Abstract
Hoxa5 is preferentially expressed in haematopoietic stem cells (HSCs) and multipotent progenitor cells (MPPs), and is more highly expressed in expanding HSCs. To date, little is known regarding the role of Hoxa5 in HSCs and downstream progenitor cells in vivo. In this study, we show that increased expression of Hoxa5 in haematopoietic stem cells leads to aberrant erythropoiesis in vivo. Hoxa5 differentially modifies the cell cycle of HSCs and lineage committed progenitor cells, depending on the cellular context. Hoxa5 drives HSCs, but not MPPs, through the cell cycle and arrests erythroid progenitor cells in G0 phase. Although the HSC pool shrinks after overexpression of Hoxa5, HSCs sustain the abilities of self-renewal and multipotency. In vivo, Hoxa5 has two effects on erythropoiesis: it causes a predominance of mature erythroid lineage cells and the partial apoptosis of erythroid progenitors. RNA-seq indicates that multiple biological processes, including erythrocyte homeostasis, cell metabolism, and apoptosis, are modified by Hoxa5. The results of this study indicate that Hoxa5 is a key regulator of the HSC cell cycle, and the inappropriate expression of Hoxa5 in lineage-committed progenitor cells leads to aberrant erythropoiesis.
Keywords: apoptosis; cell cycle; erythropoiesis; haematopoietic stem cells, Hoxa5
Abbreviations
- BFU-E
burst-forming unit-erythroid
- CFU-G
colony forming unit-granulocyte
- CFU-GM
colony forming unit-granulocyte macrophage
- CMP
common myeloid progenitor
- GMP
granulocyte monocyte progenitor
- HSC
haematopoietic stem cell
- LSK
lineage negative, Sca1 positive, cKit positive
- MEP
megakaryocyte-erythroid progenitor
- MP
myeloid progenitor
- MPP
multipotent progenitor
Introduction
Among the haematopoietic hierarchy, only HSCs possess both long-term self-renewal and haematopoietic multipotency. Upon asymmetric division, newborn MPPs lose this self-renewal capacity, but they sustain their haematopoietic multipotency. The intrinsic regulatory mechanism for haematopoietic stemness remains elusive. Several Hox genes, including Hoxa10, Hoxa9, Hoxb4, and Hoxb3, have been shown to be involved in hematopoiesis.1,2 Hox genes are thought to be involved in hematopoiesis regulation mainly through their differential expression in HSCs and haematopoietic progenitors.1 Of note, Hoxb4 is predominantly expressed in HSCs rather than MPPs, and it is essential to HSC expansion.3 A newly engineered fusion gene, Nup98-Hoxa10, causes the expansion of HSCs over a thousand fold, and this expansion is associated with the upregulation of Hoxa5 expression.4 Thus, evaluating the role of Hoxb5 in HSCs and MPPs is necessary.
In Trp53 null mice, the loss of Hoxa5 increases the incidence of mammary tumors.5 In vitro, the activation of HOXA5 leads to the apoptosis of breast cancer cells through the activation of the Trp53 apoptosis pathway,6 which indicates a tumor suppressor role of Hoxa5 in solid tissues. However, there is no evidence that Hoxa5 has a tumor suppressor role in the haematopoietic system.7 However, the activation of Hoxa5 signaling has been reported during leukemogenesis. For example, the upregulation of Hoxa5 by epigenetic factor hDOT1L is associated with leukemic transformation by CALM-AF10.8 Hoxa5 is also upregulated in AML cells transformed with the NUP98-HHEX fusion gene and in bone marrow cells overexpressing Nup98-NSD1.9,10 Recently, in vitro expanded HSCs were also shown to have upregulated Hoxa5 expression.4,11 Thus, we hypothesize that the activation of Hoxa5 signaling plays an important role in hematopoiesis at the level of HSCs and progenitors.
Early in vitro studies demonstrated that Hoxa5 blocks erythropoiesis and enhances myelopoiesis,12,13 but the mechanism behind that is elusive. Here, we show in vivo data that the overexpression of Hoxa5 in HSCs leads to predominant erythropoiesis in vivo, which is contradictory to the phenotype observed by an in vitro CFU assay. Hoxa5 drives HSCs, but not MPPs, through the cell cycle. Further analysis demonstrates that Hoxa5 has two effects on erythropoiesis: it causes a predominance of mature erythroid lineage cells and the partial apoptosis of erythroid progenitors. Further analysis at the progenitor cell level indicated that Hoxa5 modifies multiple processes, including erythrocyte homeostasis, cell metabolism, apoptosis, and lineage differentiation.
Results
HSCs show higher expression of Hoxa5
To investigate the role of Hoxa5 in hematopoiesis, we measured the expression pattern of Hoxa5 in highly purified HSCs and MPPs. We first chose an RNA-seq approach to obtain the global transcriptional information of HSCs and MPPs. To separate bulk HSCs, we first enriched Sca1+ cells from 20 B6 mice at 6–8 weeks of age. With a strict gating and sorting strategy (Fig. S1), we obtained over 10,000 HSCs and 20,000 MPPs from each of the 20 mice. To guarantee consistency, 2 ng RNA from each cell sample were used for the sample preparation for RNA-seq. To confirm our RNA-seq results, we first evaluated their quality by examining the expression of genes that are well-known to be differentially expressed in mouse HSCs and MPPs.14 Consistent with previous reports, all of the chosen genes were differentially expressed in the HSCs used for our RNA-seq analysis (Supplemental Table 1). After confirming the quality of our data, we focused on analyzing Hoxa5 gene expression because recent studies have shown that Hoxa5 might play an important role in HSC function.2,4,15 As expected, Hoxa5 is highly expressed in HSCs (Fig. 1A). Then, we used real time PCR to verify the expression pattern of Hoxa5 in HSCs and MPPs. Consistent with our RNA-seq data, Hoxa5 was highly expressed in HSCs by real time PCR (Fig. 1B). Thus, we investigated the biological effect of Hoxa5 in haematopoietic stem/progenitor cells.
Table 1.
