Abstract
Type 1 narcolepsy, an autoimmune disease affecting hypocretin (orexin) neurons, is strongly associated with HLA-DQB1*06:02. Among polymorphisms associated with the disease is single-nucleotide polymorphism rs2305795 (c.*638G>A) located within the P2RY11 gene. P2RY11 is in a region of synteny conserved in mammals and zebrafish containing PPAN, EIF3G and DNMT1 (DNA methyltransferase 1). As mutations in DNMT1 cause a rare dominant form of narcolepsy in association with deafness, cerebellar ataxia and dementia, we questioned whether the association with P2RY11 in sporadic narcolepsy could be secondary to linkage disequilibrium with DNMT1. Based on genome-wide association data from two cohorts of European and Chinese ancestry, we found that the narcolepsy association signal drops sharply between P2RY11/EIF3G and DNMT1, suggesting that the association with narcolepsy does not extend into the DNMT1 gene region. Interestingly, using transethnic mapping, we identified a novel single-nucleotide polymorphism rs3826784 (c.596–260A>G) in the EIF3G gene also associated with narcolepsy. The disease-associated allele increases EIF3G mRNA expression. EIF3G is located in the narcolepsy risk locus and EIF3G expression correlates with PPAN and P2RY11 expression. This suggests shared regulatory mechanisms that might be affected by the polymorphism and are of relevance to narcolepsy.
Introduction
Narcolepsy with cataplexy (Type 1 narcolepsy) affects 1 in 3000 individuals and is caused by the loss of around 70 000 hypocretin- (hcrt, also known as orexin) producing neurons in the hypothalamus. The disease is strongly associated with HLA-DQB1*06:02 and DQA1*01:02,1 the T-cell receptor alpha locus2 and polymorphisms in other immune-related genes.3 The loss of hcrt-producing cells is hypothesized to have an autoimmune basis.4
One of the genetic associations in narcolepsy is rs2305795, a polymorphism located in the purinergic receptor P2RY11.5, 6 The disease-associated allele of rs2305795 decreases P2RY11 expression and increases the sensitivity of CD8+ T cells to cell death induced by ATP. Interestingly, we also recently found that missense mutation in exon 21 of the DNA methyltransferase 1 (DNMT1) gene causes a rare hereditary form of narcolepsy-associated deafness, cerebellar ataxia and eventually dementia (ADCA-DN).7 As the P2RY11 and DNMT1 genes are located in close proximity (within 18 kb) on chromosome 19, we hypothesized that the two signals could be related at the pathophysiological level. In support of this hypothesis, Kornum et al.5 observed a correlation between P2RY11 and DNMT1 expression in peripheral blood mononuclear cells (PBMCs) of both patients and healthy controls. These findings made us question whether the association with P2RY11 in spontaneous narcolepsy could be secondary to linkage disequilibrium (LD) with DNMT1. In the original study, Kornum et al.5 fine-mapped the locus using seven single-nucleotide polymorphisms (SNPs) and reported an association between DNMT1 and narcolepsy in individuals of European ancestry but not Asians. In Asians, LD pattern pointed towards an association with P2RY11.5
One of the biggest challenges of association studies is to identify which genetic variant is causal within a given locus. To address this challenge, increased sample size, functional studies and transethnic mapping may be beneficial. In transethnic mapping, different LD patterns in the disease-associated locus can be explored to narrow down the associated genetic region. This approach has previously been shown to be very powerful in narcolepsy and other diseases.1, 8, 9 In this study, we reinvestigated SNPs in the P2RY11-DNMT1 region in narcolepsy across three different ethnic backgrounds. Based on genome-wide association study (GWAS) data available in Chinese and Caucasians, we observed that the narcolepsy association signal drops sharply between P2RY11 and DNMT1 in both cohorts. Based on LD patterns, we next selected four SNPs for genotyping in a cohort of African Americans. Following genotyping, we identified a novel SNP in EIF3G, rs3826784, best associated with narcolepsy across all three cohorts. EIF3G is located between P2RY11 and DNMT1. To further explore association between these different genes and narcolepsy, we finally also examined gene expression patterns in the entire region.
Materials and methods
Subjects
Narcolepsy with cataplexy cases were selected based on documented hypocretin deficiency or clearcut cataplexy and HLA-DQB1*06:02.2, 6 The initial European ancestry GWA data set was comprised of 807 cases and 1074 DQB1*06:02-positive controls: 415 cases and 753 controls were recruited from the United States and Canada; 392 cases and 321 controls were recruited from European centers. The Chinese GWA data set was comprised of 1078 cases and 1903 controls. Analysis of the GWA data (549 596 SNPs in Europeans and 603 382 SNPs in Chinese) was performed as described.2, 6 The African-American sample contained 1297 individuals (249 cases, 1048 controls). The controls were recruited from the National Institute of Mental Health (NIMH).
For the expression quantitative trait locus (eQTL) analysis, we used a large case–control study of major depression, for which RNA-sequencing data from whole blood together with genotype data were available for 922 individuals of European ancestry.
Imputation of Chinese and European cohorts
Imputation for the PPAN-DNMT1 region (from rs7250025 to rs2043305) was performed in the European and Chinese cohorts separately, using Beagle v.3.2.110 against reference genotypes from the 1000 genomes integrated data set. Chinese genotypes, originally obtained through the Affymetrix Axiom CHP array, were imputed against the CHB reference population. European ancestry genotypes from the Affymetrix 6.0 array were imputed using four European populations as a reference (CEU, TSI, GBR, IBS). In the paper by Kornum et al.,5 SNPs rs2305795 and rs12460842 were fine typed in many of the Caucasian samples. Further, rs2305795 was also retyped in the Chinese cohort6 and all the results were consistent with imputed results. The imputation quality score for rs3826784 was consistently above 0.8 and the overall association on pooled imputed genotypes is considered valid. Therefore, SNP markers with poor imputation quality score in either Chinese or Europeans (r2<0.8) were excluded from all further analysis.
The data of this study is submitted to the ClinVar Database (http://www.ncbi.nlm.nih.gov/clinvar/; accession numbers: SCV000196702−SCV000196705).
Fine mapping of the chromosome 19 locus, and replication of published SNPs
We used 1297 African-American individuals (249 cases, 1048 controls) for genotyping in the region of association. Based on LD and r2 data from the International HapMap Project (Hapmap.org), we observed that the LD pattern between the SNPs found from the GWAS was different in African Americans.
