Abstract
In about 50% of sporadic cases of retinoblastoma, no constitutive RB1 mutations are detected by conventional methods. However, recent research suggests that, at least in some of these cases, there is somatic mosaicism with respect to RB1 normal and mutant alleles. The increased availability of next generation sequencing improves our ability to detect the exact percentage of patients with mosaicism. Using this technology, we re-tested a series of 40 patients with sporadic retinoblastoma: 10 of them had been previously classified as constitutional heterozygotes, whereas in 30 no RB1 mutations had been found in lymphocytes. In 3 of these 30 patients, we have now identified low-level mosaic variants, varying in frequency between 8 and 24%. In 7 out of the 10 cases previously classified as heterozygous from testing blood cells, we were able to test additional tissues (ocular tissues, urine and/or oral mucosa): in three of them, next generation sequencing has revealed mosaicism. Present results thus confirm that a significant fraction (6/40; 15%) of sporadic retinoblastoma cases are due to postzygotic events and that deep sequencing is an efficient method to unambiguously distinguish mosaics. Re-testing of retinoblastoma patients through next generation sequencing can thus provide new information that may have important implications with respect to genetic counseling and family care.
Introduction
Retinoblastoma (RB; OMIM#180200) is an intraocular malignancy that occurs in children, usually before age 5 years, with a reported prevalence of 1 out of 15 000–28 000 live births.1 RB is responsible for 1% of childhood cancer deaths and 5% of childhood blindness.2 A two-step inactivation of both alleles of the RB1 gene is required for tumor initiation, but additional mutational events usually accompany malignancy.3, 4, 5 In hereditary RB, the initial RB1 mutation (M1) occurs in the germline cells, while the second mutation (M2) occurs in the retinal cells.6 RB patients transmit their predisposition as an autosomal-dominant trait with high penetrance (>90%).6 All bilateral RBs (40%) are heritable, while unilateral RBs (60%) are heritable in only a small percentage of cases (7%).7 Patients with hereditary RB also show an increased risk of developing non-ocular tumors.8 In non-hereditary RB, mutations of both RB1 alleles occur in the retinal cells, determining unilateral tumors in children.6
The majority of children with RB have newly acquired RB1 mutations. De novo mutations can arise in the parental germ cell (usually in the paternal germ cell), or at some point during embryogenesis, resulting in mosaicism.9, 10, 11, 12, 13 If the alteration occurs during the first embryonic cell divisions, it can be confused with a mutation originated in parental germline cells, but if it occurs later, during the embryonic development, it can be tissue-specific or tissue-limited.14 One previous study showed that in 10% of RB families the first mutation was in mosaic state, either in the proband or in one of the proband's parents.11 However, because of the unavailability of key family members, the authors hypothesized that mosaicism could be even more frequent.11 Underestimation of mosaicism at that time was also due to the limited sensitivity of traditional methods employed for RB1 mutation analysis (SSCP, Sanger sequencing and Southern blotting). In more recent years, the detection rate of RB1 gene mutations has been greatly increased by the introduction of highly sensitive allele-specific PCR (AS-PCR).12 In particular, using this technique, it was possible to identify even low-level RB1 mosaicisms (frequency <15% of the normal allele). However, this study was limited to only 11 mutational ‘hot spots', while >50% of RB1 mutations are private mutations.15 The introduction of unbiased next generation (or deep) sequencing technology, reporting exactly how many molecules have been sequenced and the exact percentage of variants, has offered an excellent opportunity to detect mosaicism in different diseases.16, 17, 18, 19 Recently, Chen et al,13 analyzing by deep sequencing (Ion Torrent Personal Genome Machine, Life Technologies, Carlsbad, CA, USA) lymphocyte DNA from 90 RB cases where Sanger sequencing excluded the presence of mutations, were able to identify 30 and 6% low-level RB1 mosaic mutations in bilateral and unilateral cases, respectively.