Genes dysregulated by ectopic expression of Hoxa5 in erythroid and myeloid progenitors
| Gene Symbol | Accession | log2 Fold | p-Value | Biological Process |
|---|---|---|---|---|
| Pou2af1 | NM_011136 | 4.13 | 0.040 | Regulation of transcription |
| CD19 | NM_009844 | 4.06 | 0.015 | Signal transduction |
| Pax5 | NM_008782 | 3.79 | 3E-04 | B cell differentiation/development |
| Cxcl12 | NM_001012477 | 2.04 | 0.013 | Signal transduction/development |
| Slamf1 | NM_013730 | 1.80 | 0.032 | Signal transduction |
| Tgm2 | NM_009373 | 1.49 | 0.036 | Metabolism/Signal transduction/Cell adhesion |
| Rtn4r | NM_022982 | 1.28 | 0.021 | Cell anchor |
| Dok2 | NM_010071 | 1.18 | 0.002 | Signal transduction |
| Wfs1 | NM_011716 | −2.16 | 0.028 | Metabolism/apoptosis |
| Klf4 | NM_010637 | −1.70 | 0.002 | Cell differentiation/cell growth |
| Zfp26l2 | NM_001001806 | −1.52 | 9E-13 | T Cell differentiation/cell division |
| HoxA9 | NM_001277238 | −1.49 | 8E-04 | Transcription/Development |
| Aldh6a1 | NM_134042 | −1.49 | 5E-04 | Metabolism/cell differentiation |
| IL7 | NM_008371 | −1.35 | 0.028 | T cell commitment/cell proliferation |
| Cdkn1a | NM_001111099 | −1.33 | 0.014 | Cell cycle/cell proliferation |
| Egr1 | NM_007913 | −1.29 | 0.014 | Apoptosis /T Cell differentiation |
| Irf8 | NM_008320 | −1.25 | 5E-07 | Cell differentiation/signal transduction |
| Fos | NM_010234 | −1.24 | 1E-06 | Signal transduction/development/metabolism |
| Klf2 | NM_008452 | −1.21 | 0.020 | Erythrocyte homeostasis/cell growth |
| Cdk5rap1 | NM_025876 | −1.21 | 0.005 | Cell cycle |
Figure 1.
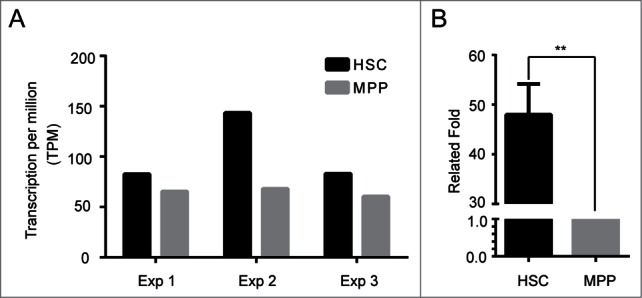
Hoxa5 is predominantly expressed in haematopoietic stem cells. (A) RNA-seq showed higher expression level of Hoxa5 in HSCs. Murine HSCs are defined as Lin (CD2, CD3, CD4, CD8, Mac1, Gr1, Ter119, B220)-, CD48-, Sca1+, cKit+, and CD150+. MPPs are defined as Lin (CD2, CD3, CD4, CD8, Mac1, Gr1, Ter119, B220)-, CD48-, Sca1+, cKit+, and CD150-. Total RNA of 10,000 sorted HSCs and MPPs was extracted using RNeasy micro kit. Two nanograms of HSC and MPP RNA were used as the RNA input for RNA-seq sample preparation. Three independent experiments are shown. (B) Real time PCR confirmed the expression pattern of Hoxa5 in HSCs and MPPs. Two nangrams of RNA from sorted HSCs and MPPs were linearly amplified. Ten nanograms of cDNA were used as the input for q-RT-PCR. The relative fold increase was calculated by a value of 2^(-ΔΔCt). The value of MPP was normalized to 1 for comparison. Error bars indicate standard error. (**) P < 0.01.
HSCs sustain self-renewal and multipotency after overexpressing Hoxa5
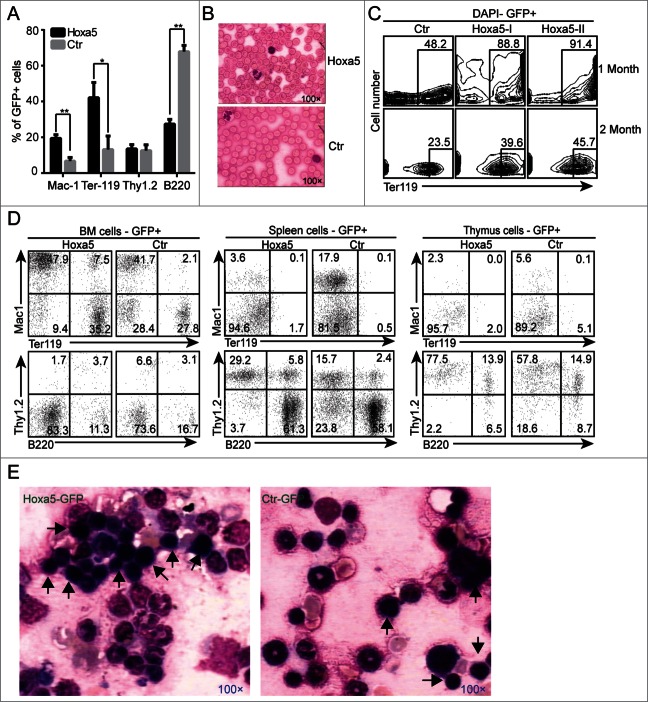
To investigate the role of Hoxa5 in hematopoiesis in vivo, we chose a classic retro-viral expression system to increase Hoxa5 expression in mouse haematopoietic stem/progenitor cells (Fig. S2A). After two rounds of viral transduction through spin infection, the transduction efficiencies of the lineage negative cells reached over 20% (Fig. S2B). Two hundred HSCs (Lin-cKit+Sca1+Mac1+GFP+) were sorted and transplanted into each sublethally irradiated recipient. Because only long-term HSCs can only contribute multiple lineages to the peripheral blood for 16–20 weeks after transplantation,16 we analyzed the engraftment of HSCs overexpressing either Hoxa5 (Hoxa5-HSCs) or GFP control (control-HSCs) for 24 weeks after transplantation. Our results showed that Hoxa5-HSCs reconstituted myeloid, erythroid and lymphoid lineages in the peripheral blood of the recipients. Compared to the control HSCs, Hoxa5-HSCs preferentially differentiated to erythroid (Ter119+) and myeloid (Mac1+) fates (Fig. 2A). To investigate the maturation status of red blood cells in the peripheral blood (PB), we performed a blood smear analysis (Fig. 2B), which showed enucleated red blood cells in the PB of Hoxa5-HSCs recipients. To determine if the overexpression of Hoxa5 conferred an erythroid differentiation bias for HSCs, we analyzed the erythroid lineages in the secondary recipients (Fig. 2C). We detected predominantly erythroid cells in the PB for the first 2 months after transplantation, which confirmed the erythroid phenotype in the primary recipients. To confirm that the haematopoietic effect was indeed the result of the overexpression of Hoxa5, we further verified the