The selected SNPs were rs1551570: NM_020230.5:c.190−151C>T; rs12460842: NM_002566.4:c.−203A>G; rs2305795: NM_002566.4:c.*638G>A; and rs3826784: NM_003755.3:c.596−260A>G. For genotyping, we used predesigned Taqman SNP genotyping assays (Applied Biosystems, Carlsbad, CA, USA) to validate the observed LD pattern from the International HapMap project in a cohort of African-American narcolepsy patients and controls. Genotyping was performed at Stanford University. SNPs rs1551750, rs12460842 and rs3826784 are all located within introns of P2RY11 and EIF3G, respectively. SNP rs2305795 is located in the 3′-UTR of P2RY11. Genotype results were analyzed (allelic associations, LD calculations and analysis) and standard r2 plots were generated using Haploview 4.2.11
RNA sequencing and genotyping of 922 individuals of European ancestry
The complete procedure for whole blood processing, RNA sequencing and genotyping is described in Battle et al.12 Briefly, whole blood was collected in PAXGene tubes for RNA and in acid-citrate-dextrose tubes for DNA. Tubes were stored at −80 °C. Total RNA was extracted using the GLOBINclear Kit (Invitrogen, Carlsbad, CA, USA). Libraries for RNA sequencing was performed using Illumina TruSeq protocol. DNA was genotyped on the Illumina HumanOmni1-Quad BeadChip (Illumina, San Diego, CA, USA).
Statistical analysis
Genome-wide association analysis of the European and Chinese ancestry has been described previously.2, 6 Statistical analysis of genotyped and imputed genotype data were performed using the PLINK software package (v.1.07) (http://pngu.mgh.harvard.edu/purcell/plink/),13 and in Haploview 4.2.11
Associations within ethnic groups were performed with basic allelic χ2 statistic tests. For associations combining multiple ethnic groups, the Mantel–Haenszel test was used together with the Breslow Day test of homogeneity of the odds ratio (both implemented in Plink).
The details of the cis-eQTL analysis are described in Battle et al.12 Briefly, expression data was first normalized to remove the effect of technical factors, population structure and ‘hidden' factors (which have been shown to reduce the statistical power to identify cis-eQTLs14). Such normalization approaches can significantly increase power for detecting cis-eQTLs; however, in doing so, such approaches also remove broader expression patterns that may be driven by trans-regulation (as a trans-acting factor may affect the expression levels of multiple target genes) or reflect coregulation. As a result, previous work has shown that separate normalization of gene expression data may be appropriate for detecting cis versus trans associations.12, 14 Therefore, we quantified coexpression levels between genes using the normalization approach optimized for detecting trans-eQTLs. For each SNP, a P-value for association with gene expression level of each nearby gene (1 Mb from the transcription start site) was computed based on the Spearman's correlation coefficient. Significant associations were reported using 0.05 FDR (false discovery rate).
PPAN: NM_020230.4; P2RY11: NM_002566.4; EIF3G: NM_003755.3; DNMT1: NM_001130823.1.
Rs1551570: NM_020230.5:c.190−151C>T; rs12460842: NM_002566.4:c.−203A>G; rs2305795: NM_002566.4:c.*638G>A; rs3826784: NM_003755.3:c.596−260A>G.
SNPs reported in Table 1: rs1551570: NM_020230.5:c.190−151C>T; rs12460842: NM_002566.4:c.−203A>G; rs2010353: NM_002566.4:c.19+142G>T; rs3826784: NM_003755.3:c.596−260T>C; rs2305795: NM_002566.4:c.*638G>A; rs12462506: NM_002566.4:c.20−178T>G; rs3826785: NM_003755.3:c.595+327G>A; rs73011220: NM_002566.4:c.19+880G>A; rs112647895: NM_002566.4:c.19+446G>A; rs3745601: NM_002566.4:c.259G>A; rs2305789: NM_020230.5:c.291+50A>G; rs6511570: NM_001130823.1:c.2895−855C>T; rs2288349: NM_001130823.1:c.2721−45C>T; rs35693490: NM_001130823.1:c.2721−959G>A; rs8101626: NM_001130823.1:c.4773+383C>T; rs4804122: NC_000019.10:g.10131268C>T; 9 rs2114724: NM_001130823.1:c.1832+14A>G; rs2290684: NM_001130823.1:c.3394+34T>C; rs8112801: NM_001130823.1:c.3117−203A>C; rs11880553: NM_001130823.1:c.3117−660G>A; rs11880388: NM_001130823.1:c.3117−677C>T; rs11085587: NC_000019.10:g.10124423G>C; rs35374357: NC_000019.10:g.10129267T>C; rs7710: NM_003755.3:c.846T>C; rs870612: NC_000019.10:g.10120814A>C; rs4555265: NC_000019.10:g.10121508G>A; rs11667630: NM_003755.3:c.300+47G>T; rs2290687: NM_003755.3:c.240+23G>A; rs55752217: NM_020230.5:c.292−46C>T; rs2305792: NM_020230.5:c.902−10C>G; rs1037686: NM_002566.4:c.19+419T>A; rs3745600: NM_002566.4:c.237C>T; rs34484805: NM_002566.4:c.20−882A>G; rs7401: NM_002566.4:c.*363A>G; rs11666402: NM_020230.5:c.513+266T>C; rs10414661: NC_000019.10:g.10131561T>A; rs12462004: NC_000019.10:g.10127588T>G; rs10404209: NC_000019.10:g.10126751T>C; rs9305012: NM_001130823.1:c.2266−225A>G; rs10407514: NM_001130823.1:c.2895−449G>C; rs10418707: NM_001130823.1:c.2894+810C>T; rs12611113: NM_001130823.1:c.2721−212A>G; rs60565702: NM_001130823.1:c.2720+999C>T; rs72620548: NM_001130823.1:c.4490−306C>T; rs10854076: NM_001130823.1:c.4293+238C>G; rs4804489: NM_001130823.1:c.2265+418T>C; rs57366074: NM_001130823.1:c.2586+99G>T; rs10414537: NC_000019.10:g.10126048C>T.
Table 1. SNP markers located in the PPAN-P2RY11-EIF3G-DNMT1 locus and their association with Type 1 narcolepsy.
