Abstract
The dual-specificity tyrosine-phosphorylation-regulated kinase 1A (DYRK1A) gene, located on chromosome 21q22.13 within the Down syndrome critical region, has been implicated in syndromic intellectual disability associated with Down syndrome and autism. DYRK1A has a critical role in brain growth and development primarily by regulating cell proliferation, neurogenesis, neuronal plasticity and survival. Several patients have been reported with chromosome 21 aberrations such as partial monosomy, involving multiple genes including DYRK1A. In addition, seven other individuals have been described with chromosomal rearrangements, intragenic deletions or truncating mutations that disrupt specifically DYRK1A. Most of these patients have microcephaly and all have significant intellectual disability. In the present study, we report 10 unrelated individuals with DYRK1A-associated intellectual disability (ID) who display a recurrent pattern of clinical manifestations including primary or acquired microcephaly, ID ranging from mild to severe, speech delay or absence, seizures, autism, motor delay, deep-set eyes, poor feeding and poor weight gain. We identified unique truncating and non-synonymous mutations (three nonsense, four frameshift and two missense) in DYRK1A in nine patients and a large chromosomal deletion that encompassed DYRK1A in one patient. On the basis of increasing identification of mutations in DYRK1A, we suggest that this gene be considered potentially causative in patients presenting with ID, primary or acquired microcephaly, feeding problems and absent or delayed speech with or without seizures.
Introduction
Intellectual disability (ID) is clinically defined by the childhood onset (prior to age 18) of impaired cognitive functioning (intelligence quotient (IQ) below 70) and limited adaptive skills and behaviors. The causes of ID are heterogeneous and complex, with both environmental and genetic factors well recognized. Most known causes of severe ID are genetic, with about 25% of cases resulting from monogenic anomalies, copy-number variants and chromosomal abnormalities.1, 2 Efforts to identify causative genes implicated in monogenic forms of ID have produced multiple candidates, with a significant proportion of these genes (>100) mapping to the X chromosome.3, 4, 5 In recent years, the implementation of high-throughput technologies, such as next-generation sequencing (NGS), array comparative genomic hybridization and single-nucleotide polymorphism (SNP) arrays, has facilitated and expedited the discovery of numerous genes that cause or contribute to various forms of ID.6, 7
Out of the autosomal genes known to have a role in severe ID, the dual-specificity tyrosine-phosphorylation-regulated kinase (DYRK1A) gene (OMIM 600855) has been one of the most extensively studied.8, 9, 10, 11 Interest in DYRK1A originated because of its genomic localization within the Down syndrome critical region (DSCR) on chromosome 21q22, a chromosomal region that is presumed to be largely responsible for the neuropathologic abnormalities observed in Down syndrome.12 DYRK1A exhibits an extraordinarily high degree of protein sequence conservation in animals across all vertebrates,13 and animal models have been instrumental in determining the functions of the encoded proline-directed serine/threonine kinase. Some of the well-delineated roles of DYRK1A are in neuronal development, particularly in regulating neuronal proliferation, differentiation, plasticity and death.14, 15, 16, 17 Drosophila mutants lacking the DYRK1A ortholog minibrain (mnb) have reduced brain size and abnormalities of olfaction and vision.18 DYRK1A haploinsufficiency in mice results in a smaller brain size, developmental delay,19 decreased motor function,20 cognitive impairment and adverse stress coping behavior.21
Patients with partial monosomy 21 or rearrangements that involve DYRK1A and other contiguous genes have been described with phenotypes similar to the ones found in DYRK1A ortholog-deficient animal models including microcephaly, ID and developmental delay.22, 23, 24, 25, 26 Seven previously reported patients with chromosomal rearrangements, microdeletions or truncating mutations that disrupted specifically DYRK1A had ID, microcephaly, speech delay/absence and feeding dysfunction.22, 25, 27, 28, 29, 30, 31 Here, we describe an additional 10 unrelated patients with mutations involving DYRK1A, and further delineate a recurrent pattern of clinical features.
Subjects and methods
Most probands had been previously evaluated with extensive single gene testing, karyotyping, microarray analysis and/or metabolic testing.
