Abstract
Tourette syndrome is a heritable neurodevelopmental disorder whose pathophysiology remains unknown. Recent genome-wide association studies suggest that it is a polygenic disorder influenced by many genes of small effect. We tested whether these genes cluster in cellular function by applying gene-set analysis using expert curated sets of brain-expressed genes in the current largest available Tourette syndrome genome-wide association data set, involving 1285 cases and 4964 controls. The gene sets included specific synaptic, astrocytic, oligodendrocyte and microglial functions. We report association of Tourette syndrome with a set of genes involved in astrocyte function, specifically in astrocyte carbohydrate metabolism. This association is driven primarily by a subset of 33 genes involved in glycolysis and glutamate metabolism through which astrocytes support synaptic function. Our results indicate for the first time that the process of astrocyte-neuron metabolic coupling may be an important contributor to Tourette syndrome pathogenesis.
Introduction
Tourette syndrome is a childhood-onset neuropsychiatric disorder characterized by chronic, repetitive involuntary movements and vocalizations, that is, motor and vocal tics. Although genetic factors play an important role in the etiology of Tourette syndrome, and results from twin and family studies have indicated strong familiality,1 the underlying pathophysiology is still unclear.2 Identifying genetic factors and associated biological mechanisms would be a major step forward, and could provide putative hallmarks for treatment.
To date, only one Tourette syndrome genome-wide association study (GWAS) has been published.3 Their top signal was in the COL27A1 gene with P=1.85 × 10−6, and there were no genetic variants that reached genome-wide significance. In addition, candidate genes from earlier, smaller-scaled candidate gene studies were not replicated, suggesting that these genes are either not causally related to Tourette syndrome or are only important in specific subtypes of Tourette syndrome. A recent study demonstrated that Tourette syndrome is polygenic and likely influenced by hundreds, possibly thousands, of genetic variants with small effects, and that >75% of Tourette syndrome heritability is captured by common genetic variants included in GWAS chips.4 An important question that arises from this polygenic nature is whether these thousands of genes of small effect cluster across cellular function or whether they are distributed randomly across function. Gene-set analysis, which evaluates the combined effect of multiple genetic variants, has been proposed as an efficient method to test functional clustering by identifying sets of functionally related genes underlying polygenic disorders.5 In the present study, we applied gene-set analysis for Tourette syndrome using the current largest available Tourette syndrome GWAS data set to elucidate the genetic factors involved in Tourette syndrome. As Tourette syndrome is assumed to be a brain disorder, we restricted ourselves to cellular function related to genes expressed in the brain, and tested sets of genes involved in specific synaptic, astrocytic, oliogodendrocyte and microglial functions.
Materials and methods
Subjects and quality control
The gene-set analysis was performed on the raw GWAS genotype data as described in Scharf et al.3 Subject inclusion criteria required a Tourette syndrome Classification Study Group diagnosis of definite Tourette syndrome (a DSM-IV-TR diagnosis of Tourette syndrome plus tics observed by an experienced clinician),6 and available genomic DNA were extracted either from blood or cell lines. Exclusion criteria consisted of a history of intellectual disability, tardive tourettism or other known genetic, metabolic or acquired tic disorders. European ancestry controls were derived primarily from cohorts of previously genotyped, unselected population controls, as previously described.3
Principal components computed from the data were used to control for population stratification. After quality control, the full data set contained 1285 cases and 4964 controls, divided into three samples according to genetic ancestry: European ancestry, non-isolates (778 cases, 4414 controls) from North America and Europe; Ashkenazi Jewish (242 cases, 354 controls) from the US and Israel; and French Canadian (265 cases, 196 controls). Quality control was the same as for Scharf et al, except with more stringent SNP filters (removing SNPs with: MAF<0.01 or HWE P-value<1e-4 for the European non-isolate sample; MAF<0.05 or HWE P-value<1e-3 for the Ashkenazi/French Canadian isolate samples).