In the present study, using next generation sequencing (454 GS Junior System, Roche, Life Sciences, Indianapolis, IN, USA), we re-analysed a series of 40 sporadic RB cases (11 bilateral and 29 unilateral). Previous analysis by traditional methods (Sanger sequencing and Multiplex Ligation Probe Amplification, MLPA) had shown constitutive RB1 mutations in apparent heterozygosis in 10 patients (9 bilateral and 1 unilateral), whereas there was no detectable mutation in the remaining 30 patients. In particular, mutations were analysed in tissues other than blood, namely ocular tissues (tumor, muscle, retina and sclera), urine and oral mucosa, whenever available. Present data could have important implications for genetic counseling, because they can alter diagnosis and family care.
Materials and methods
Patient samples
We collected 40 patients with diagnosis of sporadic RB, which was bilateral in 11 cases and unilateral in 29. In the last screening group of our diagnostic service, traditional methods (Sanger sequencing and MLPA) had previously identified 10 heterozygous sequence variants: 9 were in patients with bilateral RB and 1 in a patient with unilateral RB (Table 1). Clinical diagnosis was made by physicians with long-lasting experience in Ophthalmology and Ocular Oncology, belonging to the ‘Retinoblastoma Referral Center' of the General Hospital of Siena. All patients underwent genetic counseling and had their blood and tissue samples collected and analysed for RB1 variants at the Medical Genetics Unit of the University of Siena. Blood samples were collected in EDTA-containing tubes. Urine samples and oral swabs were also collected during genetic couselling. Formalin-fixed paraffin-embedded ocular samples from enucleated RB patients were obtained from the archives of the Pathology Unit of the University of Siena. After surgery, enucleated eyes were immersion-fixed in buffered formalin for 48 h. After fixation, sampling, paraffin embedding and cutting were carried out according to the usual pathological methods. Informed consent was obtained from all adult patients and parents of all affected children.
Table 1. RB1 variants detected in patients with sporadic retinoblastoma.
| Patients' ID | Gender | Current age (years) | UL/BL | Foci | Age of diagnosis (months) | R-E | ABC | Relapses | Therapy | RB1 variant | Forward % (seq n) | Reverse % (seq n) | WM % (seq n) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 390 | F | 35 | UL | Uni | 6 | NA | NA | No | E (RE) | c.1215+1G>A | 8.2 (1138) | 8.8 (1588) | 8.6 (2726) |
| 973/2013 | F | 31 | BL | Multi | 36 | IVB (RE) VB (LE) | NA | NA | E (LE) RT—FT (RE) | c.596T>G (p.Leu199*) | 23.3 (597) | 25.2 (934) | 24.4 (1531) |
| 39/2013 | M | 5 | UL | Uni | 49 | VB (RE) | D (RE) | No | IACT (2 cycles M+T) IVM (8 injections) SCC (1 injection)—E (RE) | c.1735C>T (p.Arg579*) | 23.0 (724) | 26.5 (614) | 24.7 (1338) |
| 768 | F | 4 | UL | Uni | 29 | VB (RE) | D (RE) | No | E (RE) | c.1301_1302dup (p.Gly435Trpfs*23) | 38.9 (955) | 32.8 (579) | 36.6 (1534) |
| 762 | F | 2 | BL | Multi | 4 | VB (RE) VE (LE) | C (RE) E (LE) | Yes | E (LE) SCT (6 cycles C+Et) IACT (1 cycle M+T) IVM (2 injections)—FT (RE) | c.2212-1G>T | 34.7 (1068) | 43.8 (491) | 37.6 (1559) |
| 795 | F | 2 | BL | Multi | At birth | IIA (RE) IIA (LE) | A (RE) A (LE) | Yes | SCT (2 cycles C+Et+V and 2 cycles C+Et)—FT (RE and LE) | c.1644dup (p.His549Thrfs*6) | 47.4 (410) | 43.5 (1872) | 44.2 (2282) |
