an Agilent microarray platform (Agilent Technologies Inc., Santa Clara, CA USA) with an Agilent’s human 4 × 180 K CGH slide. Sample preparation, labelling, and microarray hybridization were performed according to the Agilent CGH Enzymatic Protocol version 7.3. Slides were scanned using the Agilent G2565CA scanner and analysed using the Agilent Feature Extraction 10.7.3.1 software. The a-CGH profile was extrapolated using the Agilent Genomic Workbench 6.5.0.18 software with the following parameters: ADM-2 threshold 6, Fuzzy Zero ON, and Centralization OFF. Coverage plots were generated using the Circos visualization tool compared with the 2.2 × 109 genome covered by the Agilent probes.
Hypoxic treatment
Before administering the hypoxia treatment, the HeLaP and HeLaSR cells were maintained at 70–80% confluence. The medium was refreshed before the hypoxia treatment. Cells (two flasks per cell line) were maintained under hypoxic (1% O2, 5% CO2, 94% N2) conditions or normoxic (95% air and 5% CO2) conditions for 24 h.
RNA extraction
Total RNA was purified from the HeLa cells using the RNeasy Plus kit (Qiagen). RNA was quantified using an ND-1000 UV-Vis spectrophotometer (Thermo Scientific, Wilmington, DE, USA), and the integrity of the RNA was assessed with the Agilent 2100 Bioanalyzer (Agilent Technologies Inc.) according to the manufacturer’s instructions. All of the RNA samples used in this study exhibited a 260/280 ratio above 1.9 and an RNA Integrity Number (RIN) above 9.0.
Microarray expression profiling
The microarray experiment included two biological replicates per treatment. All sample-labelling, hybridization, washing, and scanning steps were conducted according to the manufacturer’s specifications. Briefly, Cy3-labelled cRNA was generated from 50 ng input total RNA using the One Color Quick Amp Labelling Kit (Agilent Technologies Inc.). For every sample, 600 ng cRNA from each labelling reaction (with a specific activity above 9.0) was hybridized using the Gene Expression Hybridization Kit (Agilent Technologies Inc.) to the Agilent Whole Human Genome Oligo Microarray (Agilent Technologies Inc.), which is in a 4 × 44k 60-mer slide format, where each of the 4 arrays represents approximately 41,000 unique genes and transcripts. After hybridization, the slides were washed and then scanned with the Agilent G2565BA Microarray Scanner (Agilent Technologies Inc.). The fluorescence intensities of the scanned images were extracted and pre-processed using the Agilent Feature Extraction Software (10.7.3.1).
Gene expression data analysis
Quality control and array normalization were performed in the R statistical environment using the Agi4 × 44PreProcess (v 1.18.0) package, which was downloaded from the Bioconductor web site. The normalization and filtering steps were based on those described in the Agi4 × 44PreProcess reference manual. Briefly, the Agi4 × 44PreProcess options were set to use the Mean Signal and the BG Median Signal as foreground and background signals, respectively. The data were normalized between arrays using the quantile method. Genes with a fold change greater than 1 log2 were designated as modulated. All of the above computations were conducted using the R statistical programming environment. Principal-component analysis (PCA) was performed on all genes under investigation to determine their expression trends within the dataset. PCA is a useful technique to reduce the dimensionality of large data sets. The expression analysis systematic explorer (EASE) biological theme analysis of the regulated genes was conducted online using DAVID.
Quantitative real-time PCR validation of microarray data
A real-time quantitative PCR (qRT-PCR) analysis was performed, in triplicate, on the same RNA samples that were used for the microarray hybridization to validate the microarray results. One μg of total RNA was retro-transcribed using SuperScript II (Life Technologies, Carlsbad, CA, USA). qRT-PCR was performed using the iQ SYBR® Green supermix (Bio-Rad, Hercules, CA, USA,) on the ABI Prism 7000 platform (Applied Biosystems) with the following thermal cycling protocol: denaturation for 1 min at 95 °C followed by 40 cycles of 15 sec at 95 °C and 1 min at 60 °C. The primers are listed in Supplementary Table S3. A relative quantitative analysis was performed using the 2−ΔΔCt method.
