Abstract
Echinoderm microtubule-associated protein (EMAP)-like (EML) family proteins are microtubule-associated proteins that have a conserved hydrophobic EMAP-like protein (HELP) domain and multiple WD40 domains. In this study, we examined the role of EML4, which is a member of the EML family, in cell division. Time-lapse microscopy analysis demonstrated that EML4 depletion induced chromosome misalignment during metaphase and delayed anaphase initiation. Further analysis by immunofluorescence showed that EML4 was required for the organization of the mitotic spindle and for the proper attachment of kinetochores to microtubules. We searched for EML4-associating proteins by mass spectrometry analysis and found that the nuclear distribution gene C (NUDC) protein, which is a critical factor for the progression of mitosis, was associated with EML4. This interaction was mediated by the WD40 repeat of EML4 and by the C-terminus of NUDC. In the absence of EML4, NUDC was no longer able to localize to the mitotic spindle, whereas NUDC was dispensable for EML4 localization. Our results show that EML4 is critical for the loading of NUDC onto the mitotic spindle for mitotic progression.
Keywords: cytokinesis, EML4, kinetochore, mitosis, NUDC, spindle
Introduction
The proliferation of living cells requires an accurate distribution of duplicated genetic materials to daughter cells. The failure of this process produces aneuploidy and induces cellular apoptosis or transformation.1-2 A dynamic structural arrangement of microtubules is initiated at the onset of mitosis to generate the mitotic spindle to achieve the high fidelity distribution of the genome. The mitotic spindle is a bipolar array of microtubules with minus ends that are concentrated at the MTOC (microtubule organizing center) or centrosomes and with plus ends that extend outward from the MTOC.3 In metaphase cells, spindle microtubules that are attached to the kinetochore, which is a large multi-protein complex on each chromosome, are bundled and stabilized to form kinetochore fibers (k-fibers).4 Once all the kinetochores are properly attached to the spindle microtubules, then the spindle assembly checkpoint (SAC) is inactivated to initiate the separation of chromosomes to either side of spindle poles; this separation is mediated by the depolymerization and shrinkage of the spindle microtubules.5,6 With the separation of chromosomes to each pole, some spindle microtubules further extend to the opposite poles and interdigitate at the center of 2 separating cells to form a structure called the central spindle. The central spindle is a critical structure for the determination of the site of furrow ingression.7,8 With the progression of furrow ingression, microtubules and associated proteins are tightly packed to form an electron dense structure called the midbody.9 These dynamic rearrangements of the mitotic spindle are regulated by several associating proteins and by their post-translational modification, such as phosphorylation. Although extensive studies have been performed to identify these microtubule-associated proteins, not all their functions are understood in detail.
Echinoderm microtubule-associated protein (EMAP) is the most abundant microtubule-associated protein in sea urchin.10-12 EMAP is localized to the interphase microtubules and to the mitotic spindle and regulates microtubule dynamics.13-15 The EML (EMAP-like) protein family is composed of mammalian homologs of EMAP. Six EML proteins (EML1-6) have been identified, and all the EML proteins have a HELP (hydrophobic EMAP-like protein) domain, which is unique to this family, and multiple WD40 domains.16–;20 Although EML proteins are highly similar in sequence and localize to the microtubules, these proteins have little sequence homology with other microtubule-associated proteins, and their exact functions are largely unknown. EML2 promotes microtubule catastrophes and may be a microtubule destabilizer;16 however, whether other EML proteins have similar functions has not been determined. Some studies have reported that EML proteins are required for the spindle function during mitosis. EML3 is localized to the nucleus during interphase; however, once the nuclear membrane has broken down for mitosis, EML3 accumulates to the mitotic spindle.19 EML3 depletion induced chromosome misalignment during metaphase and delayed anaphase initiation. EML4 is also localized to the mitotic spindle, and its depletion inhibited cell proliferation.18 These studies have shown that EML proteins are localized to the mitotic spindle and necessary for the correct spindle function; however, a detailed understanding of EML functions during mitosis is lacking. In this report, we studied the role of EML4 in mitotic progression. We demonstrated that EML4 is required for the organization of the mitotic spindle and for the proper attachment of spindle microtubules to kinetochores. We also showed that EML4 is essential for the recruitment of NUDC, which is a critical factor for mitotic progression, to the mitotic spindle.
Results
EML4 knockdown delays the progression of mitosis
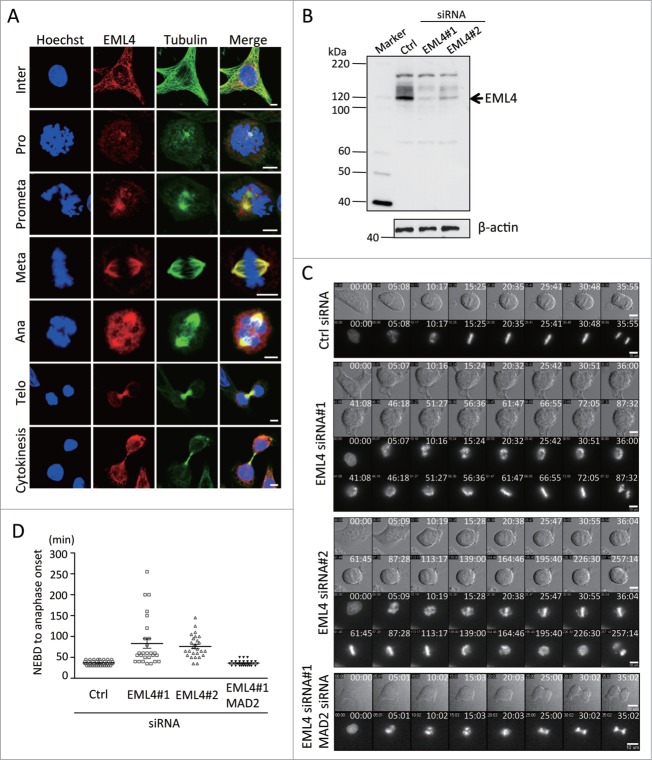
First, we examined the subcellular localization of EML4 during mitosis by indirect immunofluoresence analysis to obtain further insight into the role of EML4 in cell division. EML4 accumulated at the MTOC during prophase and localized to the mitotic spindle during metaphase. During telophase, EML4 concentrated to the either side of the midbody where microtubule bundles accumulated (Fig. 1A). We observed EML4 siRNA-transfected cells using time-lapse microscopy to characterize the defects induced by EML4 knockdown during mitosis. We used 2 different siRNAs (siRNA#1 and siRNA#2) that efficiently depleted EML4 (Fig. 1B). HeLa cells that constitutively expressed GFP-histone H2B were transfected with control or EML4 siRNAs, and 24 h later, the cells were observed by time-lapse microscopy for an additional 48 h. We observed a significant defect in the chromosome alignment of EML4-knockdown cells. Control siRNA-transfected cells showed prompt chromosome alignment at the division plane during metaphase (Fig. 1C). In contrast, the chromosomes in EML4-depleted cells were unstable, and fractions of chromosomes were dispersed during metaphase (Fig. 1C). Most of the control cells entered anaphase within 50 min after nuclear envelope breakdown (NEBD) (Fig. 1D). However, some EML4-knockdown cells took more than 100 min for anaphase initiation (Fig. 1D). EML4-knockdown cells finally completed cytokinesis; however, some cells eventually underwent apoptosis after cytokinesis completion. The delay in the initiation of anaphase indicated that EML4-knockdown induced the spindle assembly checkpoint (SAC). We co-depleted EML4 and MAD2, which is a critical component for the SAC, and observed the cells using time-lapse microscopy (Fig. 1C). The time duration from the NEBD to the anaphase initiation of EML4-knockdown cells was significantly reduced by the co-depletion of MAD2 (Fig. 1D). These results indicated that EML4 knockdown delayed proper chromosome alignment and induced the SAC.