| European | Chinese | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Position (bp) | Major allele | Minor allele | Freq cases | Freq cont. | P-value | OR (95% CI) | Imp. quality | r2 | Major allele | Minor allele | Freq cases | Freq cont. | P-value | OR (95% CI) | Imp. quality | r2 |
| PPAN | Rs1551570 | 10 218 030 | T | C | 0.42 | 0.48 | 4.16E−04 | 0.79 (0.69–0.90) | 0.84 | 0.98 | T | C | 0.25 | 0.30 | 1.20E–05 | 0.77 (0.68–0.86) | 1.00 | 0.94 |
| P2RY11 | Rs12460842 | 10 222 195 | G | A | 0.42 | 0.48 | 3.45E–04 | 0.79 (0.69–0.90) | 0.86 | 0.99 | G | A | 0.25 | 0.30 | 1.47E–05 | 0.77 (0.68–0.87) | 0.98 | 0.94 |
| P2RY11 | Rs2010353 | 10 222 558 | T | G | 0.42 | 0.48 | 3.45E–04 | 0.79 (0.69–0.90) | 0.86 | 0.99 | T | G | 0.25 | 0.30 | 1.47E–05 | 0.77 (0.68–0.87) | 0.97 | 0.94 |
| EIF3G | Rs3826784 | 10 227 010 | G | A | 0.38 | 0.44 | 1.28E–04 | 0.77 (0.68–0.88) | 0.84 | 0.87 | G | A | 0.25 | 0.30 | 1.47E–05 | 0.77 (0.68–0.87) | 0.93 | 0.94 |
| P2RY11 | Rs2305795 | 10 226 052 | A | G | 0.42 | 0.48 | 2.31E–04 | 0.78 (0.69–0.89) | 0.89 | 1.00 | A | G | 0.26 | 0.31 | 5.99E–05 | 0.79 (0.70–0.88) | 0.93 | 1.00 |
| P2RY11 | Rs12462506 | 10 224 131 | T | G | 0.14 | 0.11 | 4.72E–03 | 1.33 (1.09–1.61) | 0.87 | 0.11 | T | G | 0.46 | 0.42 | 2.23E–03 | 1.18 (1.06–1.31) | 0.95 | 0.28 |
| EIF3G | Rs3826785 | 10 227 149 | C | T | 0.14 | 0.11 | 3.55E–03 | 1.33 (1.10–1.62) | 0.99 | 0.11 | C | T | 0.44 | 0.40 | 4.06E–03 | 1.17 (1.05–1.30) | 0.94 | 0.31 |
| P2RY11 | Rs73011220 | 10 222 862 | G | A | 0.14 | 0.11 | 4.72E–03 | 1.33 (1.10–1.63) | 0.97 | 0.11 | G | A | 0.44 | 0.40 | 4.68E–03 | 1.17 (1.05–1.30) | 0.97 | 0.31 |
| P2RY11 | Rs112647895 | 10 223 296 | G | A | 0.14 | 0.11 | 3.63E–03 | 1.34 (1.09–1.61) | 0.94 | 0.11 | G | A | 0.44 | 0.40 | 4.68E–03 | 1.17 (1.05–1.30) | 0.97 | 0.31 |
| P2RY11 | Rs3745601 | 10 224 548 | G | A | 0.14 | 0.11 | 4.72E–03 | 1.33 (1.09–1.61) | 0.96 | 0.11 | G | A | 0.44 | 0.40 | 4.68E–03 | 1.17 (1.05–1.30) | 0.95 | 0.31 |
| PPAN | Rs2305789 | 10 218 332 | A | G | 0.14 | 0.11 | 3.63E–03 | 1.34 (1.10–1.63) | 0.84 | 0.11 | A | G | 0.44 | 0.41 | 7.40E–03 | 1.16 (1.04–1.29) | 1.00 | 0.31 |
| DNMT1 | Rs6511570 | 10 246 029 | A | G | 0.39 | 0.45 | 2.35E–04 | 0.78 (0.68–0.89) | 0.98 | 0.62 | A | G | 0.24 | 0.27 | 2.29E–02 | 0.87 (0.77–0.98) | 0.97 | 0.51 |
| DNMT1 | Rs2288349 | 10 255 518 | G | A | 0.39 | 0.45 | 1.88E–04 | 0.78 (0.69–0.89) | 0.98 | 0.61 | G | A | 0.24 | 0.27 | 2.29E–02 | 0.87 (0.77–0.98) | 0.97 | 0.51 |
| DNMT1 | Rs35693490 | 10 257 245 | G | A | 0.39 | 0.45 | 1.88E–04 | 0.78 (0.68–0.89) | 0.95 | 0.61 | G | A | 0.24 | 0.27 | 2.29E–02 | 0.87 (0.77–0.98) | 0.97 | 0.51 |
| DNMT1 | Rs8101626 | 10 258 159 | C | T | 0.39 | 0.45 | 1.88E–04 | 0.78 (0.68–0.89) | 0.97 | 0.61 | C | T | 0.24 | 0.27 | 2.29E–02 | 0.87 (0.77–0.98) | 0.92 | 0.51 |
| Intergenic | Rs4804122 | 10 241 944 | T | C | 0.40 | 0.46 | 4.78E–05 | 0.76 (0.67–0.87) | 0.97 | 0.65 | T | C | 0.26 | 0.28 | 2.78E–02 | 0.87 (0.78–0.99) | 0.96 | 0.55 |
| DNMT1 | Rs2114724 | 10 265 248 | T | C | 0.46 | 0.50 | 9.66E–03 | 0.84 (0.74–0.96) | 0.99 | 0.50 | C | T | 0.30 | 0.29 | 1.79E–01 | 1.08 (0.96–1.22) | 1.00 | 0.10 |
| DNMT1 | Rs2290684 | 10 251 747 | G | A | 0.46 | 0.51 | 1.02E–02 | 0.84 (0.74–0.96) | 0.98 | 0.50 | A | G | 0.30 | 0.29 | 1.84E–01 | 1.08 (0.96–1.21) | 0.99 | 0.10 |
| DNMT1 | Rs8112801 | 10 253 099 | G | T | 0.46 | 0.51 | 1.02E–02 | 0.84 (0.74–0.96) | 0.98 | 0.50 | T | G | 0.30 | 0.29 | 1.84E–01 | 1.08 (0.96–1.21) | 0.99 | 0.10 |
| DNMT1 | Rs11880553 | 10 253 556 | T | C | 0.46 | 0.51 | 1.02E–02 | 0.84 (0.74–0.96) | 0.98 | 0.50 | C | T | 0.30 | 0.29 | 1.84E–01 | 1.08 (0.96–1.21) | 0.99 | 0.10 |
| DNMT1 | Rs11880388 | 10 253 573 | A | G | 0.46 | 0.51 | 1.02E–02 | 0.84 (0.74–0.96) | 0.97 | 0.50 | G | A | 0.30 | 0.29 | 1.84E–01 | 1.08 (0.96–1.21) | 0.99 | 0.10 |
| Intergenic | Rs11085587 | 10 235 099 | C | G | 0.46 | 0.51 | 8.90E–03 | 0.84 (0.74–0.96) | 0.98 | 0.50 | G | C | 0.31 | 0.29 | 1.96E–01 | 1.08 (0.96–1.21) | 0.91 | 0.10 |
| Intergenic | Rs35374357 | 10 239 943 | T | C | 0.43 | 0.41 | 1.69E–01 | 1.10 (0.96–1.25) | 0.91 | 0.35 | T | C | 0.30 | 0.29 | 2.08E–01 | 1.08 (0.96–1.21) | 0.94 | 0.10 |
| EIF3G | Rs7710 | 10 226 256 | A | G | 0.44 | 0.41 | 6.65E–02 | 1.13 (0.99–1.29) | 0.88 | 0.63 | A | G | 0.31 | 0.29 | 2.24E–01 | 1.07 (0.96–1.21) | 0.98 | 0.13 |