ectopic expression level of Hoxa5. Q-RT-PCR using sorted cells from recipient bone marrow demonstrated that the expression level of Hoxa5 in Hoxa5-GFP transduced cells was 40-fold more than in the GFP control cells (Fig. S2C). To investigate whether Hoxa5 overexpression modified the long-term self-renewal capacity of HSCs, we isolated 0.25 million GFP+ bone marrow cells from primary recipients, and then transplanted the cells into secondary recipients. Twenty-four weeks later, flow cytometry analysis showed that the Hoxa5-HSCs successfully contributed to myeloid, lymphoid and erythroid lineages in the peripheral blood, bone marrow, spleen and thymus of the secondary recipients (Fig. S2D, Fig. 2D). Furthermore, Giemsa-Wright staining of bone marrow cells from the secondary recipients showed more erythroid progenitors (Fig. 2E) in the recipient mice transplanted with Hoxa5-HSCs. Further analysis of the HSC pools from the recipient mice showed that the contribution of Hoxa5-HSCs decreased, which indicated that Hoxa5 might disturb the homeostasis of HSCs (Fig. S3A, S3B). Thus, serial transplantations demonstrated that Hoxa5-HSCs sustained self-renewal and multipotency and predominantly reconstituted erythroid and myeloid lineages in vivo.
Figure 2.
HSCs overexpressing Hoxa5 maintain self-renewal and multipotency. (A) Primary recipients engrafted with HSCs expressing Hoxa5 showed higher ratios of Mac1+ and Ter119+ cells in peripheral blood. Lineage cells not expressing Hoxa5-GFP (GFP-) were used as internal control for each single mouse (n = 3). (B) Blood smear of PB showed that red blood cells were mature. (C) Secondary transplant showed predominant red blood cells in PB during the first 1–2 months. (D) Secondary transplant showed that HSCs overexpressing Hoxa5 still possessed properties of self-renewal and multipotency. A half million sorted cells (GFP positive) from primary recipients were transplanted into each secondary recipient. Twenty-four weeks after transplantation, myeloid (Mac1+), erythroid (TER119+), T lymphoid (Thy1.2+), and B lymphoid (B220+) lineages in peripheral blood were analyzed by flow cytometry. The results from one representative recipient are shown. (E) Giemsa-Wright staining of bone marrow cells from secondary recipients showed more erythroid progenitors (black arrow) in recipients transplanted with Hoxa5 HSCs compared to those transplanted with GFP control.
Accumulation of Megakaryocyte-Erythroid progenitor cells after ectopic expression of Hoxa5
To investigate the underlying mechanism by which Hoxa5 preferentially pushed HSCs toward erythropoiesis and myelopoieis in vivo, we analyzed the 3 progenitor components, common myeloid progenitor cells (CMP), granulocyte/macrophage lineage-restricted progenitor cells (GMP), and megakaryocyte erythroid progenitor cells (MEP) in the bone marrow of Hoxa5-HSCs recipients. Flow cytometry analysis showed that the percentage of MEP increased significantly in the bone marrow of Hoxa5-HSCs recipients (Fig. 3A, B) compared to that in control cells (GFP-, derived from Hoxa5 negative HSCs) in the same animal. In addition, increased expression of Hoxa5 significantly decreased the GMP proportion in the bone marrow (Fig. 3A, B). To directly evaluate the effects of Hoxa5 on GMP and MEP, We sorted 500 progenitor cells (Lin-IL7R-sca1-cKit+) from the bone marrow of the Hoxa5-HSCs recipients and performed a colony forming (CFU) assay in vitro. Interestingly, the total colony numbers planted with Hoxa5-GFP cells sharply decreased. The total bone marrow cells from the Hoxa5-HSCs recipients showed a similar phenotype in the CFU assay (Fig. S3C). To investigate if Hoxa5 overexpression blocks the differentiation of erythroid progenitors, we analyzed erythroid blasts at different stages based on CD71 and Ter119 markers. Consistent with the phenotype of mature red blood cells, overexpression of Hoxa5 showed no effect on erythroid differentiation (Fig. 3C). In contrast to the higher proportion of MEP in vivo, CFU-Es from Hoxa5-HSCs cells were nearly undetectable. Of note, CFU-GMs from Hoxa5 expressing cells were predominant, though their colony sizes shrank (Fig. 3D, E). Consistent with previous observations,12,13 our in vitro data indicated that Hoxa5 overexpression causes functional defects in MEPs and GMPs.
Figure 3.