Figure 1.
Depletion of EML4 inhibits chromosome alignment during metaphase and activates the SAC. (A) HeLa cells were immunostained for EML4, α-tubulin and nuclear material (Scale bar = 5 μm). (B) HeLa cells were transfected with siRNAs, and EML4 protein expression was examined by immunoblot 72 h later. (C) siRNA-transfected cells were observed by time-lapse microscopy. Representative images of each siRNA-transfected cell group are shown (Scale bar = 10 μm). (D) The graph shows the time duration from the NEBD to the initiation of anaphase of each siRNA-transfected cells. Three independent experiments were performed, and more than 25 cells in total were evaluated (mean ± SEM).
EML4 depletion inhibits spindle organization and kinetochore-microtubule attachment
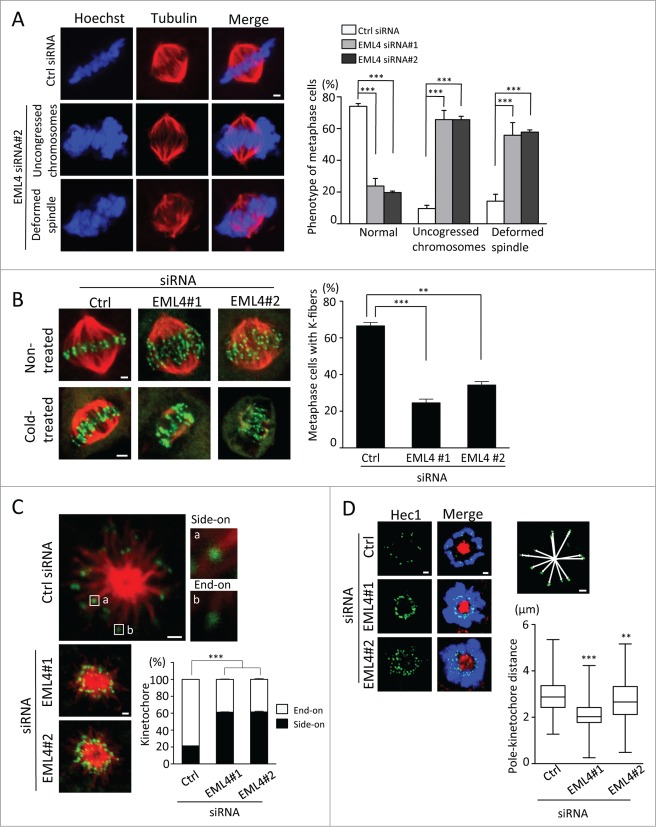
EML4 siRNA-transfected cells were fixed and immunostained with anti-tubulin antibody and Hoechst to further investigate the mitotic defects induced by EML4 depletion. Most of the control siRNA-transfected cells showed an organized bipolar spindle and properly aligned chromosomes during metaphase. In contrast, a significant number of EML4-depleted cells displayed disorganized mitotic spindles (Fig. 2A). In addition, more than 60% of EML4-knockdown cells showed uncongressed chromosomes during metaphase (Fig. 2A). Impaired chromosome alignment is often induced by defects in the attachment of kinetochores and microtubules. Kinetochore attachment to the ends of spindle microtubules organizes k-fibers that are stable under cold temperatures.21 We incubated siRNA-transfected cells on ice for 10 min and then fixed the cells to observe k-fiber formation to determine whether EML4 depletion induced the disruption of microtubule-kinetochore attachment. As shown in Fig. 2B, EML4 depletion significantly reduced the number of metaphase cells with stable mitotic spindles. We observed the kinetochore-microtubule attachment of monastrol-induced monopolar cells to corroborate these results further. A previous study showed that monopolar cells allowed the easy evaluation of side-on or end-on kinetochore attachment.22 Most of the kinetochores in control siRNA-transfected cells were attached to the ends of microtubules (Fig. 2C). In contrast, we observed that many kinetochores in EML4-depleted cells were attached to the sides of microtubules (Fig. 2C). Consistent with the increase in side-on attached kinetochores, the distance from the pole to each kinetochore significantly decreased in EML4-depleted monopolar cells (Fig. 2D). These results indicate that EML4 is required for the organization of the mitotic spindle and for the proper attachment of spindle microtubules to kinetochores.
Figure 2 (See previous page).