Additional Information
How to cite this article: Frattini, A. et al. High variability of genomic instability and gene expression profiling in different HeLa clones. Sci. Rep. 5, 15377; doi: 10.1038/srep15377 (2015).
Supplementary Material
Acknowledgments
We thank our colleagues at IRGB-CNR Paolo Vezzoni and Antonio Musio, Francesco Acquati at University of Insubria for providing HeLa cell lines and Elena Monti of University of Insubria for the hypoxia test. We thank Barbara Pressato and Martina Giaretta for the technical support in metaphases analysis. Marco Fabbri is a student of the PhD program in Biotechnology, School of Biological and Medical Sciences, University of Insubria (Italy). Elena De Paoli is a student of the PhD program in Biosciences, Biotechnologies and Surgical Technologies at the University of Insubria. We are grateful to Henrietta Lacks, now deceased, and to her surviving family members for their contributions to biomedical research. This work was supported by VNR-Regione Lombardia RSPPTECH 2013-2015 to A.F., Fondazione Monte di Lombardia and Fondazione UBI Varese to E.M.
Footnotes
Author Contributions A.F. conceived the project, designed the experiments, performed cell culture, hypoxia test and the microarray and qRT-PCR experiments; M.F. analysed the microarray and qRT-PCR data; R.V. performed the FISH and a-CGC experiment and analysed the data; G.M. and E.D.P. performed cell culture and metaphases preparation; E.M. analysed karyotypes and FISH and contributed to experimental design; L.G. contributed to study design; F.P. supervised the experimental design. M.F. and A.F. wrote the manuscript with input from all the authors. All the authors have read and approved the final manuscript.
References
- Gey G. O., Coffman W. D. & Kubicek M. T. Tissue culture studies of the proliferative capacity of cervical carcinoma and normal epithelium. Scientific Proceedings American Association for Cancer Research. Cancer Res. 12, 264–265 (1952). [Google Scholar]
- Salk J. E. Considerations in the preparation and use of poliomyelitis virus vaccine. J Am Med Assoc. 158, 1239–1248 (1955). [DOI] [PubMed] [Google Scholar]
- Berg J., Doe B., Steimer K. S. & Wabl M. HeLa-LAV, an epithelial cell line stably infected with HIV-1. J Virol Methods. 34, 173–180 (1991). [DOI] [PubMed] [Google Scholar]
- al-Allaf T. A. & Rashan L. J. Cis- and trans-platinum and palladium complexes: a comparative study review as antitumour agents. Boll Chim Farm. 140, 205–210 (2001). [PubMed] [Google Scholar]
- Khan A. A. et al. Biophysical interactions of novel oleic acid conjugate and its anticamcer potential in HeLa cells. J Fluoresc. 25, 519–525 (2015). [DOI] [PubMed] [Google Scholar]
- Murray J. I. et al. Diverse and specific gene expression responses to stresses in cultured human cells. Mol. Biol. Cell 15, 2361–2374 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider C. W. & Blackburn E. H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 43, 405–413 (1985). [DOI] [PubMed] [Google Scholar]
- Morin G. B. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 59, 521–529 (1989). [DOI] [PubMed] [Google Scholar]
- Chaudhry M. A., Chodosh L. A., McKenna W. G. & Muschel R. J. Gene expression profiling of HeLa cells in G1 or G2 phases. Oncogene. 21, 1934–1942 (2002). [DOI] [PubMed] [Google Scholar]
- Whitfield M. et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 13, 1977–2000 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnilicová J. et al. Histone deacetylase activity modulates alternative splicing. PLoS ONE. 6, e16727 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T. R., Re-evaluation of HeLa, HeLa S3, and HEp-2 karyotypes Cytogenet. Cell Genet. 48, 19–24 (1988). [DOI] [PubMed] [Google Scholar]