Genotyping and annotation
Genotyping was conducted on the Illumina Human610-Quadv1_B SNP array for the majority of the subjects and on the Illumina HumanCNV370-Duo_v1 for 148 cases. Annotation of SNPs to genes was based on NCBI human assembly build 37.3 and dbSNP release 135. SNPs were assigned to genes when they lay between the transcription start and stop sites, with no window around the gene.
Gene-set creation
Because of the neuropsychiatric nature of Tourette syndrome, the gene-set analysis focussed on brain cell-specific gene sets, which were taken from previously published, expert curated gene sets. A total of 96 gene sets containing 4666 different brain-expressed genes were used, divided into four cell-based groups representing synaptic (neuronal), astrocyte, oligodendrocyte and microglia function.
The synaptic gene sets were taken from Ruano.7 These were defined on the basis of assignment of subcellular function as determined by previous synaptic protein purification experiments and data mining for synaptic genes and gene function, where genes were considered ‘synaptic' on the basis of proteomic analysis of synaptic preparations.8, 9, 10, 11 This resulted in a subdivision into 17 functional synaptic gene sets, plus one additional gene set of otherwise unassigned synaptic genes.
Glial gene sets (oligodendrocyte, astrocyte and microglial sets) were taken from Goudriaan.12 Goudriaan et al conducted an in-depth literature study to select astrocyte, oligodendrocyte and microglia genes on the basis of microarray gene expression patterns. Specificity was further increased by removing overlap between the three glial cell types, as well as removing general neuronal genes. The resulting lists of cell-specific genes were then subdivided into gene sets using the Gene Ontology biological process annotations, resulting in 30 astrocytic, 29 oligodendrocytic and 19 microglial hierarchically organized gene sets.
Statistical analysis
The gene-set analysis was conducted using JAG.13 First, a self-contained test was performed for each gene set, testing for the evidence of association with Tourette syndrome, under the null hypothesis of no association. For gene sets found to be significant after correction for multiple testing, a competitive test was performed to test whether the observed association was stronger than expected by chance for gene sets of the same size. P-values were computed using at least 15 000 permutations for the self-contained tests, and 150 random matched gene sets (with at least 15 000 permutations each) for the competitive test. In addition, the impact of each gene on the gene-set association was assessed, by computing the change in association when removing that gene from the analysis.
Analyses were performed separately for each of the three ancestry groups described above. The resulting P-values were combined using Stouffer's Z-score method,14 weighted by the square root of the sample size. Bonferroni correction (and a significance threshold of α=0.05 for corrected P-values) was used within each of the four cell-type-based groups, to compensate for multiple testing.
Results
Gene-set analysis of the synaptic, oligodendrocytic and microglial gene sets uncovered no significant association with Tourette syndrome (Supplementary Tables S1–S3). However, within the astrocyte group, a single gene set, representing the astrocyte carbohydrate metabolism pathway, was found to be significantly associated with Tourette syndrome risk in the self-contained test (corrected P=0.04; Supplementary Table S4). The secondary competitive test was also significant (P=0.0067, based on 150 random matched gene sets).
A follow-up analysis was performed to determine whether the association signal of the astrocyte carbohydrate metabolism gene set might be concentrated within a subset of genes with more specific function. For this purpose, the 85 genes in the gene set were subjected to manual data mining based on published data. This resulted in further specification of this gene set into three specific subprocesses related to (i) astrocyte-neuron metabolic coupling (ANMC; 33 genes, coding for enzymes or transporters involved in glycolysis or glutamine metabolism), (ii) extracellular matrix (EM; 10 genes, coding for ECM proteins or proteins that modify ECM) and (iii) glycosylation (GS; 29 genes, coding for enzymes involved in biosynthesis or degradation of glycoproteins); the 15 remaining genes were combined into a fourth ‘miscellaneous' subset (see Supplementary Table S5). Gene-set analysis of these four subsets showed that the association was localized to the 33 genes comprising the ANMC gene set, with a corrected P-value of 0.011 for the self-contained test, and P=0.0067 for the competitive test (Table 1).