EML4 knockdown disrupts spindle organization and kinetochore-microtubule attachment. (A) siRNA-transfected cells were fixed and immunostained with anti-α-tubulin antibody and Hoechst. Representative images are shown (Scale bar = 1 μm). The graph indicates the percentage of metaphase cells with the indicated phenotype. Three independent experiments were performed, and more than 300 cells in total were evaluated (mean ± SEM, ***P < 0.01). (B) siRNA-transfected cells were non-treated or cold-treated and then immunostained with anti-α-tubulin and anti-CREST antibodies. Representative images are shown (Scale bar = 1 μm). The graph indicates the percentage of cold-treated cells with stable k-fibers. Three independent experiments were performed, and more than 150 cells in total were evaluated (mean ± SEM, ***P < 0.01). (C) siRNA-transfected cells were treated with monastrol and then immunostained with anti-α-tubulin and anti-CREST antibodies (Scale bar = 1 μm). The graph indicates the percentage of side-on and end-on attached kinetochores. Three independent experiments were performed, and approximately 1,000 kinetochores from 50 cells were evaluated (mean ± SEM, ***P < 0.01). (D) siRNA-transfected cells were treated with monastrol and immunostained with anti-Hec1 and anti-α-tubulin antibodies and Hoechst (Scale bar = 1 μm). Representative images are shown. The image above the graph shows how the distance from the pole to each kinetochore was measured. The graph shows the average distance of the pole to each kinetochore. Horizontal lines in the boxes indicate medians and the boxes extend from 25th to 75th percentiles. Three independent experiments were performed, and more than 600 kinetochores from 40 cells were evaluated using Image J software (***P < 0.01, **P < 0.05).
HELP domain and adjacent WD40 domains are required for mitotic progression
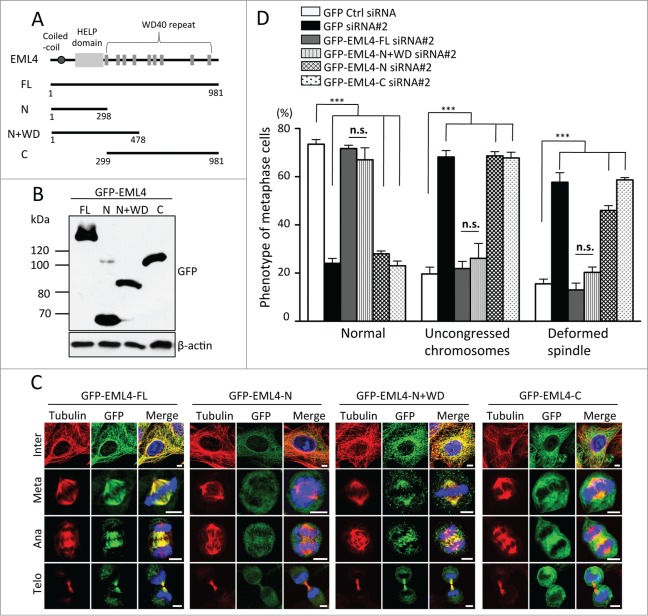
Next, we attempted to determine the region of the EML4 protein that is essential for mitotic progression. EML4 has a coiled-coil region and a HELP domain in the N-terminus, and multiple WD40 domains are in the C-terminus. As depicted in Fig. 3A, EML4 deletion mutants were generated, and the cells that constitutively expressed GFP-tagged mutants were established by retroviral infection (Fig. 3B). First, we examined the localization of mutant proteins in the absence of endogenous EML4. Each cell line was transfected with EML4 siRNA#2 that targeted the 3′ UTR of EML4 mRNA to deplete the endogenous EML4 protein specifically, and then the cells were fixed for GFP and tubulin immunostaining. Neither the fragment comprising the N-terminal region that contained the coiled-coil region and HELP domain (aa 1-298) nor the WD40 repeats (aa 299-981) alone localized to the mitotic spindle (Fig. 3C). However, an EML4 deletion mutant that contained the N-terminal region and adjacent partial WD40 repeats (aa 1-478) clearly localized to the mitotic spindle (Fig. 3C). Notably, EML4 (aa 1-298) localized to the mitotic spindle in the presence of endogenous EML4 (Fig. S1A). EML4 (aa 1-298) is able to form a homodimer with full-length EML4 (Fig. S1B); thus, EML4 (aa 1-298) seems to localize to the mitotic spindle by binding to the endogenous EML4 protein. Each cell line was transfected with EML4 siRNA#2 to determine whether these deletion mutants could rescue the mitotic defect induced by EML4 depletion. The expression of full-length EML4 clearly reduced the number of cells with disorganized spindles or with uncongressed chromosomes (Fig. 3D). Neither EML4 (aa 1-298) nor EML4 (aa 299-981) deletion mutants could rescue the mitotic defect induced by endogenous EML4 knockdown; however, the expression of EML4 (aa 1-478) clearly reduced the number of cells with disorganized spindles or with uncongressed chromosomes (Fig. 3D). These results indicated that the N-terminal HELP domain and adjacent partial WD40 repeats are required for the localization and functionality of EML4.
Figure 3.
HELP domain and N-terminal WD40 domains in EML4 are required for mitotic progression. (A) Schematic representation of EML4 deletion mutants. (B) Cells that constitutively expressed GFP-tagged EML4 deletion mutants were lysed, and the expression of each mutant was examined by immunoblot. (C) Each GFP-EML4 cell line was transfected with EML4 siRNA#2, and the cells were immunostained for GFP and tubulin 72 h later (Scale bar = 5 μm). (D) The graph shows the percentage of metaphase cells with the indicated defect. Three independent experiments were performed, and more than 150 cells in total were evaluated (mean ± SEM, ***P < 0.01, n.s; not significant compared to Ctrl siRNA).
EML4 associates with NUDC
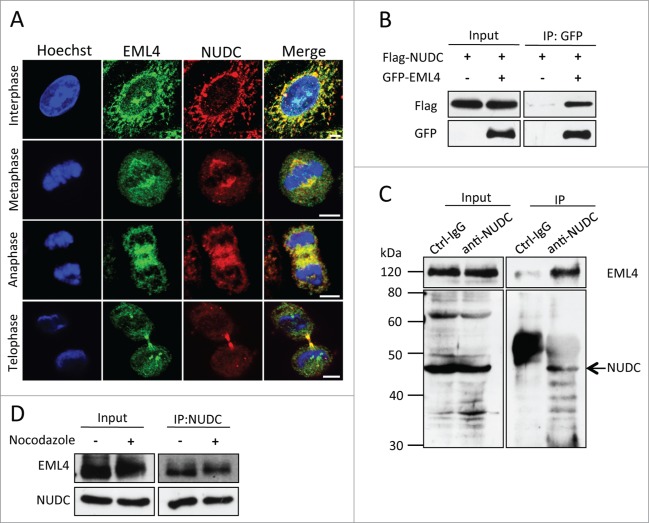
We attempted to identify EML4-associating proteins by mass spectrometry analysis to investigate the functions of EML4 in detail. Cells that constitutively expressed Flag-tagged EML4 were established, and a cell lysate was prepared from nocodazole-released cells. Flag-EML4 and its associated proteins were immunoprecipitated, eluted with Flag peptide and then identified by mass spectrometry analysis. Among the proteins that co-precipitated with Flag-EML4, we focused on NUDC, which is essential for spindle organization and for chromosome alignment.23-25 Indirect immunofluorescence analysis demonstrated that both EML4 and NUDC were localized to the mitotic spindle and concentrated to both sides of the midbody during telophase (Fig. 4A). GFP-EML4 and Flag-NUDC were transiently expressed in 293T cells to confirm the association of these 2 proteins. GFP-EML4 was immunoprecipitated from the cell lysate by anti-GFP antibody and immunoblotted with anti-Flag antibody. As shown in Fig. 4B, Flag-NUDC was co-precipitated with GFP-EML4. Next, we examined the association of endogenous proteins. Control IgG or anti-NUDC antibody was mixed with the cell lysate, and immunoprecipitated proteins were blotted for EML4. EML4 was detected in the anti-NUDC antibody immunoprecipitate, but not in the control IgG antibody immunoprecipitate (Fig. 4C). Cells were treated with nocodazole or non-treated, and endogenous NUDC was immunoprecipitated to determine whether this interaction is specific during mitosis. As shown in Fig. 4D, the interaction was observed in both non-treated cells and mitosis-arrested cells.