Whole-exome sequencing was performed on peripheral blood DNA from patients #1 and #4. The Agilent SureSelect All Exome 50 Mb kit (Agilent Technologies, Santa Clara, CA, USA) was used to capture exomes, and sequences were generated using the Illumina HiSeq 2000 platform (San Diego, CA, USA). Reads were aligned to the February 2009 human reference sequence (GRCh37/hg19) using the NextGENe software (Softgenetics, LLC State College, PA), which was also used to call and annotate small insertions/deletions and SNPs. The resulting variants were filtered to exclude common variants (frequency >5%) present in dbSNP (v.137,32 1000 Genomes Phase I (v3.20101123)33), ESP-6500 (v.0.0.14; Exome Variant Server, NHLBI GO Exome Sequencing Project, Seattle, WA, USA; http://evs.gs.washington.edu/EVS/) and an in-house database using the Cartagenia Bench lab NGS platform (Cartagenia N.V., Leuven, Belgium). We prioritized non-synonymous variants with respect to in silico prediction scores obtained from PolyPhen-2,34 SIFT35 and Mutation Taster.36
DYRK1A mutations in patients #2, #3 and #10 were identified using targeted sequencing of coding regions of 217 genes associated with ID or autism. DNA library preparation, enrichment (SureSelect, Agilent Technologies, Santa Clara, CA, USA) and sequencing (HiSeq2500, Illumina) were performed as previously described.37 Pedigree concordance was checked using highly polymorphic microsatellite markers (PowerPlex 16 HS System, Promega, Madison, WI, USA).
For patient #5, a custom NGS target enrichment system (Agilent SureSelect, Agilent Technologies, Santa Clara, CA, USA) was used to enrich coding and flanking intronic regions of 92 autosomal and X-linked genes known for their association with autism spectrum disorders. A DNA library was prepared using the Fragment Library Preparation 5500 Series SOLiD Systems (Agilent Technologies, Santa Clara, CA, USA) and loaded onto the Applied Biosystems SOLiD 5500 × l platform (Life Technologies, Carlsbad, CA, USA), as per the manufacturer's instructions. Alignment of reads, variant calling, annotating and filtering strategies were performed similarly to those for patients #1 and #4.
Comparative genomic hybridization and SNP oligonucleotide array (CGH+SNP 180 K, Agilent Technologies, Santa Clara, CA, USA) were used to identify the deletion in patient #6. The deletion was confirmed by quantitative real-time polymerase chaine reaction (PCR) analysis using SYBR Green PCR Master Mix (Applied Biosystems, Life Technologies, Carlsbad, CA, USA) with two independent pairs of primers targeting DYRK1A. Relative quantification was performed on a 7900HT Sequence detection system (Applied Biosystems, Life Technologies). Two internal reference sequences were used for normalization.
For patients #7 and #8, mutations in DYRK1A were identified by targeted NGS of 24 genes included in a panel designed for the molecular diagnosis of primary microcephalies. NGS of libraries was performed on a MiSeq platform (Illumina) after multiplex PCR enrichment on a microfluidic Access Array support (Fluidigm, San Francisco, CA, USA). Genetic variation annotation was performed with the IntegraGen in-house pipeline (IntegraGen, Evry, France).
For patient #9, whole-exome capture and sequencing were performed at Integragen (Evry, France) using the SureSelect Human All Exon V5 kit (Agilent Technologies, Santa Clara, CA, USA). The resulting libraries were sequenced on a HiSeq 2000 (Illumina) according to the manufacturer's recommendations for paired-end 76 bp reads. Over 4.8 Gbs of mappable sequences were generated per individual, resulting in a depth of coverage >10X for more than 92.8% of RefSeq-coding exons. Reads were aligned to the human genome reference sequence (GRCh37/hg19) with the Burrows–Wheeler Aligner (BWA, v0.6.2),38 and potential duplicate paired-end reads were marked using Picard 1.77 (http://broadinstitute.github.io/picard/). The Genome Analysis Toolkit (GATK) 2.6–439 was used for base quality score recalibration, indel realignment and variant discovery (both single-nucleotide variants and indels). Variants were annotated with SeattleSeq SNP Annotation. Rare variants were identified by focusing on protein-altering and splice-site changes present at a frequency below 1% in dbSNP 138 and the NHLBI GO Exome Sequencing Project, and absent from 70 local exomes of unaffected individuals.
All detected mutations and structural variants are described in accordance with the HGVS (Human Genome Variation Society) nomenclature guidelines, and are reported in ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) as ‘pathogenic' or ‘likely pathogenic' (Table 2). Exon numbering was performed using systematic numbering (as opposed to legacy numbering), numbering the first exon of the respective DYRK1A transcript NM_001396.3 as 1, as recommended.40
Table 2. Types of DYRK1A mutations identified in the present study.