Table 1. Results for association with Tourette syndrome from gene-set analyses for four specific subgroups of the astrocyte carbohydrate metabolism gene set.
| Gene set | No. of genes | No. of SNPs | Corrected self. P | Competitive P |
|---|---|---|---|---|
| Astrocyte carbohydrate metabolism | 85 | 1200 | 0.0402 | 0.0067 |
| Astrocyte-neuron metabolic coupling | 33 | 276 | 0.0106 | 0.0067 |
| Extracellular matrix | 10 | 345 | 0.117 | — |
| Glycosylation | 29 | 385 | 1 | — |
| Miscellaneous | 15 | 306 | 1 | — |
Abbreviations: corrected self. P, P-value from the self-contained test corrected for multiple testing; competitive P, P-value from competitive test. Note that competitive tests were only conducted and interpreted for gene sets that survived multiple testing on the self-contained test.
We further assessed the effect of each of the 33 genes on the gene-set association (Table 2 and Supplementary Table S6). The results show that none of the individual genes would have survived correction for multiple testing, suggesting that the association of the ANMC gene set is not driven by a single gene but rather is due to the combined effect of multiple genes of similar function.
Table 2. Results for individual genes in astrocyte-neuron metabolic coupling gene set.
| Gene Symbol | No. of SNPs | Gene P-value | Impact |
|---|---|---|---|
| ME1 | 26 | 0.00858 | 1 |
| ALDH5A1 | 8 | 0.00992 | 0.429 |
| GBE1 | 20 | 0.103 | 0.29 |
| GALM | 12 | 0.0367 | 0.269 |
| PYGL | 7 | 0.057 | 0.224 |
| CPS1 | 29 | 0.143 | 0.151 |
| PFKFB3 | 49 | 0.196 | 0.0792 |
| PYGB | 4 | 0.181 | 0.0605 |
| IDH2 | 6 | 0.165 | 0.0596 |
| ENO1 | 3 | 0.196 | 0.0441 |
| PPP1R1A | 3 | 0.525 | 0.0305 |
| MDH2 | 2 | 0.159 | 0.0211 |
| CS | 1 | 0.402 | 0.0198 |
| PYGM | 1 | 0.0659 | 0.0137 |
| PGM3 | 3 | 0.354 | 0.0014 |
| PHKG1 | 1 | 0.497 | −0.00595 |
| SLC3A2 | 3 | 0.344 | −0.00598 |
| PFKFB4 | 4 | 0.474 | −0.00728 |
| KHK | 1 | 0.506 | −0.00737 |
| LDHB | 1 | 0.442 | −0.00749 |
| PCK2 | 2 | 0.381 | −0.00955 |
| SLC2A8 | 1 | 0.527 | −0.0105 |
| PGM2 | 12 | 0.291 | −0.0183 |
| GPT | 1 | 0.594 | −0.0234 |
| AKR1B1 | 1 | 0.312 | −0.0296 |
| NANS | 3 | 0.239 | −0.0405 |
| PDK4 | 7 | 0.486 | −0.0542 |
| OGDHL | 6 | 0.606 | −0.0691 |
| DHTKD1 | 5 | 0.722 | −0.0769 |
| PFKM | 10 | 0.478 | −0.128 |
| PGM1 | 15 | 0.498 | −0.156 |
| PC | 14 | 0.62 | −0.211 |
| AGL | 15 | 0.589 | −0.326 |
Gene P-values are not corrected for multiple testing. The impact reflects the decrease in gene-set significance if that gene is removed from the gene set (positive impact means the gene-set P-value increases if the gene is removed, negative impact that the gene-set P-value decreases).