| Intergenic | Rs870612 | 10 229 297 | C | A | 0.45 | 0.42 | 5.15E–02 | 1.14 (1.00–1.30) | 0.90 | 0.62 | C | A | 0.29 | 0.28 | 3.09E–01 | 1.06 (0.95–1.19) | 1.00 | 0.16 |
| Intergenic | Rs4555265 | 10 229 521 | C | T | 0.45 | 0.42 | 5.51E–02 | 1.14 (1.00–1.30) | 0.88 | 0.62 | C | T | 0.29 | 0.28 | 3.09E–01 | 1.06 (0.95–1.19) | 1.00 | 0.16 |
| EIF3G | Rs11667630 | 10 231 490 | A | C | 0.45 | 0.42 | 5.15E–02 | 1.14 (1.00–1.30) | 0.90 | 0.62 | A | C | 0.29 | 0.28 | 3.09E–01 | 1.06 (0.95–1.19) | 1.00 | 0.16 |
| EIF3G | Rs2290687 | 10 232 184 | G | A | 0.45 | 0.42 | 5.15E–02 | 1.14 (1.00–1.29) | 0.90 | 0.62 | G | A | 0.29 | 0.28 | 3.09E–01 | 1.06 (0.95–1.19) | 1.00 | 0.16 |
| PPAN | Rs55752217 | 10 218 434 | C | T | 0.44 | 0.41 | 1.02E–01 | 1.12 (0.98–1.27) | 0.88 | 0.62 | C | T | 0.29 | 0.28 | 3.24E–01 | 1.06 (0.94–1.19) | 0.99 | 0.16 |
| PPAN | Rs2305792 | 10 221 151 | C | G | 0.44 | 0.41 | 9.99E–02 | 1.12 (0.98–1.27) | 0.87 | 0.62 | C | G | 0.29 | 0.28 | 3.33E–01 | 1.06 (0.94–1.19) | 0.99 | 0.16 |
| P2RY11 | Rs1037686 | 10 222 835 | T | A | 0.44 | 0.42 | 9.25E–02 | 1.12 (0.98–1.27) | 0.87 | 0.62 | T | A | 0.29 | 0.28 | 3.33E–01 | 1.06 (0.94–1.19) | 0.99 | 0.16 |
| P2RY11 | Rs3745600 | 10 223 427 | A | G | 0.44 | 0.41 | 4.67E–02 | 1.14 (0.99–1.28) | 0.88 | 0.62 | A | G | 0.29 | 0.28 | 3.33E–01 | 1.06 (0.94–1.19) | 0.98 | 0.16 |
| P2RY11 | Rs34484805 | 10 224 526 | C | T | 0.44 | 0.41 | 7.70E-02 | 1.13 (1.00–1.30) | 0.88 | 0.62 | C | T | 0.29 | 0.28 | 3.33E–01 | 1.06 (0.94–1.19) | 0.98 | 0.16 |
| P2RY11 | Rs7401 | 10 225 777 | A | G | 0.44 | 0.41 | 4.27E–02 | 1.14 (1.00–1.30) | 0.87 | 0.62 | A | G | 0.29 | 0.28 | 3.33E–01 | 1.06 (0.94–1.19) | 0.98 | 0.16 |
| PPAN | Rs11666402 | 10 219 076 | T | C | 0.44 | 0.42 | 9.43E–02 | 1.12 (0.98–1.27) | 0.90 | 0.62 | T | C | 0.29 | 0.28 | 3.52E–01 | 1.06 (0.94–1.19) | 0.91 | 0.16 |
| Intergenic | Rs10414661 | 10 237 427 | T | C | 0.07 | 0.06 | 3.21E–02 | 1.32 (0.99–1.66) | 0.98 | 0.01 | T | C | 0.44 | 0.43 | 4.11E–01 | 1.05 (0.94–1.16) | 0.97 | 0.15 |
| Intergenic | Rs12462004 | 10 238 264 | T | G | 0.08 | 0.06 | 3.73E–02 | 1.31 (1.02–1.70) | 0.94 | 0.01 | T | G | 0.44 | 0.43 | 4.11E–01 | 1.05 (0.94–1.16) | 0.94 | 0.15 |
| Intergenic | Rs10404209 | 10 242 237 | T | A | 0.08 | 0.06 | 6.20E–02 | 1.28 (1.02–1.71) | 0.95 | 0.01 | T | A | 0.44 | 0.43 | 4.11E–01 | 1.05 (0.94–1.16) | 0.94 | 0.15 |
| DNMT1 | Rs9305012 | 10 255 112 | C | G | 0.07 | 0.06 | 2.17E–02 | 1.36 (1.03–1.73) | 0.93 | 0.02 | C | G | 0.46 | 0.45 | 4.35E–01 | 1.04 (0.94–1.16) | 0.97 | 0.12 |
| DNMT1 | Rs10407514 | 10 260 869 | T | C | 0.07 | 0.06 | 2.99E–02 | 1.33 (1.05–1.76) | 1.00 | 0.02 | T | C | 0.46 | 0.45 | 4.35E–01 | 1.04 (0.94–1.16) | 0.97 | 0.12 |
| DNMT1 | Rs10418707 | 10 247 269 | G | A | 0.07 | 0.06 | 2.17E–02 | 1.36 (1.05–1.76) | 1.00 | 0.02 | G | A | 0.44 | 0.43 | 4.48E–01 | 1.04 (0.94–1.16) | 0.98 | 0.15 |
| DNMT1 | Rs12611113 | 10 248 270 | G | C | 0.07 | 0.06 | 2.17E–02 | 1.36 (1.05–1.76) | 1.00 | 0.02 | G | C | 0.44 | 0.43 | 4.48E–01 | 1.04 (0.94–1.16) | 0.98 | 0.15 |
| DNMT1 | Rs60565702 | 10 256 217 | G | A | 0.07 | 0.06 | 2.17E–02 | 1.36 (1.05–1.76) | 0.94 | 0.02 | G | A | 0.44 | 0.43 | 4.48E–01 | 1.04 (0.94–1.16) | 0.98 | 0.15 |
| DNMT1 | Rs72620548 | 10 257 412 | T | C | 0.07 | 0.06 | 2.17E–02 | 1.36 (1.05–1.76) | 0.99 | 0.02 | T | C | 0.44 | 0.43 | 4.48E–01 | 1.04 (0.94–1.16) | 0.98 | 0.15 |
| DNMT1 | Rs10854076 | 10 258 561 | G | A | 0.07 | 0.06 | 2.17E–02 | 1.36 (1.05–1.76) | 0.97 | 0.02 | G | A | 0.44 | 0.43 | 4.48E–01 | 1.04 (0.94–1.16) | 0.98 | 0.15 |
| DNMT1 | Rs4804489 | 10 260 030 | C | A | 0.07 | 0.06 | 2.55E–02 | 1.35 (1.05–1.76) | 0.98 | 0.02 | C | A | 0.44 | 0.43 | 4.48E–01 | 1.04 (0.94–1.16) | 0.97 | 0.15 |
| DNMT1 | Rs57366074 | 10 261 656 | A | G | 0.07 | 0.06 | 2.17E–02 | 1.36 (1.04–1.75) | 0.99 | 0.02 | A | G | 0.44 | 0.43 | 4.48E–01 | 1.04 (0.94–1.16) | 0.92 | 0.15 |
| Intergenic | Rs10414537 | 10 236 724 | C | T | 0.07 | 0.06 | 6.20E–02 | 1.28 (0.99–1.66) | 0.95 | 0.01 | C | T | 0.42 | 0.41 | 4.74E–01 | 1.04 (0.93–1.16) | 0.87 | 0.18 |
Abbreviations: bp, base pair; CI, confidence interval; freq., frequency; imp., imputation; OD, odds ratio; r2, pairwise r2 with rs2305795; SNP, single-nucleotide polymorphism.