| 721 | M | 2 | BL | Multi | 1 | IV (RE) III (LE) | B (RE) B (LE) | Yes | SCT(6 cycles C+Et)—FT (RE) SCC (1 injections LE) | c.1050-1G>C | 48.6 (806) | 41.8 (694) | 45.5 (1500) |
| 704 | M | 2 | BL | Multi | 1 | VB (RE) IB (LE) | D (RE) B (LE) | Yes | SCT (1 cycle C+Et)—E (RE) STC (5 cycles C+Et)—FT (LE) | c.1666C>T (p.Arg566*) | 44.5 (2684) | 46.7 (2576) | 45.6 (5260) |
| 779 | M | 10 | BL | Multi | 3 | II (RE) III (LE) | A (RE) B (LE) | No | SCT (6 cycles C+Et) FT (RE and LE) | c.2359C>T (p.Arg787*) | 49.4 (984) | 44.1 (719) | 47.2 (1703) |
| 714 | M | 5 | BL | Multi | 4 | IV (RE) IV (LE) | B (RE) B (LE) | Yes | SCT (6 cycles) IACT (1 cycle M+T) (LE) | c.1498+5del | 47.8 (5121) | 49.5 (7379) | 48.8 (12 500) |
| 745 | M | 15 | BL | Multi | 8 | V (RE) IV (LE) | C (RE) B (LE) | Yes | SCT (6 cycles C+Et and 2 cycles C+Et+V)—E (RE)—FT (LE) | c.1363C>T (p.Arg455*) | 48.8 (1405) | 48.9 (1095) | 48.8 (2500) |
| 777 | F | 11 | BL | Multi | At birth | I (RE) V (LE) | A (RE) D (LE) | Yes | E (LE) SCT (6 cycles C+Et)—FT (REt) | c.1363C>T (p.Arg455*) | 48.8 (944) | 52.6 (805) | 50.5 (1749) |
| 567/2013 | M | 2 | BL | Multi | 12 | IV (LE) IV (RE) | C (LE) C (RE) | Yes | SCT (7 cycles )—IACT (2 cycles M+T) | c.2211+2dup | 56.0 (613) | 48.02 (429) | 52.7 (1042) |
Abbreviations: BL, bilateral; C, carboplatin; E, enucleation; Et, etoposide; FT, focal therapy; IACT, intra-arterial chemotherapy; IVM, intravitreal melphalan; M, melphalan; Multi, multifocal; NA, not available; RT, radiotherapy; SCC, subconjunctival carbolatin; SCT, sistemic chemotherapy; T, topotecan; UL, unilateral; Uni, unifocal; V, vincristine; WM, weighted mean.
Sequencing results are shown as percentage variant of total number of reads.
Reference sequence L11910.1 (Trancript NM_000321.2).
DNA extraction
Genomic DNA was isolated from EDTA peripheral blood samples using a QIAamp DNA Blood Kit according to the manufacturer's protocol (Qiagen, http://www.qiagen.com). Urine and oral mucosa samples were freshly extracted using the QIAamp DNA Mini kit following the manufacturer's instructions (Qiagen, http://www.qiagen.com). Ocular tissues were identified on H&E-stained sections. Five-micron-thick sections were deparaffinized, rehydrated and stained with Mayer hematoxylin and yellow eosin and then dehydrated with xylene. Slides were observed using an inverted microscope (Zeiss, Telaval 31, Jena, Germany). Cells from normal eye tissues (RB, normal retina, muscle, sclera) were isolated by laser capture microdissection (Arcturus PixCell II, MWG-Biotech, Florence, Italy) as already described.5 Larger tumor areas were isolated by scraping. Genomic DNA was then extracted from ocular tissues using the QIAamp Mini kit (Qiagen, http://www.qiagen.com).
RB1 analysis by traditional methods
All 40 sporadic RB patients had been previously analysed by Sanger sequencing. Primers and PCR conditions were the same as those described for 454 GS Junior amplicon library preparation (Supplementary Table S1). PCR products were then sequenced using the PE Big Dye Terminator Cycle Sequencing Kit and an ABI Prism 310 genetic analyser (PE Applied Biosystems, Foster City, CA, USA). Sequencer software was used for data analysis.
To identify large rearrangements in the RB1 gene, MLPA had been previously performed in all 40 patients. Analysis was carried out using the SALSA MLPA kit (P047-B1 RB1) designed by MRC-Holland (Amsterdam, The Netherlands). Experiments were performed as previously described.20 Coffalyser software (MRC-Holland) was used for data analysis.