Discussion
We set out to test the hypothesis that the many genes of small effect thought to underlie Tourette syndrome are clustered across cellular function. Despite the relative modest sample size, our gene-set analysis revealed a significant association between the astrocyte carbohydrate metabolism pathway and Tourette syndrome. Competitive testing showed that this gene set was more strongly associated to Tourette syndrome than expected for a gene set of that size. This association could be further narrowed down to the ANMC subprocess, and we showed the effect of this gene set was not because of an effect of a single gene, but was because of an overall, combined effect of many genetic variants of small effect. This is the first study to point to the involvement of ANMC function in Tourette syndrome, probably through altered glycogen and glutamate/GABA metabolism, and in line with previously hypothesized mechanisms underlying Tourette syndrome pathogenesis that involve perturbations in the balance between excitatory glutamatergic and inhibitory GABAergic transmission within regulatory cortico-striato-thalamocortical circuits.15, 16, 17
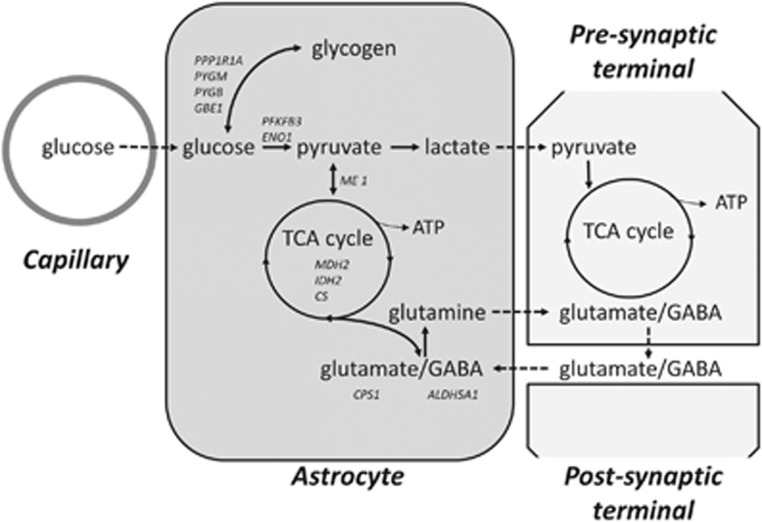
The ANMC gene set contains astrocyte-enriched genes involved in various energy metabolism processes that support synaptic function18 (Figure 1). First, whereas neurons have a low glycolytic rate, astrocytes actively take up glucose from the circulation, store it as glycogen and subsequently convert glycogen to lactate for release into neurons under neuronal command.18 The ANMC gene set contains GBE1, PGM3, PYGM and PYGB, coding for enzymes involved in glycogen storage; PPP1RA1, coding for a protein involved in hormonal control of glycogen metabolism; and PFKFB3 and ENO1, coding for glycolytic enzymes for the production of pyruvate and subsequently lactate.
Figure 1.
Schematic overview of the astrocyte-neuron metabolic coupling gene set, showing genes positively contributing to the gene-set association with Tourette syndrome. Genetic alterations in astrocyte-neuron metabolic coupling may have downstream effects on various neuronal energy metabolism processes, particularly at synapses: (1) glycolysis-dependent lactate release to the synapse where it is used for ATP generation and (2) glutamate (or GABA) uptake from the synaptic cleft by astrocytes where one part is converted to glutamine and returned to neurons for conversion back to glutamate (or GABA), and another part is used for production of pyruvate and lactate. See main text for further explanation.