P-values and imputation quality were calculated with Beagle. Pairwise LD (r2) with rs2305795 was calculated with Haploview. Bold letters indicate SNPs of interest. Only SNP marker with an imputation quality score of r2>0.8 were included. Intergenic: SNP markers located between EIF3G and DNMT1, which is ranked by statistical significance in the Chinese cohort.
PPAN: NM_020230.4; P2RY11: NM_002566.4; EIF3G: NM_003755.3; DNMT1: NM_001130823.1.
Results
Association signal within the risk locus
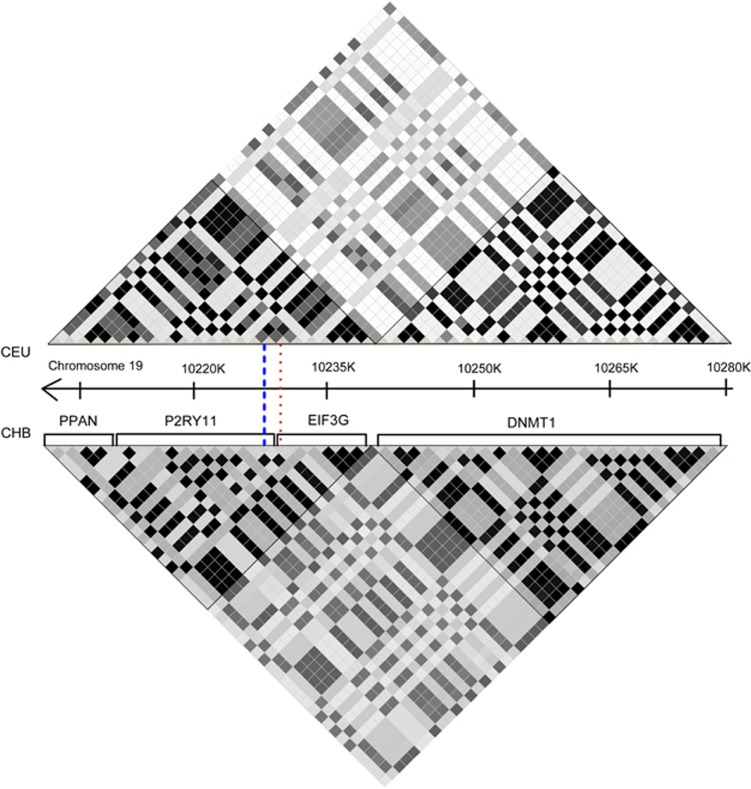
Using two GWA data sets from narcoleptic patients and controls of European and Chinese ancestry, we investigated polymorphisms in the P2RY11-DNMT1 region. The cohorts included 1881 individuals of European ancestry (807 cases and 1074 controls) and 2981 Chinese (1078 cases and 1903 controls). SNPs in the region of interest were imputed in each GWA data set, and ranked by statistical significance in the Chinese cohort (Table 1). The most significant SNPs in both GWA data sets are in high LD with rs2305795 and located in the PPAN-P2RY11-EIF3G haplotype block, and not in DNMT1 (Table 1). R2 plots clearly illustrate that SNPs in high LD with rs2305795 are restricted to the PPAN-P2RY11-EIF3G region, but not DNMT1 (Figure 1), as was also observed by Han et al.6 Based on this analysis, five SNP markers (rs1551570 (PPAN), rs12460842 (P2RY11), rs2010353 (P2RY11), rs2305795 (P2RY11) and rs3826784 (EIF3G) were significantly associated with narcolepsy in both cohorts, and were selected for further analysis. Owing to a failure of the genotyping assay, however, rs2010353 was excluded, leaving four SNPs for all subsequent analyses.
Figure 1.
Risk locus on Chr. 19q13.2. Gene organization and LD in the region of interest (10 209 865–10 276 167) compared between European (CEU) and Chinese ancestry (CHB). Top: R2-based LD plots using data from GWA data set of European ancestry (CEU). Bottom: R2-based LD plots using data from GWA data set of Chinese ancestry (CHB). Darker colors illustrate higher levels of LD. Dashed and dotted line indicates the position of rs2305795 and rs3826784, respectively.
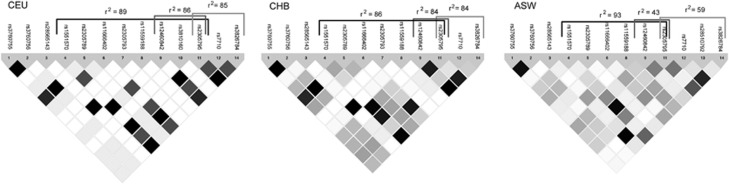
Based on data from the International HapMap Project, we next explored LD patterns for the four markers across ethnic groups. Interestingly, two SNPs rs12460842 (P2RY11) and rs3826784 (EIF3G) had a lower LD with rs1551570 (PPAN) and rs2305795 (P2RY11) in an African-American cohort (Figure 2), suggesting these to be candidates for transethnic mapping. Genotyping of these four markers in 1297 African Americans (249 cases and 1048 controls) showed that surprisingly rs3826784, and not the previously reported rs2305795, showed the strongest narcolepsy association based on odds ratio (OR) values, although none of the four SNPs reached statistical significance in this data set. A meta-analysis was performed across all ethnic groups and here we replicated the finding by Kornum et al.5 of an increased OR with the major allele rs2305795A (Table 2 shows data on minor allele). The meta-analysis further revealed that rs3826784 showed the strongest association with narcolepsy across all ethnic groups (Table 2) with a slightly higher OR for the rs3826784G risk allele and a lower P-value. There were no significant differences between the ORs from the four SNPs.
Figure 2.
R2-based LD plots for individuals of CEU, CHB and ASW. Utah residents with Northern and Western European ancestry from the CEPH collection (CEU), Han Chinese in Beijing, China (CHB) and African ancestry in Southwest USA (ASW). Curly braces indicate LD values between SNP candidates: rs1551570, rs12360842, rs2305795 and rs3826784. Darker color illustrates a higher level of LD. Chromosome 19q13.2 (10 209 865–10 230 599). See Figure 1 for entire locus.