RB1 analysis by Roche 454 GS Junior sequencing
Deep sequencing was performed to investigate the presence of RB1 mosaic variants in 40 sporadic RB cases: 30 ‘negative' and 10 ‘positive heterozygous' cases according to traditional methods (Sanger sequencing and MLPA analysis). Given that sporadic unilateral RB is predominantly associated with RB1 somatic mutations, the optimal procedure would include an initial screening in tumor DNA.21 However, traditional RB1 analysis in our unilateral samples (29 cases) did not include tumor DNA screening because of the difficulties in tumor tissue collection in routine molecular diagnosis. For the purposes of this study, we were able to collect five tumor tissues that were analysed by amplicon deep sequencing. The presence of tumor variants in matched lymphocytes of each patient was then analysed by deep sequencing of the specific exons.
To analyse the RB1 gene (Gene Bank Accession L11910.1; transcript NM_000321.2), we used a strategy based on the locus-specific amplification of genomic DNA, amplifying each amplicon separately, followed by Roche 454 resequencing. Fusion primers are reported in Supplementary Table S1. For DNA samples isolated from peripheral blood and oral mucosa, thermal cycling was performed on an Applied Biosystems 2720 (Life Technologies) using the following cycling profile: 95 °C for 5 min, followed by 35 cycles at 95 °C for 30 s, at the specific annealing temperature for 30 s, at 72 °C for 30 s, followed by a final extension step at 72 °C for 5 min (Supplementary Table S1). As DNA concentration of samples isolated from urine samples or eye tissues was low in some cases, the above described protocol was slightly changed. We elongated annealing and extension time (to 45 s) and the final extension step (to 10 min), increased the number of cycles to 40 and used 1.5 units of polymerase instead of 1 unit. Then 3 μl of amplified PCR products were electrophoretically separated on 1.2% agarose gel. For removal of small DNA fragments, samples were then purified using the AMPure PCR purification system (Agenocourt, Beverly, MA, USA). Amplicons were quantified using the Quant-iT PicoGreen dsDNA reagent (Invitrogen Corporation, Life Technologies). All amplicons were then pooled at an equimolar ratio. Subsequently, the sample pool was diluted to a final concentration of 1 × 107 PCR fragment molecules/ul.The amplicon-PCR-derived fragments were annealed to carrier beads and clonally amplified by emulsion PCR. Subsequently, the emulsions were broken by isopropanol, and the beads carrying the single-stranded DNA templates were enriched, counted and deposited into the PicoTiterPlate for pyrosequencing.22 Identified variants were submitted to LOVD database (http://rb1-lovd.d-lohmann.de).
To distinguish low-frequency mosaic variants from background errors of 454 platform, deep sequencing was performed in three healthy individuals at the same positions as mosaic variants. To detect potential mosaics among Sanger heterozygous samples, deep sequencing was performed in DNAs from ‘known' constitutional heterozygous samples (mutated offspring of familial RB cases). In particular, we selected samples with the same variants of the suspected mosaic or, when not available, samples with variants at least in the same amplicons to maintain sequencing efficiency. Each sample was analysed three times.
454 GS Junior assay validation
To determine the linearity of the deep sequencing assay and the lower limit of detection (LOD), calibration curves for two different variants (c.1363C>T and c.1072C>T) were drawn using DNA isolated from two heterozygous controls (mutated offspring of familial RB cases). Mutated DNAs were serially diluted with wild-type DNA to mimic the presence of variants at levels of 50, 33, 16.5, 8.25, 4.12 and 0%. Each sample at the different dilutions was sequenced three times. EXCEL 2013 (Microsoft, Redmond, WA, USA) was used to analyse data.
454 GS Junior sequencing data analysis
Data analysis was performed using the Roche proprietary software package for the GS Junior system. Image acquisition, image processing and signal processing were performed during the run. Post-run analysis was conducted using the latest version (2.5p1) of GS Amplicon Variant Analyzer (AVA) (http://454.com/downloads/my454/documentation/gs-junior/software-manual/454_Sequencing_Software_Manual_v2.5p1_PartD.pdf). The AVA application computes the alignment of reads from Amplicon libraries obtained on the GS Junior Instrument and identifies differences between the reads and the reference sequence. The AVA software identifies all nucleotide variants and provides read counts and frequencies. Variants are also displayed graphically with a histogram indicating the positions. The default Variant/Consensus parameters include: minimum read percentage of 0.25% (per read direction), minimum read count of two per orientation and appearing in both forward and reverse directions, and dynamic N-mer thresholding for homopolymers. However, to examine a specific sequence position analysis parameters were set to minimum (read count to ‘1', read percentage to ‘0', directional support to ‘Any' and N-Mer thresholding to ‘Fixed'). Moreover, on the basis of our experience, variants with highly unbalanced frequencies (F/R strand ratio <0.20) were considered as technical artifacts. Moreover, results in homonucleotide regions were compared with data in normal controls and excluded when present in the latter group. In silico analysis of variants' pathogenicity was performed using the interactive biosoftware Alamut v2.3 (Interactive Biosoftware, Rouen, France).