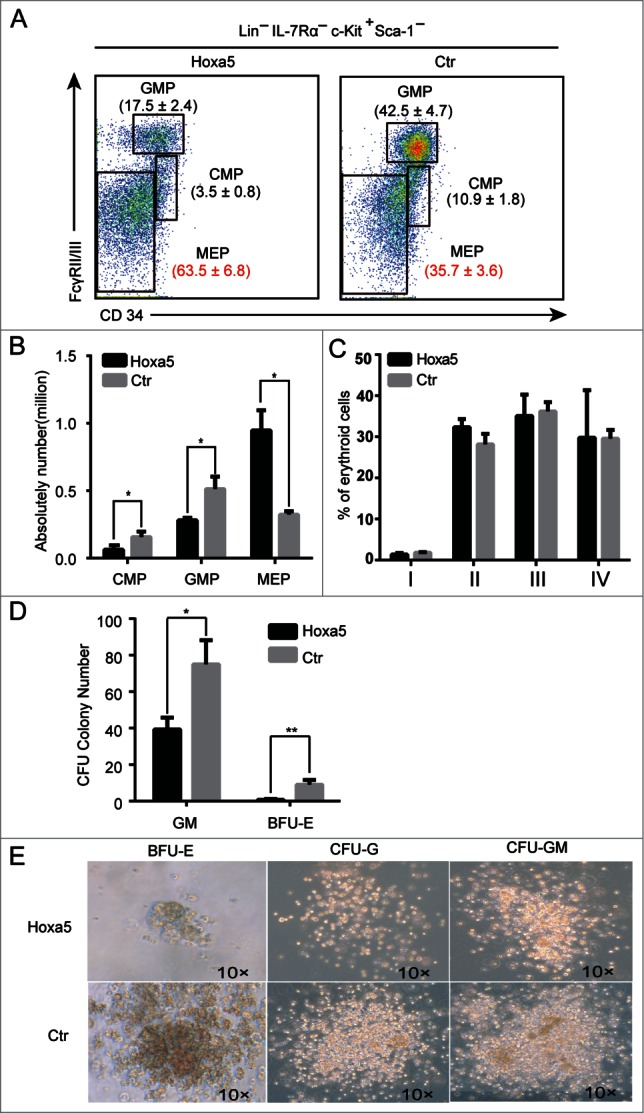
Increased expression of Hoxa5 results in accumulation of megakaryocyte-erythroid progenitor cells. (A) Flow cytometry analysis of erythroid and myeloid progenitors in bone marrow cells isolated from recipients transplanted with HSCs expressing Hoxa5-GFP and GFP control. Plots from one representative experiment showed the percentages of common myeloid progenitors (CMP; FcγRII/III low CD34+), granulocyte/macrophage lineage-restricted progenitors (GMP; FcγRII/III hi CD34+), and megakaryocyte erythroid progenitors (MEP; FcγRII/III low CD34-) relative to total progenitors. (B) Absolute numbers of GMP, CMP and MEP in recipients (n = 3, **, P < 0.01). (C) Erythroid differentiation is normal in recipients of Hoxa5-HSCs. Erythroid blasts at different stages of maturation in various populations based on CD71 and Ter119 markers. I, Ter119-, CD71 high; II, Ter119+, CD71 high; III, Ter119+, CD71 mid; IV, Ter119+, CD71 low. (D) Colony forming assay indicates that myeloid progenitor cells overexpressing Hoxa5 produce much fewer colonies in vitro. Two hundred progenitor cells (Lin-IL7R-cKit+sca1-) sorted from Hoxa5-GFP and GFP control mice were planted in semisolid medium. The data presented are colony numbers counted on day 8. Error bars indicate standard error. (n = 3, *P < 0.05; **, P < 0.01). (E) Representative colonies from cells expressing Hoxa5-GFP and control (GFP alone).
Hoxa5 differentially modifies the cell cycle of haematopoietic stem/progenitor cells depending on cellular context
To investigate the biological effects of the overexpression of Hoxa5 on haematopoietic hierarchy, we analyzed the proliferation status of HSCs, MPPs, CMPs, GMP and MEPs isolated from recipient bone marrow. In the HSC context, the overexpression of Hoxa5 pushed cells through the cell cycle; however, the HSC pool decreased (Fig. 4A, Fig. S3A, S3B). Interestingly, the ectopic expression of Hoxa5 did not change the cell cycle status of MPPs (Fig. 4A). Further analysis showed that higher proportions of MEPs, GMPs and CMPs were arrested in G0 phase (Fig. 4B). Thus, Hoxa5 regulated the proliferation status of haematopoietic stem/progenitor cells depending on cellular context.
Figure 4.
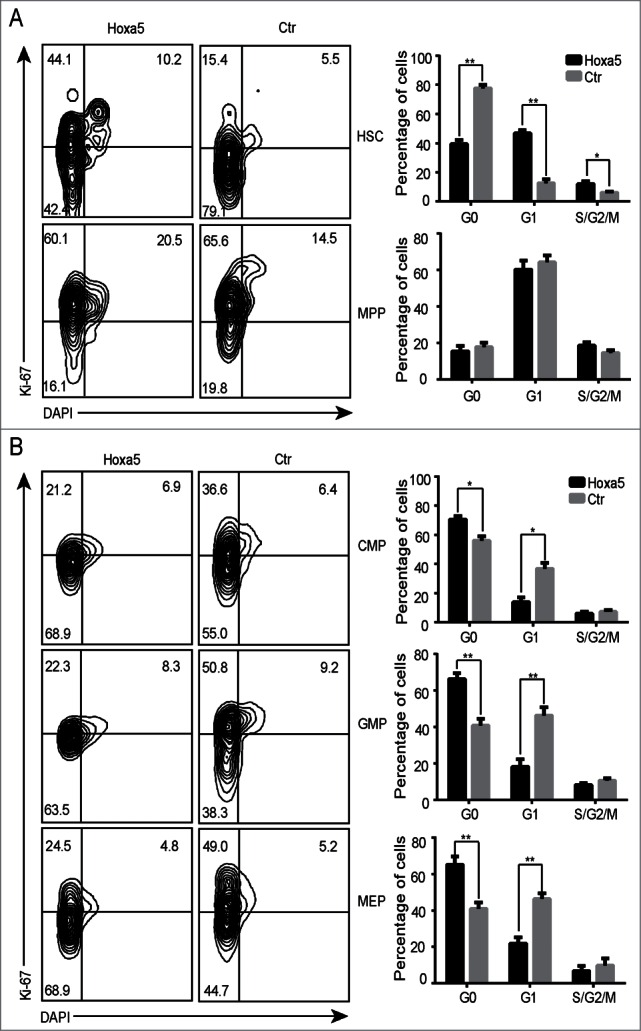
Ectopic expression of Hoxa5 differentially modifies proliferation status of haematopoietic stem/progenitor cells depending on cellular context. (A) Cell cycle analysis of HSCs and MPPs in bone marrow of GFP control and Hoxa5-HSCs recipient mice 6 months after transplantation. Cell cycle phases are defined as G0 (ki67- DAPI low), G1 (Ki67+ DAPI low), and S/G2/M (Ki-67+ DAPI high). Average values + SDs are shown in right graph, **P < 0.01 (n = 3). (B) Cell cycle analysis of CMP, GMP and MEP in bone marrow of GFP control and Hoxa5-HSCs recipient mice 6 months after transplantation. Student t test was performed (n = 3, *P < 0.05).