Figure 4.
EML4 associates with NUDC. (A) HeLa cells were immunostained with anti-EML4 and anti-NUDC antibodies (Scale bar = 5 μm). (B) 293T cells were transfected with a plasmid encoding the indicated gene, and the cells were lysed 24 h later. GFP-EML4 was immunoprecipitated with anti-GFP antibody and immunoblotted for GFP and Flag. (C) HeLa cells were lysed and immunoprecipitated with anti-NUDC antibody, followed by immunoblot with anti-NUDC and anti-EML4 antibodies. An arrow indicates endogenous NUDC. (D) Nocodazole-treated or non-treated cells were lysed, and endogenous NUDC was immunoprecipitated with anti-NUDC antibody. The immunoprecipitates were immunoblotted with anti-NUDC and anti-EML4 antibodies.
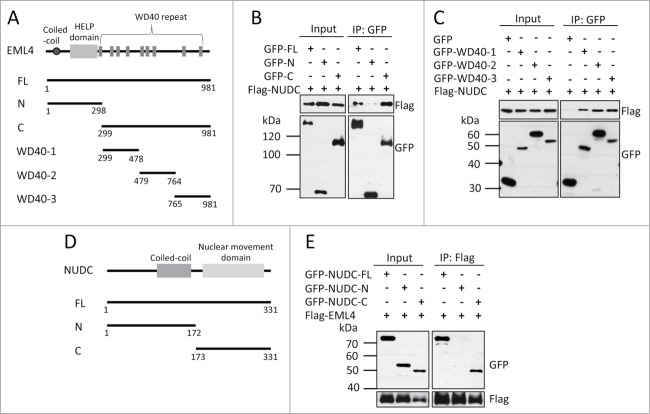
We used several EML4 deletion mutants (Fig. 5A) to determine the region of the EML4 protein that is required for the association with NUDC. First, we determined whether the N-terminal region or WD40 repeats were responsible for this association. As shown in Fig. 5B, the WD40 repeats were responsible for this interaction. Interestingly, multiple WD40 domains contributed to the binding to NUDC because the WD40 domains in the N-terminus, C-terminus and central region associated with NUDC (Fig. 5C). NUDC has a coiled-coil region in the N-terminus and a highly conserved nuclear migration domain in the C-terminus (Fig. 5D). Immunoprecipitation analysis revealed that the C-terminal region was responsible for the association with EML4 (Fig. 5E).
Figure 5.
WD40 domains of EML4 and the C-terminus of NUDC are required for the interaction between these proteins. (A) Schematic representation of EML4 deletion mutants. (B) 293T cells were transfected with a plasmid encoding each gene, and the cells were lysed 24 h later. GFP-tagged proteins were immunoprecipitated with anti-GFP antibody and immunoblotted for Flag and GFP. (C) The interaction between Flag-NUDC and GFP-tagged EML4 deletion mutants was examined. (D) Schematic representation of NUDC deletion mutants. (E) The interaction between Flag-EML4 and GFP-tagged NUDC deletion mutants was examined.
EML4 is required for the loading of NUDC to the mitotic spindle
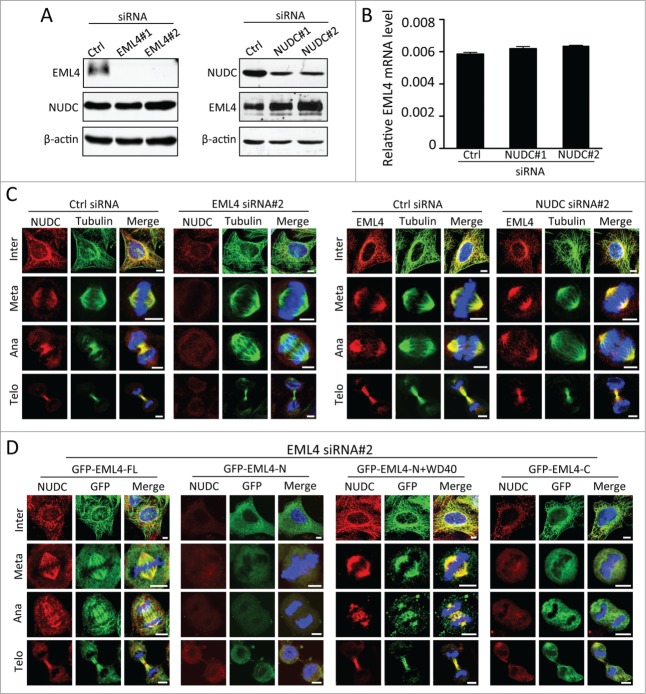
Previous studies showed that NUDC was essential for mitotic progression. Consistent with the previous report,23-25 we observed that NUDC depletion resulted in disrupted spindles and uncongressed chromosomes during metaphase (Fig. S2). The phenotype observed by EML4 depletion was similar to that observed by NUDC knockdown; thus, we speculated that the association between EML4 and NUDC is critical for the functionality of either protein. First, we tested whether this association was important for the expression of NUDC and EML4. EML4 depletion did not affect the levels of NUDC; however, EML4 expression increased in the absence of NUDC (Fig. 6A). RT-PCR analysis did not demonstrate any significant increase in EML4 mRNA (Fig. 6B). Next, we examined the localization of either protein in the absence of NUDC or EML4. EML4 localization along the mitotic spindle was clearly observed in the absence of NUDC (Fig. 6C). In contrast, NUDC localization at the spindle significantly decreased by the depletion of EML4 (Fig. 6C). We performed a rescue experiment using cells that constitutively expressed full-length or deletion mutants of EML4 to confirm that EML4 is required for the localization of NUDC to the mitotic spindle. NUDC was clearly localized to the mitotic spindle of full-length EML4-expressing cells transfected with EML4 siRNA#2 (Fig. 6D). The expression of the HELP domain (aa 1-298) or WD40 repeats (aa 299-981) of EML4 did not recover NUDC localization (Fig. 6C). However, the EML4 (aa 1-478) deletion mutant, which has the HELP domain and adjacent WD40 domains sufficient for binding to NUDC, clearly restored NUDC localization to the mitotic spindle (Fig. 6D). These results clearly demonstrated that EML4 is crucial for the loading of NUDC to the mitotic spindle.