Table 2. Association of SNP markers with Type 1 narcolepsy.
| SNP | Position | Major allele>minor allele | Freq. cases (807) | Freq. controls (1074) | P-value (MH) | OR (95% CI) | Pairwise r2 with rs2305795 |
|---|---|---|---|---|---|---|---|
| European ancestry (1881) | |||||||
| Rs1551570 | 10 218 030 | T>C | 0.42 | 0.48 | 4.00E−04 | 0.79 (0.69–0.90) | 0.98 |
| Rs12460842 | 10 222 195 | G>A | 0.42 | 0.48 | 3.00E−04 | 0.79 (0.69–0.90) | 0.99 |
| Rs2305795 | 10 226 052 | A>G | 0.42 | 0.48 | 2.00E−04 | 0.78 (0.69–0.90) | 1 |
| Rs3826784 | 10 227 010 | G>A | 0.38 | 0.44 | 1.00E−04 | 0.77 (0.68–0.88) | 0.87 |
| SNP | Position | Major allele>minor allele | Freq. cases (1078) | Freq. controls (1903) | P-value (MH) | OR (95% CI) | Pairwise r2 with rs2305795 |
| Chinese ancestry (2981) | |||||||
| Rs1551570 | 10 218 030 | T>C | 0.25 | 0.30 | 1.20E−05 | 0.77 (0.68–0.86) | 0.94 |
| Rs12460842 | 10 222 195 | G>A | 0.25 | 0.30 | 1.47E−05 | 0.77 (0.68–0.87) | 0.94 |
| Rs2305795 | 10 226 052 | A>G | 0.26 | 0.31 | 5.99E−05 | 0.79 (0.70–0.88) | 1 |
| Rs3826784 | 10 227 010 | G>A | 0.25 | 0.30 | 1.47E−05 | 0.77 (0.68–0.87) | 0.94 |
| SNP | Position | Major allele>minor allele | Freq. cases (249) | Freq. controls (1048) | P-value (MH) | OR (95% CI) | Pairwise r2 with rs2305795 |
| African Americans (1297) | |||||||
| Rs1551750 | 10 218 030 | T>C | 0.37 | 0.37 | 8.90E−01 | 0.99 (0.80–1.21) | 0.65 |
| Rs12460842 | 10 222 195 | G>A | 0.52 | 0.54 | 6.20E−01 | 0.95 (0.86–1.28) | 0.47 |
| Rs2305795 | 10 226 052 | A>G | 0.39 | 0.40 | 7.20E−01 | 0.96 (0.79–1.18) | 1 |
| Rs3826784 | 10 227 010 | G>A | 0.43 | 0.46 | 1.90E−01 | 0.87 (0.71–1.07) | 0.44 |
| SNP | Position | Major allele>minor allele | Freq. cases (2134) | Freq. controls (4025) | P-value (MH) | OR (95% CI) | CHISQ_BD | P_BD |
|---|---|---|---|---|---|---|---|---|
| All samples (6159) | ||||||||
| Rs1551750 | 10 218 030 | T>C | 0.33 | 0.37 | 1.97E−07 | 0.81 (0.74–0.88) | 4.472 | 0.1069 |
| Rs12460842 | 10 222 195 | G>A | 0.34 | 0.41 | 1.14E−07 | 0.80 (0.74–0.87) | 3.397 | 0.1829 |
| Rs2305795 | 10 226 052 | A>G | 0.34 | 0.38 | 2.94E−07 | 0.81 (0.75–0.88) | 3.33 | 0.1892 |
| Rs3826784 | 10 227 010 | G>A | 0.32 | 0.38 | 6.35E−09 | 0.79 (0.73–0.85) | 1.246 | 0.536 |
Abbreviations: BD, Breslow day; CI, confidence interval; Freq., frequency; MH, Mantel–Haenszel; OD, odds ratio; SNP, single-nucleotide polymorphism.
P-values were calculated with Beagle. Pairwise LD (r2) with rs2305795 was calculated with Haploview. Results for minor allele (for European and Chinese ancestry) are presented for each cohort.
The selected SNPs were: rs1551570:T>G), rs12460842:A>G), rs2305795:G>A) and rs3826784:A>G). The most significant SNP across all samples is indicated in bold.
PPAN: NM_020230.4; P2RY11: NM_002566.4; EIF3G: NM_003755.3; DNMT1: NM_001130823.1.
Expression quantitative trait loci analysis
To determine if rs3826784 regulates gene expression, we next performed an eQTL analysis of the genes in the risk locus. For this analysis, we used a large case–control study of major depression, for which RNA-sequencing data from whole blood together with genotype data were available for 922 individuals of European ancestry.12 After appropriate normalization, whether or not cases or controls were used it did not influence the eQTL analysis;12 thus, we performed all analysis in the combined sample.
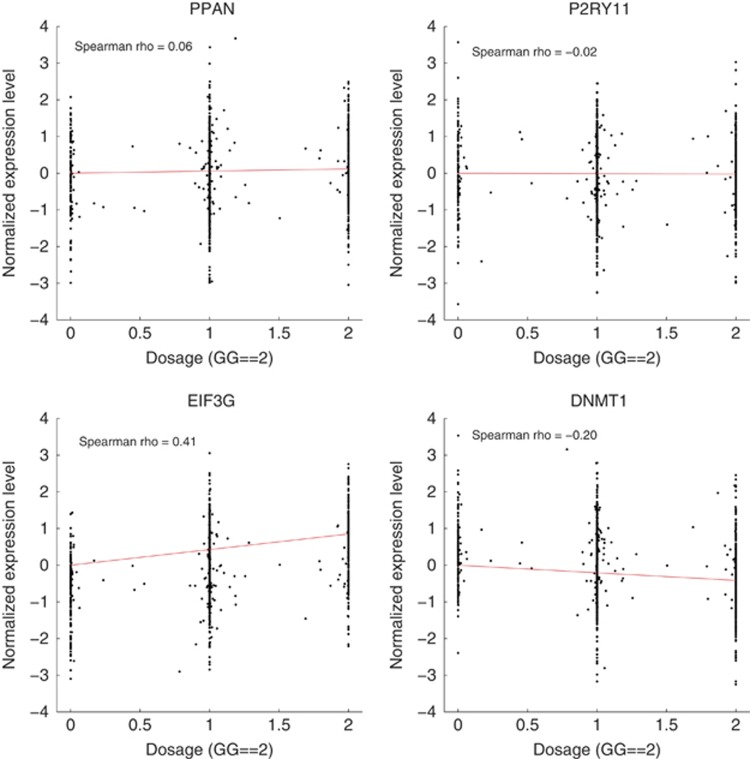
Using this data set, we found that the expression of EIF3G increased with the risk allele, DNMT1 expression decreased and the expression of PPAN and P2RY11 did not change with the risk allele (rs3826784G) (Figure 3). Furthermore, the expression of EIF3G and DNMT1, but not PPAN and P2RY11, correlated with rs3826784, the strongest association being with EIF3G (Table 3). The other four SNPs in our analysis showed equally strong association with EIF3G consistent with the high LD between these four SNPs in Caucasians (Figure 2). The expression of DNMT1 was most strongly correlated with eQTL SNP for DNMT1 rs2290684 (highlighted in Table 1). The effect of rs3826784 on DNMT1 expression is therefore likely explained by LD between rs3826784 and rs2290684 (r2=0.35). In line with this, conditional analysis showed that rs3826784 does not independently affect DNMT1 expression when the effect of rs2290684 is controlled for (P>0.1).