| Patient | Type of mutation | Nomenclaturea | Inheritance | ClinVar accession number |
|---|---|---|---|---|
| 1 | Nonsense | c.763C>T, p.Arg255a | De novo | SCV000196057 |
| 2 | Nonsense | c.613C>T, p.Arg205a | De novo | SCV000196058 |
| 3 | Frameshift | c.621_624delinsGAA, p.Glu208Asnfsa3 | De novo | SCV000196059 |
| 4 | Missense | c.1036T>C, p.Ser346Pro | De novo | SCV000196060 |
| 5 | Frameshift | c.945dupG, p.Gln316Alafsa24 | De novo | SCV000196061 |
| 6 | 1.7 Mb deletion | chr21.hg19:g.(?_38007970)_ (39747620_?)del | De novo | SCV000196062 |
| 7 | Nonsense | c.1309C>T, p.Arg437a | Parent samples not available | SCV000196063 |
| 8 | Frameshift | c.844dupA, p.Ser282Lysfsa6 | De novo | SCV000196064 |
| 9 | Missense | c.1763C>A, p.Thr588Asn | De novo | SCV000196065 |
| 10 | Frameshift | c.1232dupG, p.Arg413Thrfsa10 | De novo | SCV000196066 |
Using NCBI reference sequence NM_001396.3.
Results
The clinical findings in all 10 patients are summarized in Table 1 and more details are available in Supplementary Data S1. Seven of the patients were male. At last examination, patients' ages ranged from 2 to 32 years.
Table 1. Clinical features of patients with DYRK1A-associated ID.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7 | Patient 8 | Patient 9 | Patient 10 | Literature casesa | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | M | M | M | M | M | M | F | M | F | F | 3M+4F | 10M+9F |
| Age in yearsb | 11 | 15 | 4 | 3.5 | 6 | 3.5 | 6.5 | 3 | 32 | 5 | 2–20 | 2–32 |
| Low birth weight | o | + | o | o | o | o | o | o | + | + | 3/4 | 6/14 |
| Weight <3rd centileb | o | + | o | + | + | + | o | o | o | + | 4/4 | 9/14 |
| Height <3rd centileb | o | o | o | + | o | o | o | o | + | o | 4/4 | 6/14 |
| Microcephalyc | + | + | + | + | + | + | + | + | + | + | 6/7 | 16/17 |
| Intellectual disability | + | + | + | + | + | + | + | + | + | + | 7/7 | 17/17 |
| Autism | + | o | o | + | + | o | o | o | + | o | 4/4 | 8/14 |
| Seizures | + | o | o | + | + | + | + | + | + | + | 6/6 | 14/16 |
| Brain MRI | EV and HCC | o | Mildly EV | o | Mildly EV | o | TON | EV | N/A | EV and CA | HCC, CMA, EV | |
| Speech delay/absence | + | + | + | + | + | + | + | + | + | + | 7/7 | 17/17 |
| Feeding problem | + | + | + | + | + | + | + | + | o | + | 5/5 | 14/15 |
| Deep-set eyes | + | + | + | + | + | + | + | + | + | + | 2/4 | 12/14 |
| Hypermetropia/ myopia | + | + | + | + | + | + | o | N/A | N/A | o | 1/1 | 7/9 |
| Large/dysplastic ears | o | + | o | + | + | o | + | + | o | + | 4/5 | 10/15 |
| Pointed nasal tip | + | + | + | + | o | o | o | + | N/A | + | 2/4 | 8/13 |
| Long/flat philtrum | o | + | + | + | + | o | o | + | o | o | 2/2 | 7/12 |
| Thin upper lip | + | + | + | + | + | + | + | + | + | + | 3/5 | 13/15 |
| Micrognathia | + | + | + | + | + | + | o | + | + | + | 1/4 | 10/14 |
| Stereotypies | + | o | + | + | + | + | o | + | + | + | 4/5 | 12/15 |
| Clumsy/ incoordination | + | o | N/A | o | + | + | + | o | + | + | 3/3 | 9/12 |
| Prone to infections | + | N/A | o | + | + | o | + | o | o | o | 4/4 | 8/13 |
Abbreviations: CA, cerebellar atrophy; CMA, cortical and medullar atrophy; EV, enlarged ventricles; F, female; HCC, hypoplastic corpus collosum; M, male; MRI, magnetic resonance imaging; N/A, not available; o, absent at last examination; TON, thin optic nerves; +, present at last examination.
Patients with mutations affecting specifically DYRK1A.
At last clinical assessment.
At birth or last clinical assessment.
Some invariable features were reported in all 10 patients, including intellectual disability, absent/severely limited speech and primary or acquired microcephaly (defined as head circumference two or more standard deviations below the mean for age).41 Developmental milestones were globally delayed, with independent walking achieved after 18 months and initial words after 18 months in those who acquired speech. Intellectual disability was severe in most cases. Autism was diagnosed in four patients. Seizures were reported in eight patients.