Figure 6.
EML4 is required for NUDC localization to the mitotic spindle. (A) HeLa cells were transfected with the indicated siRNAs. Seventy-two hours later, the cells were lysed, and the expression of the indicated protein was determined by immunoblot. (B) HeLa cells transfected with control or NUDC siRNAs were lysed and level of EML4 mRNA was determined by real time PCR analysis. The graph show the relative level of EML4 mRNA normalized to the level of GAPDH mRNA. (C). Cells were transfected with siRNAs, and the cells were immunostained for NUDC or EML4 72 h later (Scale bar = 5 μm). (D) Cells that constitutively expressed each GFP-tagged EML4 deletion mutant were transfected with EML4 siRNA#2. After 72 h, the cells were fixed and immunostained for NUDC and GFP (Scale bar = 5 μm).
Discussion
EML4 has attracted many researchers since the discovery of the EML4-ALK (anaplastic lymphoma kinase) fusion oncogene for non-small cell lung cancer in 2007.26 The N-terminus of EML4 mediates the dimerization of the EML4-ALK fusion protein, which subsequently promotes the catalytic activation of the kinase for cancer progression.27 Some inhibitors for EML4-ALK are already in clinical trials, and extensive studies have been performed to elucidate the function of this fusion protein.28 However, the physiological functions of EML4 remain unclear. In this report, we showed that EML4 was required for mitotic spindle organization and for microtubule-kinetochore attachment. The depletion of EML4 by siRNAs induced cells with disorganized mitotic spindles and uncongressed chromosomes during metaphase. Uncongressed chromosomes are often observed when microtubule-kinetochore attachment is disrupted. Consistently, the stable organization of k-fibers decreased, and the side-on attachment of kinetochores to microtubules increased. These results show that EML4 has a crucial role in mitosis by contributing to mitotic spindle organization and to microtubule-kinetochore attachment.
We found that EML4 associates with NUDC and is required for the localization of NUDC to the mitotic spindle. NUDC is a highly conserved gene in a wide range of species, including filamentous fungi, plants, invertebrates and vertebrates.29 NUDC was first identified as a necessary gene for the migration of the nucleus in the filamentous fungus Aspergillus nidulans, and later studies revealed that mammalian NUDC associated with the dynein/dynactin complex and was essential for neurogenesis and for neuronal migration.30-33 Recent studies also demonstrated that NUDC is critical for cytokinetic progression. NUDC depletion induces multinuclear cells and defects in spindle organization.23 PLK1 is a mitotic kinase that regulates multiple steps of cell division.34,35 The phosphorylation of NUDC by PLK1 is essential for the end-on attachment of microtubules and kinetochores.23 In addition, NUDC is required for the accumulation of PLK1 at the kinetochore to promote chromosome congression.24 These results clearly show that NUDC is critical for spindle formation and for microtubule-kinetochore attachment. It is likely that the defect in microtubule-kinetochore attachment and spindle organization induced by EML4 knockdown results from the disruption of NUDC localization to the spindle.
All the EML family proteins contain WD40 domains in the C-terminus. WD40 domains have a β-propeller structure and mediate protein-protein interactions for many biological functions.36 Our immunoprecipitation analysis demonstrated that the association of EML4 with NUDC was dependent on the WD40 domains in EML4. Consistent with our finding, NUDC has been shown to associate with the WD40 domains of LIS1.37 Similar to NUDC, LIS1 associates with the dynein/dynactin complex and regulates nuclear motility and neuronal movement.31,38 Interestingly, a previous proteomics analysis showed that NUDC was in complex with CDC20, which is a critical regulator of mitosis and which has multiple WD40 domains.39 These results suggest that NUDC may associate with WD40 domain-containing proteins other than EML4 to regulate mitosis.
NUDCL and NUDCL2 are homologs of NUDC in mammals, and both proteins are thought to have specific roles in mitosis. NUDCL is phosphorylated during mitosis, and its expression is regulated during cell cycle progression.40 NUDCL is localized to the centrosomes and to the midbody, and the depletion of this protein induces multiple mitotic defects.41 NUDCL2 is localized to the centrosome and kinetochore during mitosis. Although both proteins can associate with LIS1 and the dynein/dynactin complex, the exact mechanisms by which NUDCL and NUDCL2 accumulate to specific sites during mitosis remain unknown.42 NUDCL and NUDCL2 appear to interact with proteins with WD40 domains because both proteins can interact with LIS1 therefore, EML family proteins may interact with NUDCL and NUDCL2 for the proper localization of both proteins.40,42 Further studies elucidating the association between EML and NUDC family proteins may reveal interesting features of the molecular mechanisms of mitosis.
Materials and Methods
Cells, antibodies, and chemicals
HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Wako, Osaka, Japan) supplemented with 10% fetal bovine serum (FBS, Equitec, Hendra, Australia). The following antibodies were used: anti-α-tubulin (T6199) and anti-β-actin (A5411) antibodies, which were purchased from Sigma-Aldrich (St. Louis, MO, USA); anti-EML4 antibodies (2428 and A310-675A), which were purchased from Cell Signaling (Beverly, MA, USA) and Bethyl Laboratories (Montgomery, TX, USA); anti-NUDC antibody (sc-135366), which was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA); anti-MAD2 antibody (610678), which was purchased from BD Biosciences (San Jose, CA, USA); anti-Flag antibody (018-22381), which was purchased from Wako (Osaka, Japan); anti-GFP antibodies (75-131 and 598), which were purchased from NeuroMab (Davis, CA, USA) and Medical and biological laboratories (MBL, Nagoya, Japan); anti-Hec1 antibody (ab3613), which was purchased from Abcam (Cambridge, UK); and FITC-conjugated anti-CREST antibody (15-234-0001), which was purchased from Antibodies Inc. (Davis, CA, USA). Anti-NUDC antibody, which was used for immunofluorescence and immunoblot analysis, was kindly provided by Dr. Yu-Lee (Baylor College of Medicine, Houston, Texas, USA). Nocodazole was purchased from Sigma-Aldrich (St. Louis, MO, USA).