- Francke U., Hammond D. S. &. Schneider J. A. The band patterns of twelve D 98-AH-2 marker chromosomes and their use for identification of intraspecific cell hybrids. Chromosoma 41, 111–121 (1973). [DOI] [PubMed] [Google Scholar]
- Kraemer P. M., Deaven L. L., Crissman H. A., Steinkamp J. A. & Petersen D. F. On the nature of heteroploidy. Cold Spring Harb. Symp. Quant. Biol. 38, 133–144 (1974). [DOI] [PubMed] [Google Scholar]
- Heneen W. K. HeLa cells and their possible contamination of other cell lines: karyotype studies. Hereditas 82, 217–248 (1976). [DOI] [PubMed] [Google Scholar]
- Nelson-Rees W. A., Hunter L., Darlington G. J. & O’Brien S. J. Characteristics of HeLa strains: permanent vs. variable features. Cytogenet. Cell Genet. 27, 216–231 (1980). [DOI] [PubMed] [Google Scholar]
- Stanbridge E. J., Flandermeyer R. R., Daniels D. W. & Nelson- Rees W. A. Specific chromosome loss associated with the expression of tumorigenicity in human cell hybrids. Somatic Cell Genet. 7, 699–712 (1981). [DOI] [PubMed] [Google Scholar]
- Ruess D., Ye L. Z. & Grond-Ginsbach C. HeLa D98/aH-2 studied by chromosome painting and conventional cytogenetical techniques. Chromosoma. 102, 473–477 (1993). [DOI] [PubMed] [Google Scholar]
- Rutledge S. What HeLa Cells Are You Using? The Winnower. (2014). Website Available at: https://thewinnower.com/papers/what-hela-cells-are-you-using (Accessed: 19th May 2015).
- Macville M. et al. Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res. 59, 141–150 (1999). [PubMed] [Google Scholar]
- Tjio J. H. & Puck T. T. Genetics of somatic mammalian cells. II. Chromosomal constitution of cells in tissue culture. J Exp Med. 108, 259–268 (1958). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. Most popular human cell in science gets sequenced. Nature News Mar. 15, 10.1038/nature.2013.12609 (2013). [DOI] [Google Scholar]
- Val Valen L. M. & Maiorana V. C. HeLa, a new microbial species. Evolutionary Theory. 10, 71–74 (1991). [Google Scholar]
- Adey A. et al. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature. 500, 207–211 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J. J. et al. The genomic and transcriptomic landscape of a HeLa cell line. G3 (Bethesda). 3, 1213–1224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M. et al. Circos: an Information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N., Rancati G. & Li R. Dr Jekyll and Mr Hyde: role of aneuploidy in cellular adaptation and cancer. Curr Opin Cell Biol. 22, 809–815 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson N. & Ratcliffe P. J. Hypoxia signalling pathways in cancer metabolism: the importance of co-selecting interconnected physiological pathways Cancer Metab. 2, 1–17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X. et al. Hypoxia-inducible factor-1α enhances the malignant phenotype of multicellular spheroid HeLa cells in vitro. Oncol Lett. 1, 893–897 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson K. On lines and planes of closest fit to systems of points in space (PDF). Philosophical Magazine. 2, 559–572 (1991). [Google Scholar]
- Lynch M. Gene duplication and evolution. Science. 297, 945–947 (2002). [DOI] [PubMed] [Google Scholar]
- Henrichsen C. N., Chaignat E. & Reymond A. Copy number variants, diseases and gene expression. Hum Mol Genet. 18, R1–R8 (2009). [DOI] [PubMed] [Google Scholar]
- Lengauer C. Aneuploidy and genetic instability in cancer. Semin Cancer Biol. 15, 1 (2005). [DOI] [PubMed] [Google Scholar]
- Loeb L. A. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 51, 3075–3079 (1991). [PubMed] [Google Scholar]
- Hartwell L. Defects in a cell cycle checkpoint may be responsible for the genomic instability of cancer cells. Cell 71, 543–546 (1992). [DOI] [PubMed] [Google Scholar]
- Mitelman F. Recurrent chromosome aberrations in cancer. Mutat Res. 462, 247–253 (2000). [DOI] [PubMed] [Google Scholar]