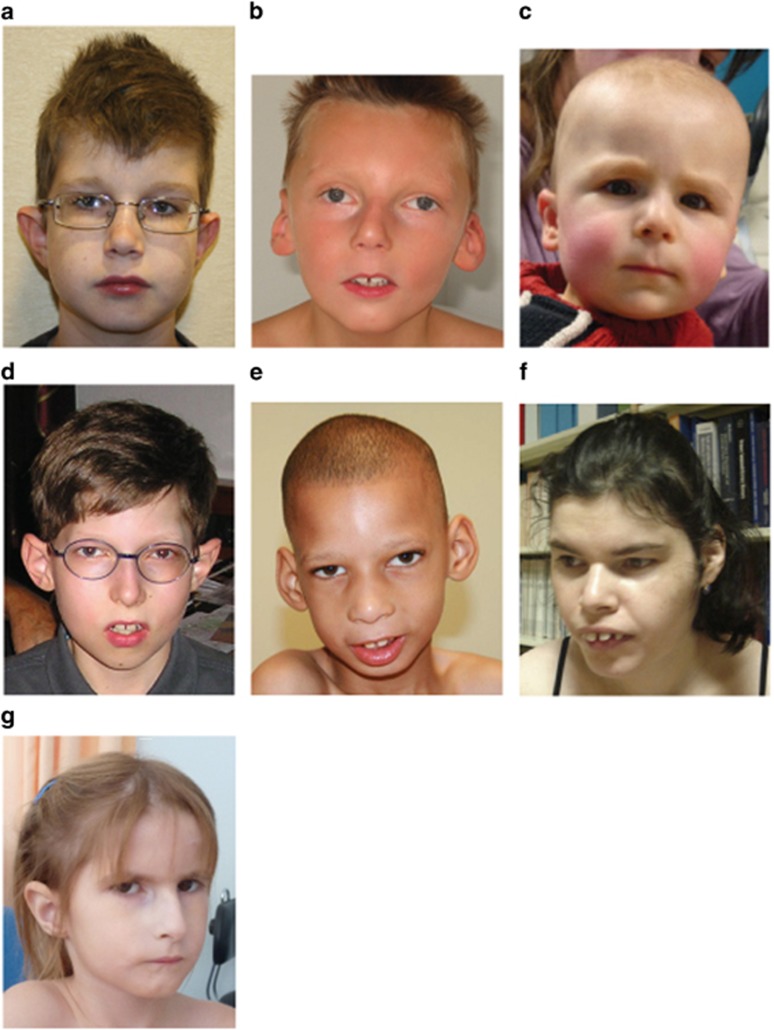
Microcephaly was congenital in 5/10 patients and developed postnatally in the other 5. However, patient #9 with initial congenital microcephaly was found to be normocephalic at her last physical assessment. Other craniofacial findings were frequent, including thin upper lip in 9/10 patients, micrognathia (7/10), large, low-set or malformed ears (6/10), long and flat philtrum (5/10), narrow and pointed nasal tip (5/10) and sparse hair (2/10). Representative facial photographs are shown in Figure 1. In addition to microcephaly, a general tendency of poor pre- and/or postnatal growth was observed in some patients, with either low birth weight (<−2 SD, three patients), poor weight gain (<−2 SD, five patients) and/or short stature (<−2 SD, two patients). Interestingly, a severe feeding disorder seems to be a prominent feature (9/10 patients), which would explain the observed problems with weight gain.
Figure 1.
Pictures of seven patients with DYRK1A-associated ID described in this study. Note the common clinical features among most of the patients, including microcephaly, deep-set eyes, protruding ears, long and flat philtrum with thin upper vermilion and micrognathia. The corresponding patient numbers are as follows: (a) #1, (b) #2, (c) #3, (d) #4, (e) #5, (f) #9 and (g) #10.
Vision abnormalities (hyperopia, myopia and exotropia) were found in 6/10 patients, with hyperopia being most common. Lastly, an apparent proneness to infections, generally of respiratory nature and limited to early childhood years, was reported in 4/10 patients.
For 9 of the 10 patients tested (#7's parental samples were unavailable), the mutations occurred de novo (see Table 2). Seven patients were found to harbor heterozygous frameshift or nonsense mutations, localized between exons 5 and 9, of DYRK1A (Table 2 and Figure 2). Two of our ten cases (patients #4 and #9) had heterozygous, non-synonymous, missense mutations. One of these (c.1036T>C, p.Ser346Pro) affects a highly conserved residue (including in invertebrates) located within the protein catalytic domain and is predicted to be deleterious by three prediction software programs (Mutation Taster: 0.99, Sift: 0 and Polyphen2 HumDiv: 1). The other missense mutation (c.1763C>A, p.Thr588Asn) affects a residue located in the more distal C-terminal portion. This residue is conserved across vertebrates and is predicted to be deleterious by two out of three prediction software programs (Mutation Taster: 0.99 and Polyphen2 HumDiv: 0.9; Sift: 0.21, ie tolerated).