DNA constructs
Human EML4 cDNA and NUDC cDNA were amplified by PCR from a HeLa cDNA library. Full-length EML4 and NUDC were cloned into a pQCXIP retrovirus vector with a Flag, GFP or HA tag on the N-terminus (Clontech, Mountain View, CA, USA). The deletion constructs for EML4 and NUDC were generated by PCR.
siRNA transfection
The sequences of the siRNAs that were used to suppress EML4 expression were 5′- CCAAAUGGCUGCAAACUAATT-3′ (EML4 siRNA#1), 5′-CCAGUGGAUGUCCAGACAUTT-3′ (EML4 siRNA#2). EML4 siRNA#2 targeted the 3′ untranslated region (UTR) of EML4 mRNA. The sequence of the MAD2 siRNA was 5′-GCUUGUAACUACUGAUCUUTT-3′, and the sequences of the NUDC siRNAs were 5′-CCAACUUCAGACGAACAGAAG-3′ (NUDC siRNA#1) and 5′-CCUACUGUUACACAUUAAAAC-3′ (NUDC siRNA#2). The sequence of the control siRNA targeting luciferase was 5′-CUUACGCUGAGUACUUCGATT-3′. All the siRNAs were obtained from Hokkaido System Science (Sapporo, Japan). HeLa cells were transfected with 40 nM siRNA using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA).
Live-cell imaging
HeLa cells that were plated on 35 mm glass-bottom dishes (Iwaki, Tokyo, Japan) were transfected with siRNAs using Lipofectamine RNAiMAX (Invitrogen Carlsbad, CA, USA). At 24 h after transfection, the cells were monitored for 48 h using a time-lapse microscope system (LCV110, Olympus, Tokyo, Japan) equipped with a Retiga EXi camera (QImaging, Tokyo, Japan). Images were analyzed using the MetaMorph Imaging System (Universal Imaging, Silicon Valley, CA, USA).
Generation of stable cell lines
HeLa cells that constitutively expressed each protein were established by retroviral infection. 293T cells were transfected with the pQCXIP retroviral vector that encoded each cDNA in combination with the pVPack-GP and pVPack-Ampho vectors (Stratagene, La Jolla, CA, USA) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). At 48 h after transfection, the supernatants were added to the cells with 2 μg/ml polybrene (Sigma-Aldrich, St. Louis, MO, USA), and infected cells were selected with 1 μg/ml puromycin for 3 days.
Immunofluorescence analysis
The cells were grown on glass coverslips coated with fibronectin, fixed with cold methanol/acetone (1:1) or 4% paraformaldehyde, and blocked with phosphate-buffered saline (PBS) containing 7% FBS for 30 min. Then, the cells were incubated with primary antibodies for 1 h, washed with PBS, and incubated with Alexa Fluor 488- or Alexa Fluor 594-labeled secondary antibodies (Invitrogen, Carlsbad, CA, USA) for 1 h. Images were acquired using a FV1000 laser scanning confocal microscope (Olympus, Tokyo, Japan).
Immunoprecipitation
The cells were washed twice with cold PBS, lysed in lysis buffer (35 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 0.1% NP-40) for 15 min on ice and centrifuged at 15,000 rpm for 20 min to obtain clear cell lysates. Then, the cell lysates were incubated with the indicated primary antibodies coupled to protein A-agarose beads (Thermo Scientific, Waltham, MA, USA) or Flag beads (Wako, Osaka, Japan) at 4°C for 3 h or overnight. The beads were washed 3 times with lysis buffer and suspended in sample buffer.
Mass spectrometry analysis
Cells that constitutively expressed Flag-EML4 were generated by retrovirus infection. The Flag-EML4-expressing cells were lysed and immunoprecipitated with an anti-Flag antibody and immunoprecipitates were mixed with a Flag peptide to elute immunoprecipitated proteins. The eluted proteins were digested with trypsin and subjected to mass spectrometry analysis using the LC-MS/MS system (Paradigm MS4, Michrom Bioresources, Sacramento, CA; HTS-PAL, CTC Analytics AG, Zwingen, Swiss; LTQ Orbitrap XL, Thermo Scientific). The proteins were identified using the Mascot software package (Matrix Science, London, UK).
Cold treatment
HeLa cells were grown on glass coverslips and transfected with EML4 siRNA. Sixty hours later, the cells were washed twice with cold medium, placed on ice for 10 min, fixed in 4% paraformaldehyde for 7 min, incubated with 0.3% Triton X-100 in PBS for 5 min and blocked with 7% FBS in PBS for 30 min. The cells were immunostained with anti-α-tubulin antibody and FITC-conjugated anti-CREST antibody for 1 h and then analyzed using a FV1000 laser scanning confocal microscope (Olympus, Tokyo, Japan). Then, the number of cells with stable k-fibers was counted.
Monopolar cell assessment by monastrol treatment
HeLa cells that were plated on glass coverslips were transfected with EML4 siRNA, and 24 h later, the cells were treated with the Eg5 inhibitor monastrol at 100 μM for 14 h. Next, the cells were fixed with ice-cold methanol/acetone (1:1) for 10 min and stained with anti-α-tubulin antibody and FITC-conjugated anti-CREST antibody. Last, the cells were analyzed using a confocal microscope.
Statistical analysis
Statistical analyses were done by one way or 2 way analysis of variance (ANOVA) using GraphPad Prism 5. P value < 0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank the members of the Division of Cancer Biology for technical assistance and Dr. Yu-Lee (Baylor College of Medicine, Houston, Texas, USA) for anti-NUDC antibodies. We gratefully acknowledge Mr. Kentaro Taki (Division for Medical Research Engineering, Nagoya University Graduate School of Medicine) for assistance in mass spectrometry analysis.
Funding
This research was funded by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (25650063 and 2306 (Nanomedicine Molecular Science) to TS).