Erythroid progenitors undergo apoptosis after overexpressing Hoxa5
Hoxa5-HSCs produced higher ratios of erythroid lineages in vivo, consistent with the observed higher percentage of MEPs in recipient bone marrow. In contrast, the results of our in vitro CFU assay indicated that Hoxa5 caused defects in MEPs and GMPs, demonstrated by the presence of few CFU-Es and even fewer CFU-GMs with smaller colony sizes (Fig. 3D, E). We hypothesized that Hoxa5 caused partial defects in MEPs and GMPs in vivo. To test our hypothesis, we performed an apoptosis analysis of MEPs, GMPs and CMPs isolated directly from recipients. Consistent with our hypothesis, a proportion of these cells were undergoing apoptosis (Fig. 5A). There were more CMPs undergoing apoptosis than GMPs and MEPs (Fig. 5B). Interestingly, approximately 20% of MEPs were dead (Fig. 5B). Thus, Hoxa5 confers defects in erythroid cells, presented by cell accumulation at G0 phase and partial apoptosis.
Figure 5.
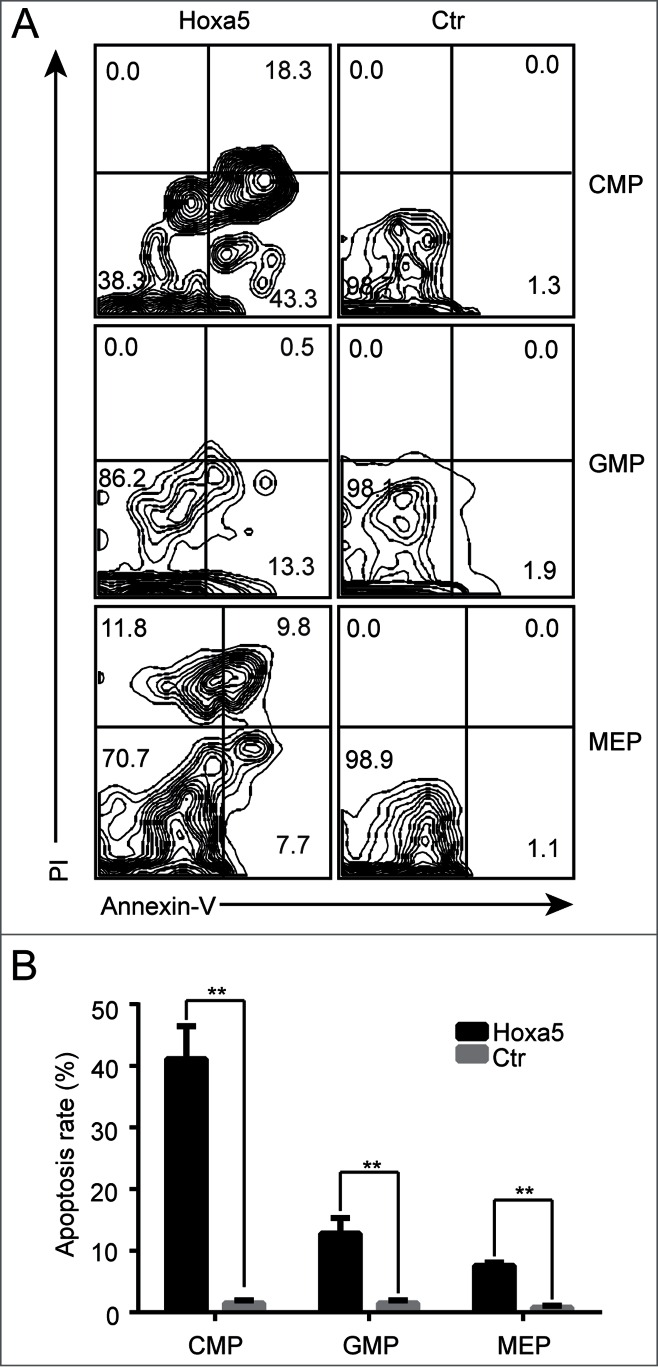
Accumulated megakaryocyte-erythroid progenitor cells expressing Hoxa5 are partially undergoing apoptosis. (A) Apoptosis analysis of CMP, GMP and MEP in bone marrow of GFP control and Hoxa5-GFP recipient mice 6 months after transplantation. Annexin-V+, PtdIns- cells represent apoptotic cells. Annexin-V+, PI+ cells represent dead cells. Plots from one representative experiment are shown. (B) Statistical analysis of apoptosis of CMP, GMP and MEP (n = 3, **P < 0.01).
Genes targeted by Hoxa5 in progenitor cells
To investigate the gene effectors of Hoxa5 in erythroid progenitor cells, we carried out RNA-seq of sorted progenitor cells (lin-IL7R-sca1-cKit+) from recipients transplanted with Hoxa5-HSCs or GFP control. As shown in Table 1, several genes involved in multiple processes of hematopoiesis were dysregulated by Hoxa5. Hoxa5 downregulated the expression levels of Wfs1, Klf4, Zfp2612, Aldh6a1, Irf8 and Fos, which have been shown to be involved in cell proliferation, differentiation and metabolism. Of note, Wfs1 and Egr1 also play an important role in apoptosis. Consistent with the observation that erythroid progenitor cells showed difficulty in forming CFU-Es in vitro, Hoxa5 downregulated the erythrocyte homeostasis gene Klf2. Thus, the ectopic expression of Hoxa5 leads to a hematopoiesis defect at the progenitor cell level by dysregulating genes involved in multiple processes of hematopoiesis and common cell biology.