Results
In the present study, 40 patients with a diagnosis of sporadic RB (which was bilateral in 11 cases and unilateral in 29 cases) were re-analysed for RB1 sequence variants by amplicon next generation sequencing (454 GS Junior System, Roche). In these patients, Sanger sequencing on lymphocytes had previously identified apparent RB1 heterozygous variants in 10 cases: 9 bilateral and 1 unilateral. MLPA analysis in these samples had not revealed the presence of RB1 large rearrangements. For amplicon deep sequencing, coverage of the target region (promoter and 27 exons of the RB1 gene) was 100%. Different amplicons showed different sequencing efficiencies. The average depth and the balance between forward/reverse reads across individual RB1 amplicons were thus not uniform. In one 454 GS Junior sequencing run, we obtained an average of 2500 reads per sample (minimum 1002, maximum 4716 mapped reads per sample).
454 GS Junior assay validation
To determine linearity of the assay and LOD of RB1 mosaic variants, two DNA samples with different heterozygous variants were serially diluted with wild-type DNA and analysed by deep sequencing. Supplementary Figure S1 shows the calibration curves and establishes the LOD values as 5.0% (c.1363C>T) and 3.9% (c.1072C>T).
454 GS Junior analysis in mutation-negative cases
Amplicon deep sequencing was performed in five available unilateral tumors. We identified a total of seven truncating variants, including two in apparent homozygosis (Supplementary Table S2). The presence of these variants in matched lymphocytes of each patient was excluded by deep sequencing of the specific amplicons (Supplementary Table S2).
In the remaining 23 unilateral and 2 bilateral patients, next generation sequencing detected 3 cases (samples 390, 973/2013 and 39/2013) with RB1 variants varying in frequencies from 8.6 to 24.7% (Table 1). These frequencies were confirmed by independent next generation sequencing experiments (Table 2). To rule out the possibility that the variant calls were due to sequencing errors of the 454 Roche platform, data were compared with frequencies obtained in three healthy individuals at the same positions as the mosaic variants (Table 2). Statistical analysis performed using t-test indicated that the variants in the three cases were called with high confidence with respect to background (Table 2). To confirm mosaicism, we collected additional tissues from these patients, including eye (tumor, muscle, retina and sclera), oral mucosa and urine, and we analysed the specific amplicon containing the variant by deep sequencing (Supplementary Table S3; Table 3). Different tissues showed variable frequencies of variants, confirming mosaicism (Supplementary Table S3; Table 3).
Table 2. t-Test of variant frequencies for three suspected low-level mosaic RB cases against background noises from runs of three healthy controls.
| RB1 variants (ID of patient) | Blood % of RB cases (CI 95%) | Blood % of healthy controls (CI 95%) | t-Test | ||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | ||
| c.1215+1G>A (390) | 8.6 (7.6–9.7) | 8.5 (7.8–9.2) | 9.4 (8.8–10.0) | 0.1 (0.0–0.2) | 0.1 (0.0–0.3) | 0.0 (–) | P<0.0001 |
| c.596T>G (973/2013) | 24.4 (22.1–26.7) | 23.3 (21.4–25.2) | 25.4 (22.8–28.0) | 0.7 (0.3–1.1) | 0.5 (0.2–0.8) | 0.7 (0.3–1.1) | P<0.0001 |
| c.1735C>T (39/2013) | 24.7 (22.4–27.0) | 27.4 (25.1–29.7) | 26.5 (24.3–28.7) | 0.1 (0.0–0.2) | 0.1 (0.0–0.2) | 0.2 (0.0–0.4) | P<0.0001 |
Abbreviation: CI, confidence interval. Variant frequencies, expressed in percentages, represent the weighted mean values calculated on the number of forward and reverse reads for each sample.