SUPPLEMENTAL MATERIAL
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Fang X. and Zhang P.. Aneuploidy and tumorigenesis. Semin Cell Dev Biol 2011; 22:595-601; PMID:21392584; http://dx.doi.org/ 10.1016/j.semcdb.2011.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet 2012; 13:189-203; PMID:22269907 [DOI] [PubMed] [Google Scholar]
- 3.Helmke KJ, Heald R, Wilbur JD. Interplay between spindle architecture and function. Int Rev Cell Mol Biol 2013; 306:83-125; PMID:24016524; http://dx.doi.org/ 10.1016/B978-0-12-407694-5.00003-1 [DOI] [PubMed] [Google Scholar]
- 4.Rieder CL. Kinetochore fiber formation in animal somatic cells: dueling mechanisms come to a draw. Chromosoma 2005; 114:310-8; PMID:16270218; http://dx.doi.org/ 10.1007/s00412-005-0028-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kops GJ, Shah JV. Connecting up and clearing out: how kinetochore attachment silences the spindle assembly checkpoint. Chromosoma 2012; 121:509-25; PMID:22782189; http://dx.doi.org/ 10.1007/s00412-012-0378-5 [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Jin F, Higgins R, McKnight K. The current view for the silencing of the spindle assembly checkpoint. Cell Cycle 2014; 13:1694-701; PMID:24776751; http://dx.doi.org/ 10.4161/cc.29027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douglas ME, Mishima M. Still entangled: assembly of the central spindle by multiple microtubule modulators. Semin Cell Dev Biol 2010; 21:899-908; PMID:20732438; http://dx.doi.org/ 10.1016/j.semcdb.2010.08.005 [DOI] [PubMed] [Google Scholar]
- 8.White EA, Glotzer M. Centralspindlin: at the heart of cytokinesis. Cytoskeleton (Hoboken) 2012; 69:882-92; PMID:22927365; http://dx.doi.org/ 10.1002/cm.21065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elad N, Abramovitch S, Sabanay H, Medalia O. Microtubule organization in the final stages of cytokinesis as revealed by cryo-electron tomography. J Cell Sci 2011; 124:207-15; PMID:21187346; http://dx.doi.org/ 10.1242/jcs.073486 [DOI] [PubMed] [Google Scholar]
- 10.Suprenant KA, Dean K, McKee J, Hake S. EMAP, an echinoderm microtubule-associated protein found in microtubule-ribosome complexes. J Cell Sci 1993; 104:445-50; PMID:9867489 [DOI] [PubMed] [Google Scholar]
- 11.Li Q, Suprenant KA. Molecular characterization of the 77-kDa echinoderm microtubule-associated protein. Homology to the beta-transducin family. J Biol Chem 1994; 269:31777-84; PMID:7989351 [PubMed] [Google Scholar]
- 12.Suprenant KA, Tuxhorn JA, Daggett MA, Ahrens DP, Hostetler A, Palange JM, VanWinkle CE, Livingston BT. Conservation of the WD-repeat, microtubule-binding protein,EMAP, in sea urchins, humans, and the nematode C. elegans. Dev Genes Evol 2000; 210:2-10; PMID:10603080; http://dx.doi.org/ 10.1007/PL00008183 [DOI] [PubMed] [Google Scholar]
- 13.Brisch E, Daggett MA, Suprenant KA. Cell cycle-dependent phosphorylation of the 77 kDa echinoderm microtubule-associated protein (EMAP) in vivo and association with the p34cdc2 kinase. J Cell Sci 1996; 109:2885-93; PMID:9013336 [DOI] [PubMed] [Google Scholar]
- 14.Hamill DR, Howell B, Cassimeris L, Suprenant KA. Purification of a WD repeat protein, EMAP, that promotes microtubule dynamics through an inhibition of rescue. J Biol Chem 1998; 273:9285-91; PMID:9535922; http://dx.doi.org/ 10.1074/jbc.273.15.9285 [DOI] [PubMed] [Google Scholar]
- 15.Eichenmüller B, Ahrens DP, Li Q, Suprenant KA. Saturable binding of the echinoderm microtubule-associated protein (EMAP) on microtubules, but not filamentous actin or vimentin filaments. Cell Motil Cytoskeleton 2001; 50:161-72; PMID:11807937; http://dx.doi.org/ 10.1002/cm.10002 [DOI] [PubMed] [Google Scholar]
- 16.Eichenmuller B, Everley P, Palange J, Lepley D, Suprenant KA. The human EMAP-like protein-70 (ELP70) is a microtubule destabilizer that localizes to the mitotic apparatus. J Biol Chem 2002; 277:1301-9; PMID:11694528; http://dx.doi.org/ 10.1074/jbc.M106628200 [DOI] [PubMed] [Google Scholar]
- 17.O'Connor V, Houtman SH, De Zeeuw CI, Bliss TV, French PJ. Eml5, a novel WD40 domain protein expressed in rat brain. Gene 2004; 336:127-37; PMID:15225882; http://dx.doi.org/ 10.1016/j.gene.2004.04.012 [DOI] [PubMed] [Google Scholar]
- 18.Pollmann M, Parwaresch R, Adam-Klages S, Kruse ML, Buck F, Heidebrecht HJ. Human EML4, a novel member of the EMAP family, is essential for microtubule formation. Exp Cell Res 2006; 312:3241-51; PMID:16890222; http://dx.doi.org/ 10.1016/j.yexcr.2006.06.035 [DOI] [PubMed] [Google Scholar]
- 19.Tegha-Dunghu J, Neumann B, Reber S, Krause R, Erfle H, Walter T, Held M, Rogers P, Hupfeld K, Ruppert T, et al.. EML3 is a nuclear microtubule-binding protein required for the correct alignment of chromosomes in metaphase. J Cell Sci 2008; 121:1718-26; PMID:18445686; http://dx.doi.org/ 10.1242/jcs.019174 [DOI] [PubMed] [Google Scholar]
- 20.Hueston JL, Herren GP, Cueva JG, Buechner M, Lundquist EA, Goodman MB, Suprenant KA. The C. elegans EMAP-like protein, ELP-1 is required for touch sensation and associates with microtubules and adhesion complexes. BMC Dev Biol 2008; 8:110; PMID: 19014691; http://dx.doi.org/ 10.1186/1471-213X-8-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Euteneuer U, McIntosh JR. Structural polarity of kinetochore microtubules in PtK1 cells. J Cell Biol. 1981; 89:338-45; PMID:7251657; http://dx.doi.org/ 10.1083/jcb.89.2.338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shrestha RL, Draviam VM. Lateral to end-on conversion of chromosome-microtubule attachment requires kinesins CENP-E and MCAK. Curr Biol 2013; 23:1514-26; PMID:23891108; http://dx.doi.org/ 10.1016/j.cub.2013.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aumais JP, Williams SN, Luo W, Nishino M, Caldwell KA, Caldwell GA, Lin SH, Yu-Lee LY. Role for NudC, a dynein-associated nuclear movement protein, in mitosis and cytokinesis. J Cell Sci 2003; 116:1991-2003; PMID:12679384; http://dx.doi.org/ 10.1242/jcs.00412 [DOI] [PubMed] [Google Scholar]