Figure 3.
Gene expression in whole blood with various rs3826784 genotypes. (a and b) The expression of PPAN and P2RY11 did not change with the risk allele (rs3826784G). (c) The expression of EIF3G increased with the risk allele (rs3826784GG). (d) The expression of DNMT1 decreased with the risk allele (rs3826784GG). The data are reported as normalized effect size, which equals the correlation coefficient (Spearman's ρ correlation coefficient is used as a measure of change in expression level on the y axis).
Table 3. Association between SNPs and eQTLs in the Type 1 narcolepsy risk locus (negative log10 P-values for association between genotype and expression levels).
| Gene/SNP name | Rs12460842 (genotyped) | Rs2305795 (genotyped) | Rs3826784 (imputed) | Rs1551570 (genotyped) |
|---|---|---|---|---|
| PPAN | 0.94 | 1.01 | 1.10 | 1.00 |
| P2YR11 | 0.14 | 0.22 | 0.27 | 0.18 |
| EIF3G | 42.94 | 43.95 | 38.83 | 43.25 |
| DNMT1 | 10.48 | 10.86 | 9.10 | 10.12 |
PPAN: NM_020230.4; P2RY11: NM_002566.4; EIF3G: NM_003755.3; DNMT1: NM_001130823.1.
The effect of rs2305795 on P2RY11 was also re-examined. Using our data, we found that rs2305795 was not an eQTL for P2RY11 (see Supplementary Table S1). In addition, as rs2305795 is located in the 3′-UTR of P2RY11, it was possible to analyze allele-specific association for this SNP. Using a test of association between rs2305795 and the allelic ratio of P2RY11 (as an eQTL), we did not observe a significant association between rs2305795 and the allelic expression of P2RY11 in the human whole blood data set in contrast to earlier reports in PBMCs.5
Gene expression correlation
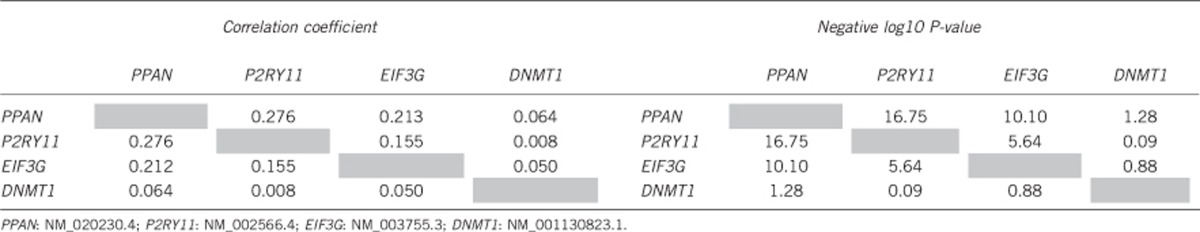
We finally investigated the possibility of coregulation and/or regulatory relationships between the four genes in our locus by analyzing correlations between expression of PPAN, P2RY11, EIF3G and DNMT1. PPAN and P2RY11 were found to be expressed at very low levels overall in whole blood cells, which correlate with the expression in the brain (Supplementary Tables S2+S3). Expression of EIF3G correlated with the expression of PPAN in normalized data optimized for detecting trans-eQTLs but not in data optimized for detecting cis-eQTLs (for more information see Materials and methods). Finally, the expression level of EIF3G was not correlated with DNMT1 (Table 4). We also did not observe a significant correlation between P2RY11 and DNMT1 in the normalized data for detecting cis-eQTL or trans-eQTL in contrast to what has earlier been reported for PBMCs.5
Table 4. Correlation coefficient between gene expression levels of each gene pair was measured using the Spearman's correlation coefficient.
Discussion
In this study, we found rs3826784, a SNP located within an intron of the EIF3G gene, to be the most significantly associated SNP of the region. Transethnic mapping further revealed that rs3826784 could be a better marker for the genetic association with narcolepsy than the previously identified rs2305795. However, the magnitude of the association is very similar to that of the previously reported SNP rs2305795.5 The new SNP, rs3826784, is in high LD with SNPs located in PPAN and P2RY11, but not in DNMT1. Genetic LD between P2RY11 and DNMT1 is weak in narcolepsy and control cohorts, confirming the observation by Han et al.6 Importantly, however, elements located within the P2RY11 gene may still regulate DNMT1 expression. We also found that the expression of EIF3G increased with the risk allele, DNMT1 expression decreased and there was no effect of rs3826784G on PPAN or P2RY11 mRNA levels in whole blood. The results suggest that the narcolepsy-associated SNP in this locus affects EIF3G expression rather than P2RY11.
To gain insight into gene regulation in the entire risk locus, we investigated gene expression correlations. In whole blood, expression of EIF3G strongly correlated with the expression of PPAN and P2RY11, and less with DNMT1. These results were consistent with strong LD found with SNPs located in the EIF3G, P2RY11 and PPAN region, an effect not extending to DNMT1. Surprisingly, however, expression of P2RY11 and DNMT1 did not correlate as earlier discovered in PBMCs.5 PPAN and P2RY11 were found to be expressed at very low levels in whole blood cells. The discrepancy in P2RY11 expression is most likely caused by the different input material in the two studies. Kornum et al.5 showed that P2RY11 expression was predominantly seen in CD8+ cells. These account for ~20% of PBMCs. However, whole blood also contains granulocytes, which accounts for 65% of all cells, and may thus mask the expression of P2RY11 in the CD8+ cells. In agreement with our results, it has previously been reported that EIF3G is highly expressed in blood cells.15 These findings illustrate the complexity of interpreting eQTL studies in whole blood, considering heterogeneity of the effect of regulatory regions on gene expression in various white blood cell subsets. Indeed, cell-specific regulatory mechanisms in the PPAN-P2RY11-EIF3G region are likely present. Future studies will need to address the regulation of the PPAN-P2RY11-EIF3G narcolepsy risk locus within distinct sub-populations of white blood cells. That cell-specific effects are present is not surprising considering the importance of the regulation of apoptosis in various immune cells types in reaction to changes in the environmental milieu.