Table 3. Frequencies of RB1 variants in additional tissues of Sanger mutation-negative cases.
| Eye tissues | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient ID | Retina | Sclera | Oral mucosa | Urine | ||||||||
| 1 (seq n) | 2 (seq n) | WM (CI 95%) | 1 (seq n) | 2 (seq n) | WM (CI 95%) | 1 (seq n) | 2 (seq n) | WM (CI 95%) | 1 (seq n) | 2 (seq n) | WM (CI 95%) | |
| 390 | — | — | — | — | — | — | 12.7 (1611) | 11.2 (1680) | 11.9 (10.8–13.0) | 11.4 (1532) | 13.5 (1549) | 12.4 (11.2–13.6) |
| 973/2013 | — | — | — | — | — | — | 16.8 (1914) | 18.8 (2800) | 18.0 (16.9–19.1) | 23.8 (1584) | 25.6 (1074) | 24.5 (22.9–26.1) |
| 39/2013 | 6.2 (3064) | 4.9 (1904) | 5.7 (5.1–6.3) | 3.4 (2663) | 4.1 (4177) | 3.8 (3.4–4.3) | 1.0 (1050) | 1.2 (1551) | 1.2 (0.8–1.6) | 6.8 (1081) | 7.8 (2397) | 7.5 (6.6–8.4) |
Abbreviations: CI, confidence interval; WM, weighted mean. Variant frequencies, expressed in percentages, represent the weighted mean values calculated on the number of forward and reverse reads. Each amplicon has been sequenced twice (1, 2). Highlighted in grey, the weighted mean values calculated on the total of reads number of the two experiments.
454 GS Junior analysis in heterozygous cases
To detect possible mosaicism among the 10 patients (9 bilateral and 1 unilateral) interpreted as heterozygotes after Sanger sequencing on lymphocytes, next generation sequencing was performed. To investigate whether detected variant frequencies were statistically different from those of a ‘known' constitutional heterozygote, data were compared with frequencies obtained in mutated offspring of familial RB cases with the same variant of the mosaic variant or, when not available, a different variant present at least in the same amplicon of the suspected mosaic (Table 4). Unfortunately, for variants in exons 13 (sample 768) and 23 (sample 779), ‘known' heterozygous controls were not available. For variants c.1666C>T in exon 17 of patient 704 and c.2212-1G>T in exon 22 of patient 762, t-test demonstrated that the differences were statistically significant (Table 4). Suspected mosaicism of both samples was then confirmed by indipendent experiments of next generation sequencing in additional tissues (Supplementary Table S3; Table 5). Analysis in additional tissues also confirmed that sample 768, showing a read percentage of 36.6% in blood DNA, was a mosaic case (Supplementary Table S3; Table 5). Four other cases showed a range of frequency values compatible with an heterozygous sample (Supplementary Table S3; Table 5).
Table 4. t-Test of variant frequencies of RB cases against frequencies of heterozygous controls.