- 24.Zhou T, Aumais JP, Liu X, Yu-Lee LY, Erikson RL. A role for Plk1 phosphorylation of NudC in cytokinesis. Dev Cell 2003; 5:127-38; PMID:12852857; http://dx.doi.org/ 10.1016/S1534-5807(03)00186-2 [DOI] [PubMed] [Google Scholar]
- 25.Nishino M, Kurasawa Y, Evans R, Lin SH, Brinkley BR, Yu-LeeL Y. NudC is required for Plk1 targeting to the kinetochore and chromosome congression. Curr Biol 2006; 16:1414-21; PMID:16860740; http://dx.doi.org/ 10.1016/j.cub.2006.05.052 [DOI] [PubMed] [Google Scholar]
- 26.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al.. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007; 448:561-6; PMID:17625570; http://dx.doi.org/ 10.1038/nature05945 [DOI] [PubMed] [Google Scholar]
- 27.Mano H. Non-solid oncogenes in solid tumors: EML4-ALK fusion genes in lung cancer. Cancer Sci 2008; 99:2349-55; PMID:19032370; http://dx.doi.org/ 10.1111/j.1349-7006.2008.00972.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esfahani K, Agulnik JS, Cohen V. A Systemic Review of Resistance Mechanisms and Ongoing Clinical Trials in ALK-Rearranged Non-Small Cell Lung Cancer. Front Oncol 2014; 4:174; PMID: 25101240; http://dx.doi.org/ 10.3389/fonc.2014.00174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riera J, Lazo PS. The mammalian NudC-like genes: a family with functions other than regulating nuclear distribution. Cell Mol Life Sci 2009; 66:2383-90; PMID:19381437; http://dx.doi.org/ 10.1007/s00018-009-0025-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osmani AH, Osmani SA, Morris NR. The molecular cloning and identification of a gene product specifically required for nuclear movement in Aspergillus nidulans. J Cell Biol 1990; 111:543-51; PMID:2199460; http://dx.doi.org/ 10.1083/jcb.111.2.543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aumais JP, Tunstead JR, McNeil RS, Schaar BT, McConnell SK, Lin SH, Clark GD, Yu-Lee LY. NudC associates with Lis1 and the dynein motor at the leading pole of neurons. J Neurosci 2001; 21: RC187; PMID: 11734602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamada M, Toba S, Takitoh T, Yoshida Y, Mori D, Nakamura T, Iwane AH, Yanagida T, Imai H, Yu-Lee LY, et al.. mNUDC is required for plus-end-directed transport of cytoplasmic dynein and dynactins by kinesin-1. EMBO J 2010; 29:517-31; PMID:20019668; http://dx.doi.org/ 10.1038/emboj.2009.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cappello S, Monzo P, Vallee RB. NudC is required for interkinetic nuclear migration and neuronal migration during neocortical development. Dev Biol 2011; 357:326-35; PMID:21771589; http://dx.doi.org/ 10.1016/j.ydbio.2011.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petronczki M, Lénárt P, Peters JM. Polo on the Rise-from Mitotic Entry to Cytokinesis with Plk1. Dev Cell 2008; 14:646-59; PMID:18477449; http://dx.doi.org/ 10.1016/j.devcel.2008.04.014 [DOI] [PubMed] [Google Scholar]
- 35.Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol 2009; 10:265-75; PMID:19305416; http://dx.doi.org/ 10.1038/nrm2653 [DOI] [PubMed] [Google Scholar]
- 36.Stirnimann CU, Petsalaki E, Russell RB, Müller CW. WD40 proteins propel cellular networks. Trends Biochem Sci 2010; 35:565-74; PMID:20451393; http://dx.doi.org/ 10.1016/j.tibs.2010.04.003 [DOI] [PubMed] [Google Scholar]
- 37.Helmstaedt K, Laubinger K, Vosskuhl K, Bayram O, Busch S, Hoppert M, Valerius O, Seiler S, Braus GH. The nuclear migration protein NUDF/LIS1 forms a complex withNUDC and BNFA at spindle pole bodies. Eukaryot Cell 2008; 7:1041-52; PMID:18390647; http://dx.doi.org/ 10.1128/EC.00071-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morris SM, Albrecht U, Reiner O, Eichele G, Yu-Lee LY. The lissencephaly gene product Lis1, a protein involved in neuronal migration, interacts with a nuclear movement protein,NudC. Curr Biol 1998; 8:603-6; PMID:9601647; http://dx.doi.org/ 10.1016/S0960-9822(98)70232-5 [DOI] [PubMed] [Google Scholar]
- 39.Hutchins JR, Toyoda Y, Hegemann B, Poser I, Hériché JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, et al.. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 2010; 328:593-9; PMID:20360068; http://dx.doi.org/ 10.1126/science.1181348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou T, Zimmerman W, Liu X, Erikson RL. A mammalian NudC-like protein essential for dynein stability and cell viability. Proc Natl Acad Sci U S A 2006; 103:9039-44; PMID:16754861; http://dx.doi.org/ 10.1073/pnas.0602916103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai Y, Yang Y, Shen M, Zhou T. Inhibition of cytokinesis by overexpression of NudCL that is localized to the centrosome and midbody. Cell Res 2009; 19:1305-8; PMID:19806165; http://dx.doi.org/ 10.1038/cr.2009.118 [DOI] [PubMed] [Google Scholar]
- 42.Yang Y, Yan X, Cai Y, Lu Y, Si J, Zhou T. NudC-like protein 2 regulates the LIS1/dynein pathway by stabilizing LIS1 with Hsp90. Proc Natl Acad Sci U S A 2010; 107:3499-504; PMID:20133715; http://dx.doi.org/ 10.1073/pnas.0914307107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.