- Sandberg A. A. & Meloni-Ehrig A. M. Cytogenetics and genetics of human cancer: methods and accomplishments Cancer Genetics and Cytogenetics. 203, 102–126 (2010). [DOI] [PubMed] [Google Scholar]
- Jefford C. E. & Irminger-Finger I. Mechanisms of chromosome instability in cancers. Crit Rev Oncol Hematol. 59, 1–14 (2006). [DOI] [PubMed] [Google Scholar]
- Bakhoum S. F. & Swanton C. Chromosomal instability, aneuploidy, and cancer. Front Oncol. 4, 161 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman F., Johansson B. & Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 7, 233–45 (2007). [DOI] [PubMed] [Google Scholar]
- Bakhoum S. F. & Compton D. A. Chromosomal instability and cancer: a complex relationship with therapeutic potential J Clin Invest. 122, 1138–1143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C., Kinzler K. W. & Vogelstein B. Genetic instabilities in human cancers. Nature. 396, 643–649 (1998). [DOI] [PubMed] [Google Scholar]
- Ferreira D., Adega F. & Chaves R. Novel Approaches in Biomarkers Discovery and Therapeutic Targets in Cancer (ed. López-Camarillo C. & Aréchaga-Ocampo E.) 139–166 (InTech, 2013). [Google Scholar]
- Sharma S. V., Haber D. A. & Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 10, 241–253 (2010). [DOI] [PubMed] [Google Scholar]
- Scherer W. F., Syverton J. T. & Gey G. O. Studies on the propagation in vitro of poliomyelitis viruses. IV. Viral multiplication in a stable strain of human malignant epithelial cells (strain HeLa) derived from an epidermoid carcinoma of the cervix. J Exp Med. 97, 695–710 (1953). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y. Q. et al. Pathway of programmed cell death in HeLa cells induced by polymeric anti-cancer drugs. Biomaterials. 32, 3637–3646 (2011). [DOI] [PubMed] [Google Scholar]
- Sood S. & Srinivasan R. Alterations in gene promoter methylation and transcript expression induced by cisplatin in comparison to 5-Azacytidine in HeLa and SiHa cervical cancer cell lines. Mol Cell Biochem. 404, 181–191 (2015). [DOI] [PubMed] [Google Scholar]
- Tsai K. W., Kao H. W., Chen H. C., Chen S. J. & Lin W. C. Epigenetic control of the expression of a primate-specific microRNA cluster in human cancer cells. Epigenetics. 4, 587–592 (2009). [DOI] [PubMed] [Google Scholar]
- Rassmann A. et al. Identification of gene expression profiles in HeLa cells and HepG2 cells infected with Coxsackievirus B3. Journal of Virological Methods. 187, 190–194 (2013). [DOI] [PubMed] [Google Scholar]
- Chaudhry M. A., Chodosh L. A., McKenna W. G. & Muschel R. J. Gene expression profiling of HeLa cells in G1 or G2 phases. Oncogene 21, 1934–1942 (2002). [DOI] [PubMed] [Google Scholar]
- Nelson-Rees W. A., Owens R. B., Arnstein P. & Kniazeff A. J. Source, alterations, characteristics and use of a new dog cell line (Cf2Th). In Vitro. 12, 665–669 (1976). [DOI] [PubMed] [Google Scholar]
- Sakamoto J. et al. Alteration of phenotype, morphology and drug sensitivity in colon cancer cell lines under various culture conditions. Gan To Kagaku Ryoho. 4, 1864–1873 (1989). [PubMed] [Google Scholar]
- Burdall S. E., Hanby A. M., Lansdown M. R. J. & Speirs V. Breast cancer cell lines: friend or foe? Breast Cancer Res. 5, 89–95 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Staveren W. C. et al. Human cancer cell lines: experimental models for cancer cells in situ?For cancer stem cells? Biochim Biophys Acta. 1795, 92–103 (2009). [DOI] [PubMed] [Google Scholar]
- Borrell B. How accurate are cancer cell lines? Nature. 463, 858 (2010). [DOI] [PubMed] [Google Scholar]
- Domcke S., Sinha R., Levine D. A., Sander C. & Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 4, 2126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirandello L. One, No One, and One Hundred Thousand. Vol. 18 (ed. Marsilio Pub) (1990). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.