Discussion
To date, little is known about the role of the Hoxa5 transcription factor in hematopoiesis in vivo. Here, we present data showing that Hoxa5 is predominantly expressed in HSCs instead of MPPs. In this study's HSC transplantation assay, Hoxa5-HSCs demonstrated aberrant erythropoiesis and myelopoiesis but sustained self-renewal and multipotency. Interestingly, increased expression of Hoxa5 drove HSCs, but not MPPs, through the cell cycle. However, at the lineage-committed progenitor level, Hoxa5 expression caused more cells to arrest in G0 phase. Although a proportion of erythroid progenitors overexpressing Hoxa5 could differentiate into mature erythroid cells, some underwent apoptosis and were doomed to death in vivo. The RNA-seq results indicated that multiple biological processes, including erythrocyte homeostasis, cell metabolism, and apoptosis process, were modified by Hoxa5.
Under the physiological condition, adult HSCs largely remain in homeostasis (G0 phase), forming a steady HSC pool.17 However, HSCs can progress through the cell cycle under stressful conditions, such as chemical treatment and G-CSF mobilization.18 Little is known regarding the regulatory mechanism of the HSC cell cycle, which limits the ability of researchers to perform ex vivo modeling of the maintenance and expansion of HSCs. Recently, an artificial fusion gene, Nup98-Hoxa10HD, was reported to expand mouse HSCs over 10,000-fold 10 days after in vitro culture.19 Of note, the Hoxa5 transcription factor is upregulated in these expanding HSCs,4 but the related biological effects of Nup98-Hoxa10HD on HSCs remains elusive. In our study, we observed that Hoxa5 pushed more HSCs through the cell cycle. The serial transplantation assay showed that the Hoxa5-HSCs maintained self-renewal and multipotency. Interestingly, the cell cycle status of MPPs was not changed by the increased expression of Hoxa5. Given that Hoxa5 signaling is further activated in expanding HSCs,4 our study provided direct in vivo evidence that Hoxa5 mainly regulates the function of HSCs during normal hematopoiesis. Future studies on Hoxa5 signaling in HSCs, especially on the genes that regulate the cell cycle status of HSCs,20,21 would provide more insights into the molecular mechanisms of Hoxa5 in HSCs.
Consistent with our observations in the CFU assay, human HOXA5 has been reported to block erythroid differentiation in in vitro CFU assays.12,13 In contrast, our in vivo data demonstrated that a higher proportion of mature erythroid lineage cells were present in the peripheral blood of Hoxa5-HSCs recipients within the first 3 months after transplantation (Fig. 2A, C). Hoxa5 might play a role in urgent erythropoiesis, but the related mechanism remains elusive. When we performed an in vitro CFU assay using bone marrow cells and purified erythroid and myeloid progenitor cells from the recipients, the cells showed defects in forming CFU-Es. This phenotypic contradiction might be caused by the differences in the in vivo physiological condition and the in vitro CFU condition. Unfortunately, there is no report on the in vivo role of human HOXA5. Thus, further experiments on the effects of HOXA5 in hematopoiesis using humanized NSG mice might illustrate whether the physiological roles of Hoxa5 are conserved between humans and mice. In addition to the dynamic MEP pool accumulated from upstream progenitors in the haematopoietic hierarchy, a more complicated signaling network and erythropoiesis niche in live animals might facilitate the predominant erythropoiesis observed after ectopic expression of Hoxa5. Hoxa5 indeed confers defects in MEPs in vivo, indicated by cell cycle arrest at G0, partial apoptosis and cell death. Consistent with our observations, HOXA5 also induced apoptosis in breast cancer cells.6 The predominant percentage of mature erythroid cells during the reconstitution stage indicates that erythroid differentiation is normal in vivo. Clearly, Hoxa5 has dual effects on erythropoiesis in vivo.
In conclusion, the biological effects of Hoxa5 on hematopoiesis depend on cellular context. Consistent with its natural expression pattern in HSCs and MPPs, the overexpression of Hoxa5 in HSCs enhances cell proliferation by driving more cells through the cell cycle, but this is not the case for MPPs. Increased expression of Hoxa5 in lineage-committed progenitors caused aberrant erythropoiesis. While our study provides evidence that Hoxa5 is involved in regulating hematopoiesis at the level of haematopoietic stem/progenitor cells, its precise role and signaling mechanism remain elusive. Further studies using a cell-type specific expression model would help to elucidate the regulatory mechanism of Hoxa5 in hematopoiesis.
Materials and Methods
Mice
All mice used in this study were housed in the animal facility of the Guangzhou Institutes of Biomedicine and Health (GIBH). The CD45.1 strain was originally bought from Jackson lab. Donor fetal liver HSCs were isolated from C57BL/6-CD45.2 embryos (E14.5).22 All experiments were conducted with the ethical approval of the Animal Ethics Committee of GIBH.
RNA isolation, sequencing and analysis
Ten thousand cells of each HSCs (Lin-CD48-cKit+sca1+CD150+), MPPs (Lin-CD48-cKit+sca1+CD150−), and progenitor cells (Lin-IL7R-sca1-cKit+) were separately sorted into 350 ul RLT buffer. Total RNA was extracted using an RNeasy micro kit (QIAGEN). Two nanograms of RNA from each sample were used to create libraries for deep sequencing according to the manual of the TruSeq RNA Sample Preparation Kit (Illumia). Samples were sequenced using the Illumina Myseq sequencer. Sequence reads were processed and analyzed with the help of the GIBH bioinformatics core.
Retroviral transduction of fetal liver Lin- cells
Hoxa5 cDNA was inserted into pMYs-IRES-EGFP to generate the pMYs-Hoxa5-IRES-EGFP vector (RTV-021, Cell Biolabs, INC). pMYs-IRES-EGFP or pMYs-Hoxa5-IRES-EGFP were transduced into Plat-E cells to produce high-titer, replication-incompetent viruses. The resulting retrovirus supernatants were collected 48–72 hours later and used immediately. Fetal liver lineage negative (FL Lin-) cells were enriched using a lineage depletion method by AutoMACS (Miltenyi Biotec). Enriched FL Lin- cells were suspended in viral supernatants (1 × 105 cells/ml) with 8 μg/ml Polybrene (Sigma Aldrich) and centrifuged at 800 g for 120 minutes. They were then cultured for 24 hours in StemSpan SFEM (09600, StemCell Technologies) in the presence of 10 μg/ml heparin, 50 ng/ml SCF (300-07, Peprotech), and 20 ng/ml TPO (300-18, Peprotech). Cells were then suspended in viral supernatant for secondary round of infection. The cells were cultured for another 24 hours before transplantation.