How could EIF3G be involved in narcolepsy? Eukaryotic initiation factors (EIF) have an important role in the initiation phase of eukaryotic translation by forming a complex with the 40S ribosomal subunit and recruiting a Met-tRNAi called the 43S to the preinitiation complex. This complex recognizes and binds the 5′ cap structure of mRNA, promoting ribosomal scanning of mRNA and regulating recognition of the AUG initiation codon.16, 17 In mammals, EIF3 is the largest scaffolding initiation factor (750 kDa), comprising 13 subunits (A–M). These subunits cluster together in subcomplexes, in which EIF3G connects to EIF3B and EIF3I. EIF3B:G contains RNA recognition motif subunits that are involved in RNA binding. Interestingly, EIF3B:G:I also interacts directly with certain viral mRNAs to promote the translation of viral proteins.18 EIF4G, which interacts with EIF3,19 contains a binding domain for the non-structural influenza protein, NS1,20 and is recruited to the viral mRNA to initiate translation of influenza virus. This is particularly interesting as recent findings have implicated a role of the H1N1 pandemic virus of 2009 as a trigger for narcolepsy development.6 It has been suggested that the disruption of EIF pathways in childhood H1N1/09 influenza increases disease severity,21 so polymorphisms in this locus could indirectly change immune responses to influenza and narcolepsy risk.
Another possibility could involve regulation of cell death, a function that could be shared with P2RY11 and DNMT1, with coordinated regulation of these loci explaining the conservation of synteny. The N terminus of EIF3G indeed interacts with the C-terminal region of apoptosis-inducing factor (AIF). AIF is a ubiquitous FAD-binding flavoprotein that has an important role in caspase-independent apoptosis. Mature AIF inhibits newly translated protein synthesis via its interactions with EIF3G. Additionally, mature AIF overexpression specifically activates caspase-7, thereby amplifying the inhibition of protein synthesis including EIF3G cleavage.22, 23 Additionally, EIF3G is expressed in the brain and the lateral hypothalamus (Allen Brain Atlas: www.brain-map.org).15 Higher EIF3G expression with the narcolepsy risk allele could cause a dysregulated cell death, which again could have an important role in white blood cells in the context of autoimmunity, or in the brain in the context of hypocretin cell loss.
In conclusion, using transethnic mapping, we found a novel genetic association of Type 1 narcolepsy with rs3826784, an SNP located in EIF3G. The new SNP is in high LD with the previously reported P2YR11 rs2305795 SNP in individuals of European and Chinese ancestry, but displays lower LD in African Americans. As the association signal from rs3826784 is nominally better compared with rs2305795 in all three cohorts, rs3826784 may be a better marker for the genetic association with narcolepsy, although this conclusion is tentative considering sample size of the African-American Cohort. Further confirmation is needed through extended GWA studies that combine multiple ethnic groups, and local resequencing. A complex correlation between the expression of three genes in the region suggests that a shared regulatory mechanism exists and might be affected by the polymorphism.
Acknowledgments
We are indebted to all the narcoleptic patients of the study. Without their contributions this study would not have been possible. This work was supported by the Lundbeck Foundation and NIH Grant P50 NS23724.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- 1Matsuki K, Grumet FC, Lin X et al: DQ (rather than DR) gene marks susceptibility to narcolepsy. Lancet 1992;339:1052. [DOI] [PubMed] [Google Scholar]
- 2Hallmayer J, Faraco J, Lin L et al: Narcolepsy is strongly associated with the T-cell receptor alpha locus. Nat Genet 2009; 41: 708–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3Faraco J, Lin L, Kornum BR et al: ImmunoChip study implicates antigen presentation to T cells in narcolepsy. PLoS Genet 2013; 9: e1003270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Partinen M, Kornum BR, Plazzi G et al: Narcolepsy as an autoimmune disease: the role of H1N1 infection and vaccination. Lancet Neurol 2014; 13: 600–613. [DOI] [PubMed] [Google Scholar]
- 5Kornum BR, Kawashima M, Faraco J et al: Common variants in P2RY11 are associated with narcolepsy. Nat Genet 2011; 43: 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Han F, Faraco J, Dong XS et al: Genome wide analysis of narcolepsy in China implicates novel immune loci and reveals changes in association prior to versus after the 2009 H1N1 influenza pandemic. PLoS Genet 2013; 9: e1003880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Winkelmann J, Lin L, Schormair B et al: Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. Hum Mol Genet 2012; 21: 2205–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Mignot E, Lin L, Rogers W et al: Complex HLA-DR and -DQ interactions confer risk of narcolepsy–cataplexy in three ethnic groups. Am J Hum Genet 2001; 68: 686–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Wu Y, Waite LL, Jackson AU et al: Trans-ethnic fine-mapping of lipid loci identifies population-specific signals and allelic heterogeneity that increases the trait variance explained. PLoS Genet 2013; 9: e1003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Browning SR, Browning BL: Rapid and accurate haplotype phasing and missing data inference for whole genome association studies by use of localized haplotype clustering. Am J Hum Genet 2007; 81: 1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Barrett JC, Fry B, Maller J et al: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 12Battle A, Mostafavi S, Zhu X et al: Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res 2014; 24: 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Purcell S, Neale B, Todd-Brown K et al: PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Stegle O, Parts L, Durbin R, Winn J: A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput Biol 2010; 6: e1000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Wu C, Orozco C, Boyer J et al: BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 2009; 10: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Zhou M, Sandercock AM, Fraser CS et al: Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc Natl Acad Sci USA 2008; 105: 18139–18144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Hinnebusch AG: eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci 2006; 31: 553–562. [DOI] [PubMed] [Google Scholar]
- 18Park HS, Himmelbach A, Browning KS, Hohn T, Ryabova LA: A plant viral 'reinitiation' factor interacts with the host translational machinery. Cell 2001; 106: 723–733. [DOI] [PubMed] [Google Scholar]
- 19Spahn CM, Kieft JS, Grassucci RA et al Hepatitis C virus IRES RNA-induced changes in the conformation of the 40 s ribosomal subunit. Science 2001;291:1959–1962. [DOI] [PubMed] [Google Scholar]
- 20Aragón T, de la Luna S, Novoa I, Carrasco L, Ortín J, Nieto A: Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol Cell Biol 2000; 20: 6259–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Herberg JA, Kaforou M, Gormley S et al: Transcriptomic profiling in childhood H1N1/09 influenza reveals reduced expression of protein synthesis genes. J Infect Dis 2013; 208: 1664–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Kim JT, Kim KD, Song EY et al: Apoptosis-inducing factor (AIF) inhibits protein synthesis by interacting with the eukaryotic translation initiation factor 3 subunit p44 (eIF3g). FEBS Lett 2006; 580: 6375–6383. [DOI] [PubMed] [Google Scholar]
- 23Kim JT, Lee SJ, Kim BY et al: Caspase-mediated cleavage and DNase activity of the translation initiation factor 3, subunit G (eIF3g). FEBS Lett 2013; 587: 3668–3674. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.