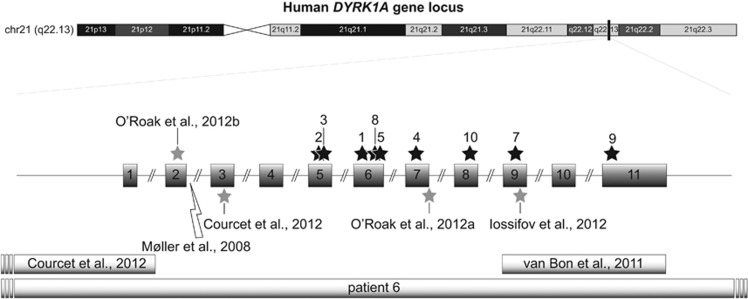
Figure 2.
Schematic depicting the approximate locations of all the truncating and missense mutations, deletions and chromosomal rearrangements affecting DYRK1A (NM_001396.3). Truncating and missense mutations (gray stars), translocation breakpoints (lightning symbols) and microdeletions (rectangles) of the previously published cases with DYRK1A syndrome. Black-colored stars represent the approximate location of mutation sites within the DYRK1A locus of patients presented in this paper. Idiogram of human chromosome 21 (GRCh37/hg19 assembly) was obtained from the UCSC genome browser website.52
Patient #6 from this study has a heterozygous 1.7 Mb interstitial deletion that includes DYRK1A and an additional 11 genes (SIM2, HLCS, RIPPLY3, PIGP, TTC3, DSCR3, KCNJ6, DSCR4, DSCR8, KCNJ15 and 3′ end of ERG; see Table 2). Heterozygous deletion of these other 11 genes did not appear to increase the severity of this patient's phenotype.
Overall, no specific genotype-to-phenotype correlation could be drawn from these patients. For example, patients with missense mutations or truncating mutations near the terminal end of the gene did not present with milder phenotypes as compared with patients harboring early-truncating DYRK1A mutations or large deletions encompassing several genes.
Discussion
DYRK1A has received attention in the past decade because of its association with early-onset cognitive impairment resulting from complete or partial duplication/deletion of chromosome 21. Here, we report 10 novel patients with mutations affecting DYRK1A, compare them with those previously described and highlight recurrent clinical manifestations in those patients.
Remarkably, all patients with DYRK1A-associated ID from this and previous studies present with similar clinical manifestations, regardless of the type of mutation affecting DYRK1A function. This study expands the spectrum of genetic aberrations affecting the DYRK1A locus that have been described to date, which now includes large deletions, chromosomal translocations and truncating and missense point mutations.
Given the locations of the observed missense mutations along with the associated patients' phenotypes (eg similar to patients carrying null mutations), it is likely that these are loss-of-function mutations that alter protein folding and/or the protein's kinase activity.42, 43 The truncating mutations described in this study presumably promote nonsense-mediated decay of the DYRK1A mRNA. All point mutations (non-synonymous and truncating mutations) affect residues widespread within the protein, located either in the DYRK1A-protein catalytic domain or the DYRK1A-C-terminal portion.
There are a number of reported cases with large deletions or chromosomal translocations disrupting the DYRK1A locus.22, 23, 24, 25, 26 Most of these patients exhibited a constellation of phenotypes with variable severity, which could be explained by the deletion of additional genes from 21q or imbalances in other regions of the genome. For example, two previously reported cases have chromosomal translocations with breakpoints within DYRK1A.22 Although these two patients shared many similar clinical features, such as microcephaly and feeding difficulties, one of them was more severely affected with more severe developmental delay, features attributed to a chromosomal breakpoint within the LRP1B gene and duplication of a region at Xp22 including the GEMIN8 gene. In our study, a microdeletion of DYRK1A and surrounding genes did not seem to influence the severity of the patient's (#6) phenotype, providing further evidence that DYRK1A is the primary gene within the DSCR that is responsible for the clinical features reported in patients with DYRK1A-associated ID.