Second, astrocytes take up glutamate (or to a lesser extent GABA) from the synaptic cleft using astrocyte-specific glutamate transporters. A small portion of this glutamate is used in the astrocyte TCA cycle for oxidative energy metabolism and for the production of pyruvate and lactate, in a manner proportional to extracellular glutamate concentration.19 The larger portion of glutamate is converted to glutamine and shuttled back to neurons for conversion into glutamate (or GABA), independent of extracellular glutamate concentrations and astrocyte energy status.20 The ANMC gene set also contains CPS1 and ALDH5A1, coding for enzymes involved in glutamine and GABA metabolism, respectively; the genes coding for TCA cycle enzymes MDH2, CS and IDH2; and for the key enzyme ME1, which links the TCA cycle with the glycolytic pathway. Interestingly, astrocyte glutamate uptake is known to drive glycolysis and subsequent shuttling of lactate to neurons.6
Tight regulation of neuronal energy supply by astrocytes in response to synaptic activity is crucial for proper neuronal function.18, 20 Thus, genetic alterations in glycolysis and glutamate metabolism can have profound influences on astrocyte modulation of synapse function. Such perturbations in the balance between excitatory glutamatergic and inhibitory GABAergic transmission within regulatory cortico-striato-thalamocortical circuits have long been hypothesized as a core defect in Tourette syndrome pathogenesis.15, 16, 17 Taken together, our findings highlight an often underestimated function of astrocytes in supporting synaptic function and suggest that abnormalities in this process may contribute to the etiology of Tourette syndrome.
Acknowledgments
We are grateful to all the patients with Tourette syndrome who generously agreed to participate in this study. Furthermore, the members of the Tourette Syndrome Association International Consortium for Genetics are deeply indebted to the Tourette Syndrome Association for their guidance and support. This work was supported by a grant from the David Judah Fund, NIH grants NS40024 to DLP, JMS and the Tourette Syndrome Association International Consortium for Genetics, NIH grant NS16648 and a grant from the Tourette Syndrome Association to DLP, American Recovery and Reinvestment Act (ARRA) Grants NS40024-07S1 to DLP/JMS and NS16648-29S1 to DLP, NIH grant NS037484 to NBF and NIH grant MH085057 to JMS. The Broad Institute Center for Genotyping and Analysis was supported by grant U54 RR020278 from the National Center for Research Resources. Support was also provided by the New Jersey Center for Tourette Syndrome & Associated Disorders (through New Jersey Department of Health and Senior Services: 08-1827-FS-N-0) to GAH and JAT. Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative (GEI) (U01 HG004422). SAGE is one of the GWAS funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for the collection of data sets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract ‘High throughput genotyping for studying the genetic contributions to human disease' (HHSN268200782096C). The data sets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000092.v1.p1 through dbGaP accession number phs000092.v1.p. Statistical analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org), which is financially supported by the Netherlands Scientific Organization (NWO 480-05-003). We gratefully acknowledge financial support by NWO via the Complexity project (645.000.003) and TOP ZonMW (40-00812-98-07-032). ABS and DP received funding from the HEALTH-2009-2.1.2-1 EU-FP7 Synsys project (grant number 242167).
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Drs Scharf and Mathews have received research support from the NIH and the Tourette Syndrome Association (TSA) on behalf of the TSA International Consortium for Genetics (TSAICG). Drs Scharf and Mathews have received honoraria and travel support from the TSA and are members of the TSA Medical Advisory Board (CAM) and Scientific Advisory Board (JMS).
None of the funding agencies for this project (NINDS, NIMH, the Tourette Syndrome Association and the David Judah Fund) had any influence or played any role in (a) the design or conduct of the study; (b) management, analysis or interpretation of the data; (c) preparation, review or approval of the manuscript. The remaining authors declare no conflict of interest.