Antibodies
CD2 (RM2-5), CD3 (145-2C11), CD4 (RM4-5), CD8 (53-6.7), Gr1 (RB6-8C5), Mac1 (M1/70), IgM (II/41), Thy1.2 (53-2.1), B220 (6B2), IL7Rα (B12-1), cKit (2B8), Sca1 (E13-161.7), CD34 (RAM34), FcγRII/III (2.4 G2), CD150 (TC15-12F12.2), and Ki67 (SolA15) antibodies conjugated with biotin or related dyes, as required, were purchased from eBioscience.
HSC transplantation
Adult C57BL/6 recipient mice (CD45.1+, 8–10 weeks old) were irradiated with 2 doses of 550 rads (RS 2000, Rad Sourse) for a 2 hour interval. Two hundred sorted GFP+ LSK cells (CD45.2+) with 0.25 million Sca1 depleted helpers were injected into the retro-orbital venous sinus of the irradiated CD45.1-recipients. The transplanted mice were maintained on trimethoprim-sulfamethoxazole-treated water for 2 weeks. After transplantation, blood was obtained from the retro-orbital venous sinus as needed for flow cytometric analysis.
Cell-cycle analysis
The methods of cell cycle analysis were described previously.23 Briefly, for the HSCs/MPP cell cycle analysis, we first stained the total bone marrow cells with biotin-labeled CD2, CD3, CD4, CD8, Mac1, Gr1, B220, and TER119, and with CD48. After washing, PE-CF594-cKit-, PerCP-Sca1-, APC-CD150-, and APC-efluor780-conjugated streptavidin were added. Then, the cells were fixed with 4% PFA. Finally, the fixed cells were permeabilized with 0.1% saponin in PBS and were simultaneously stained with PE-ki67 (eBioscience) and DAPI (Invitrogen) for 45 minutes. For the erythroid and myeloid progenitor cell cycle assay, antibodies recognizing the lineage markers CD2, CD3, CD4, CD8, Mac1, Gr1, B220, TER119 and IgM and recognizing IL-7Rα were added first. After washing, CD16/32-PE and APC-efluor 780 conjugated to streptavidin and PE-CF594-cKit, PE-Cy7-Sca1, and PerCP-Cy5.5-CD34 antibodies were added. The subsequent steps were the same as for the cell cycle analysis of HSCs/MPPs. The stained cells were analyzed using AriaII (BD Bioscience), and the data were analyzed using Flowjo software (FlowJo).
Apoptosis analysis
Biotin-labeled antibodies recognizing the lineage markers CD2, CD3, CD4, CD8, Mac1, Gr1, B220, TER119 and IgM and recognizing IL-7Rα were added to Fc-blocked total bone marrow cells. After washing, a second round of antibody cocktails, including APC-efluor 780 conjugated to streptavidin, PE-CF594-cKit, PE-Cy7-Sca1, and PerCP-Cy5.5-CD34, were added. The cells were washed in 1× DPBS and 1× binding buffer (eBioscience) and were then incubated with APC-conjugated Annexin V for 15 minutes in 1× binding buffer. Finally, PI was added to 1× binding buffer before FACS analysis (AriaII; BD Biosciences). All data were analyzed using Flowjo software (Tree Star Inc.).
Quantitative real-time PCR analysis
To confirm the mRNA expression level of Hoxa5 in HSCs and MPPs by quantitative real time polymerase chain reaction (RT-PCR), total RNA was isolated from 10,000 purified HSCs and MPPs with an RNeasy micro kit (QIAGEN), and the RNA was digested with DNase I to remove any genomic DNA contamination. Then, 2 ng of RNA was used for linear amplification, according to the manufacturer's instructions (3302–12, Ovation Pico WTA System V2, NuGEN Technologies, Inc.). Ten ng cDNA was used as the template according to the q-PCR protocol in CFX-96 (Bio-Rad). The primers for Hoxa5 were 5’-CGCAAGCTGCACATTAGTCACG-3′ and 5′-GAGAGGCAAAGGGCATGAGCTA-3′. HSP90ab1 was used as the control for HSCs and, especially, MPPs, with the following primers: (Fw: 5′-CCTGAAGGTCATCCGCAAGAAC-3′; Rev: 5′-GGCGTCGGTTAGTGGAATCTTC-3′). All operation were performed as manufacturer's protocol(Bio-Rad). For the mRNA transcript expression analysis of Hoxa5 in recipient haematopoietic tissues, GAPDH (Fw: 5′-CCTGGAGAAACCTGCCAAGTATG-3′; Rev: 5′-AGAGTGGGAGTTGCTGTTGAAGTC-3′) was used as the control. Fold expression relative to the reference gene was calculated using the comparative method 2-ΔΔCt, and the values were normalized to 1 for comparison.