Despite the phenotypic variability apparent in patients with DYRK1A-associated ID, there are five cardinal clinical features that are found almost universally among patients with DYRK1A mutations. These include moderate-to-severe ID (100%), absent or delayed speech (100%), primary or acquired microcephaly (94.1%), feeding issues (93.3%) and seizures (87.5%). The prevalent appearance of these clinical features supports the essential roles of DYRK1A in brain development and function. DYRK1A has been shown to positively regulate brain size by controlling neuronal precursor proliferation, neuronal differentiation and synaptogenesis and survival, particularly in the optic lobes, superior colliculus, central brain hemispheres and hippocampus.18, 19, 21, 44, 45 The reduced brain size may be partially explained by the dual role of DYRK1A in inhibiting the proliferation of neuronal progenitors via suppression of NOTCH signaling and Cyclin D1 while also promoting neurogenesis by stabilizing the cyclin-dependent kinase inhibitor p27(KIP1).46, 47 In addition, DYRK1A interacts with the microtubule and actin cytoskeletal networks to regulate neurite outgrowth and synaptogenesis.48, 49, 50 Through these mechanisms, haploinsuffciency of DYRK1A presumably leads to augmented neuronal precursor proliferation and apoptosis, decreased neurogenesis and abnormal neuritogenesis. A molecular mechanism that may explain the suppressed appetite in patients with DYRK1A-associated ID has recently been elucidated in flies and mice. Dyrk1A is part of a positive feedback loop that regulates neuropeptide Y levels and consequently food intake.51
Although many of the five principal clinical features found in patients with DYRK1A defects are also typically present in a number of other monogenic (eg Rett, Christianson and FOXG1 syndromes) and chromosomal disorders (eg 1p36 deletion syndrome), patients with DYRK1A-associated ID can be distinguished based on other commonly observed manifestations. Often these patients also present with thin upper lip vermilion border (86.7%), deep-set eyes with somewhat full/hooded upper lids (85.7%), stereotypies (80%), hyperopia/myopia (77.8%), clumsy or incoordinated movement (75%), and micrognathia (71.4%). The appearance around the eyes and noses of some cases is reminiscent of the velocardiofacial syndrome due to microdeletion at 22q11.2. (Figure 1, cases D, E and G)
In conclusion, this study describes nine unrelated patients with disease-causing point mutations in DYRK1A, and one with similar phenotypic features caused by a microdeletion involving this gene. Together with previously described cases, these findings produce a more detailed picture of the recurring phenotype and types of mutations in patients with DYRK1A-associated ID. Given the increasing frequency of identification of mutations in DYRK1A, we suggest that this gene be considered potentially causative in patients presenting with intellectual disability, primary or acquired microcephaly, feeding problems and absent or delayed speech with or without seizures.
Acknowledgments
The authors thank the parents for consenting to the clinical and molecular studies and presentation of these findings. The authors also thank the international consortium of clinical and laboratory personnel involved in carrying out these studies. Partial support was provided by the South Carolina Department of Disability and Special Needs and the Greenwood Genetic Center Foundation, and by grants from Fondation Jerome Lejeune, Agence de Biomedecine and CREGEMES to the Strasbourg team.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- 1van Bokhoven H: Genetic and epigenetic networks in intellectual disabilities. Annu Rev Genet 2011; 45: 81–104. [DOI] [PubMed] [Google Scholar]
- 2Srivastava AK, Schwartz CE: Intellectual disability and autism spectrum disorders: causal genes and molecular mechanisms. Neurosci Biobehav Rev 2014; 46: 161–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3Lubs HA, Stevenson RE, Schwartz CE: Fragile X and X-linked intellectual disability: four decades of discovery. Am J Hum Genet 2012; 90: 579–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Bassani S, Zapata J, Gerosa L, Moretto E, Murru L, Passafaro M: The neurobiology of X-linked intellectual disability. Neuroscientist 2013; 19: 541–552. [DOI] [PubMed] [Google Scholar]
- 5Piton A, Redin C, Mandel JL: XLID-causing mutations and associated genes challenged in light of data from large-scale human exome sequencing. Am J Hum Genet 2013; 93: 368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Gilissen C, Hehir-Kwa JY, Thung DT et al: Genome sequencing identifies major causes of severe intellectual disability. Nature 2014; 511: 344–347. [DOI] [PubMed] [Google Scholar]