Contributor Information
Tourette Syndrome Association International Consortium for Genetics:
J M Scharf, D L Pauls, D Yu, C Illmann, L Osiecki, B M Neale, C A Mathews, V I Reus, T L Lowe, N B Freimer, N J Cox, L K Davis, G A Rouleau, S Chouinard, Y Dion, S Girard, D C Cath, D Posthuma, J H Smit, P Heutink, R A King, T Fernandez, J F Leckman, P Sandor, C L Barr, W McMahon, G Lyon, M Leppert, J Morgan, R Weiss, M A Grados, H Singer, J Jankovic, J A Tischfield, and G A Heiman
Supplementary Material
References
- 1O'Rourke JA, Scharf JM, Yu D, Pauls DL: The genetics of Tourette syndrome: a review. J Psychosom Res 2009; 67: 533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Deng H, Gao K, Jankovic J: The genetics of Tourette syndrome. Nat Rev Neurol 2012; 8: 203–213. [DOI] [PubMed] [Google Scholar]
- 3Scharf JM, Yu D, Mathews CA, Neale BM et al: Genome-wide association study of Tourette's syndrome. Mol Psychiatry 2013; 18: 721–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Davis LK, Yu D, Keenan CL et al: Partitioning the heritability of Tourette syndrome and obsessive compulsive disorder reveals differences in genetic architecture. PLoS Genet 2013; 9: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Wang L, Jia P, Wolfinger RD, Chen X, Zhao Z: Gene set analysis of genome-wide association studies: methodological issues and perspectives. Genomics 2011; 98: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6APADiagnostic and statistical manual of mental disorders, 4th edn. Washington, DC, USA: American Psychiatric Association, 2000, Text revision (DSM-IV-TR). [Google Scholar]
- 7Ruano D, Abecasis GR, Glaser B et al: Functional gene group analysis reveals a role of synaptic heterotrimeric G proteins in cognitive ability. Am J Hum Genet 2010; 86: 113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Li K, Hornshaw MP, van Minnen J, Smalla KH, Gundelfinger ED, Smit AB: Organelle proteomics of rat synaptic proteins: correlation-profiling by isotope-coded affinity tagging in conjunction with liquid chromatography-tandem mass spectrometry to reveal post-synaptic density specific proteins. J Proteome Res 2005; 4: 725–733. [DOI] [PubMed] [Google Scholar]
- 9Li K, Hornshaw MP, Van der Schors RC et al: Proteomics analysis of rat brain postsynaptic density: Implications of the diverse protein functional groups for the integration of synaptic physiology. J Biol Chem 2004; 279: 987–1002. [DOI] [PubMed] [Google Scholar]
- 10Fernández E, Collins MO, Uren RT et al: Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol Syst Biol 2009; 5: 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Emes RD, Pocklington AJ, Anderson CN et al: Evolutionary expansion and anatomical specialization of synapse proteome complexity. Nat Neurosci 2008; 11: 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Goudriaan A, de Leeuw C, Ripke S et al: Specific glial functions contribute to schizophrenia susceptibility. Schizophrenia Bull 2014; 40: 925–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Lips ES, Cornelisse LN, Toonen RF et al: Functional gene group analysis identifies synaptic gene groups as risk factor for schizophrenia. Mol Psychiatry 2012; 17: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Whitlock MC: Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J Evol Biol 2005; 18: 1368–1373. [DOI] [PubMed] [Google Scholar]
- 15Albin RL, Mink WM: Recent advances in Tourette syndrome research. Trends Neurosci 2006; 29: 175–182. [DOI] [PubMed] [Google Scholar]
- 16Singer HS, Morris C, Grados M: Glutamatergic modulatory therapy for Tourette syndrome. Med Hypotheses 2010; 74: 862–867. [DOI] [PubMed] [Google Scholar]
- 17Adamczyk A, Gause CD, Sattler R et al: Genetic and functional studies of a missense variant in a glutamate transporter, SLC1A3, in Tourette syndrome. Psychiatr Genet 2011; 21: 90–97. [DOI] [PubMed] [Google Scholar]
- 18Bélanger M, Allaman I, Magistretti PJ: Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metab 2011; 14: 724–738. [DOI] [PubMed] [Google Scholar]
- 19Schousboe A, Bak LK, Madsen KK, Waagepetersen HS: Amino acid neurotransmitter synthesis and removal; in Kettenman H, Ransom BR (eds). New York, NY, USA: Neuroglia, 2013, pp 443–456. [Google Scholar]
- 20Barros LF: Metabolic signaling by lactate in the brain. Trends Neurosci 2013; 36: 396–404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.