Statistical analysis
A minimum of 3 replicates were performed for all quantitative analyses. Data are presented as means ± SEs. Student's t tests were performed using statistic software GraphPad Prism and are indicated in the figure legends.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Major National Research Projects of China (Grant No. 2015CB964401), the National Natural Science Foundation of China (Grant No. 31271457, and 81470281), the Strategic Priority Research Program of the Chinese Academic of Sciences, Grant No. XDA01020311, and the Equipment Function Development and Technology Innovation Project of Chinese Academy of Sciences (Grant No. yg2012049).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene 2007; 26:6766-76; PMID:17934484; http://dx.doi.org/ 10.1038/sj.onc.1210760 [DOI] [PubMed] [Google Scholar]
- 2. Lebert-Ghali CE, Fournier M, Dickson GJ, Thompson A, Sauvageau G, Bijl JJ. HoxA cluster is haploinsufficient for activity of hematopoietic stem and progenitor cells. Exp Hematol 2010; 38:1074-86 e1-5; PMID:20655978; http://dx.doi.org/ 10.1016/j.exphem.2010.07.006 [DOI] [PubMed] [Google Scholar]
- 3. Sauvageau G, Thorsteinsdottir U, Eaves CJ, Lawrence HJ, Largman C, Lansdorp PM, Humphries RK. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev 1995; 9:1753-65; PMID:7622039; http://dx.doi.org/ 10.1101/gad.9.14.1753 [DOI] [PubMed] [Google Scholar]
- 4. Palmqvist L, Pineault N, Wasslavik C, Humphries RK. Candidate genes for expansion and transformation of hematopoietic stem cells by NUP98-HOX fusion genes. PloS one 2007; 2:e768; PMID:17712416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gendronneau G, Lemieux M, Morneau M, Paradis J, Tetu B, Frenette N, Aubin J, Jeannotte L. Influence of Hoxa5 on p53 tumorigenic outcome in mice. Am J Pathol 2010; 176:995-1005; PMID:20042682; http://dx.doi.org/ 10.2353/ajpath.2010.090499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen H, Chung S, Sukumar S. HOXA5-induced apoptosis in breast cancer cells is mediated by caspases 2 and 8. Mol Cell Biol 2004; 24:924-35; PMID:14701762; http://dx.doi.org/ 10.1128/MCB.24.2.924-935.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bach C, Buhl S, Mueller D, Garcia-Cuellar MP, Maethner E, Slany RK. Leukemogenic transformation by HOXA cluster genes. Blood 2010; 115:2910-8; PMID:20130239; http://dx.doi.org/ 10.1182/blood-2009-04-216606 [DOI] [PubMed] [Google Scholar]
- 8. Okada Y, Jiang Q, Lemieux M, Jeannotte L, Su L, Zhang Y. Leukaemic transformation by CALM-AF10 involves upregulation of Hoxa5 by hDOT1L. Nat Cell Biol 2006; 8:1017-24; PMID:16921363; http://dx.doi.org/ 10.1038/ncb1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood 2011; 118:6247-57; PMID:21948299; http://dx.doi.org/ 10.1182/blood-2011-07-328880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol 2007; 9:804-12; PMID:17589499; http://dx.doi.org/ 10.1038/ncb1608 [DOI] [PubMed] [Google Scholar]
- 11. Chung KY, Morrone G, Schuringa JJ, Plasilova M, Shieh JH, Zhang Y, Zhou P, Moore MA. Enforced expression of NUP98-HOXA9 in human CD34(+) cells enhances stem cell proliferation. Cancer Res 2006; 66:11781-91; PMID:17178874; http://dx.doi.org/ 10.1158/0008-5472.CAN-06-0706 [DOI] [PubMed] [Google Scholar]
- 12. Crooks GM, Fuller J, Petersen D, Izadi P, Malik P, Pattengale PK, Kohn DB, Gasson JC. Constitutive HOXA5 expression inhibits erythropoiesis and increases myelopoiesis from human hematopoietic progenitors. Blood 1999; 94:519-28; PMID:10397719 [PubMed] [Google Scholar]
- 13. Fuller JF, McAdara J, Yaron Y, Sakaguchi M, Fraser JK, Gasson JC. Characterization of HOX gene expression during myelopoiesis: role of HOX A5 in lineage commitment and maturation. Blood 1999; 93:3391-400; PMID:10233891 [PubMed] [Google Scholar]
- 14. Forsberg EC, Prohaska SS, Katzman S, Heffner GC, Stuart JM, Weissman IL. Differential expression of novel potential regulators in hematopoietic stem cells. PLoS Genet 2005; 1:e28; PMID:16151515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Doulatov S, Vo LT, Chou SS, Kim PG, Arora N, Li H, Hadland BK, Bernstein ID, Collins JJ, Zon LI, et al. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell 2013; 13:459-70; PMID:24094326; http://dx.doi.org/ 10.1016/j.stem.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity 1994; 1:661-73; PMID:7541305; http://dx.doi.org/ 10.1016/1074-7613(94)90037-X [DOI] [PubMed] [Google Scholar]
- 17. Seita J, Weissman IL. Hematopoietic stem cell: self-renewal versus differentiation. Wiley Interdiscip Rev Syst Biol Med 2010; 2:640-53; PMID:20890962; http://dx.doi.org/ 10.1002/wsbm.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morrison SJ, Wright DE, Weissman IL. Cyclophosphamide/granulocyte colony-stimulating factor induces hematopoietic stem cells to proliferate prior to mobilization. Proc Nat Acad Sci US America 1997; 94:1908-13; PMID:9050878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sekulovic S, Gasparetto M, Lecault V, Hoesli CA, Kent DG, Rosten P, Wan A, Brookes C, Hansen CL, Piret JM, et al. Ontogeny stage-independent and high-level clonal expansion in vitro of mouse hematopoietic stem cells stimulated by an engineered NUP98-HOX fusion transcription factor. Blood 2011; 118:4366-76; PMID:21865344; http://dx.doi.org/ 10.1182/blood-2011-04-350066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M, Scadden DT. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 2000; 287:1804-8; PMID:10710306; http://dx.doi.org/ 10.1126/science.287.5459.1804 [DOI] [PubMed] [Google Scholar]
- 21. Yu H, Yuan Y, Shen H, Cheng T. Hematopoietic stem cell exhaustion impacted by p18 INK4C and p21 Cip1/Waf1 in opposite manners. Blood 2006; 107:1200-6; PMID:16234365; http://dx.doi.org/ 10.1182/blood-2005-02-0685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim I, He S, Yilmaz OH, Kiel MJ, Morrison SJ. Enhanced purification of fetal liver hematopoietic stem cells using SLAM family receptors. Blood 2006; 108:737-44; PMID:16569764; http://dx.doi.org/ 10.1182/blood-2005-10-4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang J, Kong G, Liu Y, Du J, Chang YI, Tey SR, Zhang X, Ranheim EA, Saba-El-Leil MK, Meloche S, et al. Nras(G12D/+) promotes leukemogenesis by aberrantly regulating hematopoietic stem cell functions. Blood 2013; 121:5203-7; PMID:23687087; http://dx.doi.org/ 10.1182/blood-2012-12-475863 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.