- 7Henderson LB, Applegate CD, Wohler E, Sheridan MB, Hoover-Fong J, Batista DA: The impact of chromosomal microarray on clinical management: a retrospective analysis. Genet Med 2014; 16: 657–664. [DOI] [PubMed] [Google Scholar]
- 8Becker W, Sippl W: Activation, regulation, and inhibition of DYRK1A. FEBS J 2011; 278: 246–256. [DOI] [PubMed] [Google Scholar]
- 9Tejedor FJ, Hammerle B: MNB/DYRK1A as a multiple regulator of neuronal development. FEBS J 2011; 278: 223–235. [DOI] [PubMed] [Google Scholar]
- 10Wegiel J, Gong CX, Hwang YW: The role of DYRK1A in neurodegenerative diseases. FEBS J 2011; 278: 236–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Park J, Chung KC: New perspectives of Dyrk1A role in neurogenesis and neuropathologic features of Down syndrome. Exp Neurobiol 2013; 22: 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Guimera J, Casas C, Pucharcos C et al: A human homologue of Drosophila minibrain (MNB) is expressed in the neuronal regions affected in Down syndrome and maps to the critical region. Hum Mol Genet 1996; 5: 1305–1310. [DOI] [PubMed] [Google Scholar]
- 13Galceran J, de Graaf K, Tejedor FJ, Becker W: The MNB/DYRK1A protein kinase: genetic and biochemical properties. J Neural Transm Suppl 2003; 139–148. [DOI] [PubMed]
- 14Hammerle B, Elizalde C, Tejedor FJ: The spatio-temporal and subcellular expression of the candidate Down syndrome gene Mnb/Dyrk1A in the developing mouse brain suggests distinct sequential roles in neuronal development. Eur J Neurosci 2008; 27: 1061–1074. [DOI] [PubMed] [Google Scholar]
- 15Fernandez-Martinez J, Vela EM, Tora-Ponsioen M, Ocana OH, Nieto MA, Galceran J: Attenuation of Notch signalling by the Down-syndrome-associated kinase DYRK1A. J Cell Sci 2009; 122: 1574–1583. [DOI] [PubMed] [Google Scholar]
- 16Murakami N, Bolton DC, Kida E, Xie W, Hwang YW: Phosphorylation by Dyrk1A of clathrin coated vesicle-associated proteins: identification of the substrate proteins and the effects of phosphorylation. PLoS One 2012; 7: e34845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Barallobre MJ, Perier C, Bove J et al: DYRK1A promotes dopaminergic neuron survival in the developing brain and in a mouse model of Parkinson's disease. Cell Death Dis 2014; 5: e1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Tejedor F, Zhu XR, Kaltenbach E et al: Minibrain: a new protein kinase family involved in postembryonic neurogenesis in Drosophila. Neuron 1995; 14: 287–301. [DOI] [PubMed] [Google Scholar]
- 19Fotaki V, Dierssen M, Alcantara S et al: Dyrk1A haploinsufficiency affects viability and causes developmental delay and abnormal brain morphology in mice. Mol Cell Biol 2002; 22: 6636–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Fotaki V, Martinez De Lagran M, Estivill X, Arbones M, Dierssen M: Haploinsufficiency of Dyrk1A in mice leads to specific alterations in the development and regulation of motor activity. Behav Neurosci 2004; 118: 815–821. [DOI] [PubMed] [Google Scholar]
- 21Arque G, Fotaki V, Fernandez D, Martinez de Lagran M, Arbones ML, Dierssen M: Impaired spatial learning strategies and novel object recognition in mice haploinsufficient for the dual specificity tyrosine-regulated kinase-1A (Dyrk1A). PLoS One 2008; 3: e2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Moller RS, Kubart S, Hoeltzenbein M et al: Truncation of the Down syndrome candidate gene DYRK1A in two unrelated patients with microcephaly. Am J Hum Genet 2008; 82: 1165–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Oegema R, de Klein A, Verkerk AJ et al: Distinctive phenotypic abnormalities associated with submicroscopic 21q22 deletion including DYRK1A. Mol Syndromol 2010; 1: 113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Fujita H, Torii C, Kosaki R et al: Microdeletion of the Down syndrome critical region at 21q22. Am J Med Genet A 2010; 152A: 950–953. [DOI] [PubMed] [Google Scholar]
- 25Yamamoto T, Shimojima K, Nishizawa T, Matsuo M, Ito M, Imai K: Clinical manifestations of the deletion of Down syndrome critical region including DYRK1A and KCNJ6. Am J Med Genet A 2011; 155A: 113–119. [DOI] [PubMed] [Google Scholar]
- 26Valetto A, Orsini A, Bertini V et al: Molecular cytogenetic characterization of an interstitial deletion of chromosome 21 (21q22.13q22.3) in a patient with dysmorphic features, intellectual disability and severe generalized epilepsy. Eur J Med Genet 2012; 55: 362–366. [DOI] [PubMed] [Google Scholar]
- 27van Bon BW, Hoischen A, Hehir-Kwa J et al: Intragenic deletion in DYRK1A leads to mental retardation and primary microcephaly. Clin Genet 2011; 79: 296–299. [DOI] [PubMed] [Google Scholar]
- 28Courcet JB, Faivre L, Malzac P et al: The DYRK1A gene is a cause of syndromic intellectual disability with severe microcephaly and epilepsy. J Med Genet 2012; 49: 731–736. [DOI] [PubMed] [Google Scholar]
- 29O'Roak BJ, Vives L, Fu W et al: Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 2012; 338: 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30O'Roak BJ, Vives L, Girirajan S et al: Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 2012; 485: 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Iossifov I, Ronemus M, Levy D et al: De novo gene disruptions in children on the autistic spectrum. Neuron 2012; 74: 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Sherry ST, Ward MH, Kholodov M et al: dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001; 29: 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331000 Genomes Project Consortium1000 Genomes Project Consortium, Abecasis GR, Altshuler D et al: A map of human genome variation from population-scale sequencing. Nature 2010; 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Adzhubei IA, Schmidt S, Peshkin L et al: A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Kumar P, Henikoff S, Ng PC: Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 36Schwarz JM, Rodelsperger C, Schuelke M, Seelow D: MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010; 7: 575–576. [DOI] [PubMed] [Google Scholar]
- 37Redin C, Le Gras S, Mhamdi O et al: Targeted high-throughput sequencing for diagnosis of genetically heterogeneous diseases: efficient mutation detection in Bardet-Biedl and Alstrom syndromes. J Med Genet 2012; 49: 502–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Li H, Durbin R: Fast and accurate long-read alignment with Burrows-Wheeler Transform. Bioinformatics 2010; 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39McKenna A, Hanna M, Banks E et al: The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40Dalgleish R, Flicek P, Cunningham F et al: Locus Reference Genomic sequences: an improved basis for describing human DNA variants. Genome Med 2010; 2: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41Ashwal S, Michelson D, Plawner L, Dobyns WB: Quality Standards Subcommittee of the American Academy of Neurology and the Practice Committee of the Child Neurology Society: Practice parameter: evaluation of children and adolescents with recurrent headaches: report of the Quality Standards Subcommittee of the American Academy of Neurology and the Practice Committee of the Child Neurology Society. Neurology 2009; 73: 887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42Himpel S, Panzer P, Eirmbter K et al: Identification of the autophosphorylation sites and characterization of their effects in the protein kinase DYRK1A. Biochem J 2001; 359: 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43Wiechmann S, Czajkowska H, de Graaf K, Grotzinger J, Joost HG, Becker W: Unusual function of the activation loop in the protein kinase DYRK1A. Biochem Biophys Res Commun 2003; 302: 403–408. [DOI] [PubMed] [Google Scholar]
- 44Benavides-Piccione R, Dierssen M, Ballesteros-Yanez I et al: Alterations in the phenotype of neocortical pyramidal cells in the Dyrk1A+/− mouse. Neurobiol Dis 2005; 20: 115–122. [DOI] [PubMed] [Google Scholar]
- 45Guo X, Williams JG, Schug TT, Li X: DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J Biol Chem 2010; 285: 13223–13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46Hammerle B, Ulin E, Guimera J, Becker W, Guillemot F, Tejedor FJ: Transient expression of Mnb/Dyrk1a couples cell cycle exit and differentiation of neuronal precursors by inducing p27KIP1 expression and suppressing NOTCH signaling. Development 2011; 138: 2543–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47Soppa U, Schumacher J, Florencio Ortiz V, Pasqualon T, Tejedor FJ, Becker W: The Down syndrome-related protein kinase DYRK1A phosphorylates p27(Kip1) and Cyclin D1 and induces cell cycle exit and neuronal differentiation. Cell Cycle 2014; 13: 2084–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48Scales TM, Lin S, Kraus M, Goold RG, Gordon-Weeks PR: Nonprimed and DYRK1A-primed GSK3 beta-phosphorylation sites on MAP1B regulate microtubule dynamics in growing axons. J Cell Sci 2009; 122: 2424–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49Martinez de Lagran M, Benavides-Piccione R, Ballesteros-Yanez I et al: Dyrk1A influences neuronal morphogenesis through regulation of cytoskeletal dynamics in mammalian cortical neurons. Cereb Cortex 2012; 22: 2867–2877. [DOI] [PubMed] [Google Scholar]
- 50Dowjat K, Adayev T, Kaczmarski W, Wegiel J, Hwang YW: Gene dosage-dependent association of DYRK1A with the cytoskeleton in the brain and lymphocytes of down syndrome patients. J Neuropathol Exp Neurol 2012; 71: 1100–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51Hong SH, Lee KS, Kwak SJ et al: Minibrain/Dyrk1a regulates food intake through the Sir2-FOXO-sNPF/NPY pathway in Drosophila and mammals. PLoS Genet 2012; 8: e1002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52Kent WJ, Sugnet CW, Furey TS et al: The human genome browser at UCSC. Genome Res 2002; 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.