| Blood % of RB cases (CI 95%) | Blood % of heterozygous controls (CI 95%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Amplicon | Variant (patient ID) | 1 | 2 | 3 | Variant (control ID) | 1 | 2 | 3 | t-Test |
| 11 | c.1050-1G>C (721) | 45.5 (43.0–48.1) | 50.1 (48.0–52.2) | 49.7 (47.8–51.6) | c.1072C>T (1) | 50.6 (48.9–52.4) | 51.1 (49.5–52.7) | 51.1 (49.6–52.6) | P>0.05* |
| 14 | c.1363C>T (745) | 48.8 (46.8–50.8) | 49.9 (47.5–52.3) | 49.9 (46.9–51.0) | c.1363C>T (2) | 50.3 (48.2–52.4) | 50.2 (48.3–52.2) | 51.4 (49.5–53.3) | P>0.05 |
| 14 | c.1363C>T (777) | 50.5 (48.2–52.8) | 48.1 (46.1–50.1) | 50.9 (49.3–52.3) | c.1363C>T (3) | 50.3 (48.2–52.4) | 50.2 (48.3–52.2) | 51.4 (49.5–53.3) | P>0.05 |
| 15–16 | c.1498+5del (714) | 48.8 (47.9–49.7) | 45.3 (42.4–48.3) | 45.8 (43.1–48.6) | c.1399C>T (4) | 49.9 (47.0–52.8) | 49.7 (47–52.4) | 48.9 (46.2–51.6) | P>0.05 |
| 17 | c.1644dup (795) | 44.2 (42.2–46.3) | 49.3 (46.2–52.4) | 46.6 (44.2–49.1) | c.1666C>T (5) | 48.0 (45.9–50.1) | 50.5 (48.4–52.4) | 48.6 (46.2–51.0) | P>0.05 |
| 17 | c.1666C>T (704) | 45.6 (44.3–47.0) | 45.9 (43.3–48.5) | 45.9 (43.4–48.5) | c.1666C>T (5) | 48.0 (45.9–50.1) | 50.5 (48.4–52.4) | 48.6 (46.2–51.0) | P=0.0480* |
| 21 | c.2211+2dup (567/2013) | 52.7 (49.6–55.8) | 51.8 (49.0–54.6) | 48.3 (46.0–50.6) | c.2173_2174insT (6) | 51.7 (49.5–53.9) | 49.7 (47.8–51.6) | 48.7 (46.8–50.6) | P>0.05 |
| 22 | c.2212-1G>T (762) | 37.6 (35.2–40.1) | 34.0 (31.7–36.3) | 32.7 (29.9–35.4) | c.2325+1G>A (7) | 45.4 (43.6–47.3) | 48.9 (46.9–50.9) | 49.8 (47.7–51.9) | P=0.0026 |
Abbreviation: CI, confidence interval. Variant frequencies, expressed in percentages, represent the weighted mean values calculated on the number of forward and reverse reads for each sample. Each amplicon has been sequenced three times (1, 2, 3). The Fisher Snedecor F-test was performed to assess the equality of variance. If variances were not equal, the Satterthwaite's approximate t-test was used (*). Highlighted in grey, mosaic cases.
Table 5. Frequency of RB1 variants in different tissues of Sanger heterozygous cases.
| Eye tissues | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient ID | Tumor | Muscle | Retina | Sclera | Oral mucosa | Urine | ||||||||||||
| 1 %(seq n) | 2 %(seq n) | WM % (CI 95%) | 1 %(seq n) | 2 %(seq n) | WM % (CI 95%) | 1 %(seq n) | 2 %(seq n) | WM % (CI 95%) | 1 %(seq n) | 2 %(seq n) | WM % (CI 95%) | 1 %(seq n) | 2 %(seq n) | WM % (CI 95%) | 1 %(seq n) | 2 %(seq n) | WM % (CI 95%) | |
| 768 | 91.9 (3473) | 91.7 (1728) | 91.8a (91.1-92.3) | — | — | — | 10.1 (3024) | 11.3 (3796) | 10.8 (10.1–11.5) | 20.7 (2502) | 17.3 (1477) | 19.5 (18.3–20.7) | 4.0 (3730) | 4.2 (6090) | 4.1 (3.7–4.5) | 17.7 (2904) | 19.9 (5924) | 19.2 (18.4–20.0) |
| 762 | 100.0 (1314) | 100.0 (1199) | 100.0 (—) | 18.1 (1368) | 24.4 (1921) | 21.8 (20.4–23.2) | — | — | — | — | — | — | 22.8 (1321) | 33.1 (3966) | 30.5 (29.3–31.7) | 27.1 (1521) | 32.9 (3293) | 31.1 (29.8–32.4) |
| 704 | — | — | — | — | — | — | — | — | — | — | — | 38.1 (1523) | 35.9 (1518) | 37.0 (35.3–38.7) | 23.3 (1716) | 27.3 (1374) | 25.1 (23.6–26.3) | |
| 721 | — | — | — | — | — | — | — | — | — | — | — | — | 48.3 (1050) | 48.2 (1404) | 48.2 (46.2–50.2) | 46.9 (1208) | 49.3 (2564) | 48.5 (46.9–50.1) |
| 779 | — | — | — | — | — | — | — | — | — | — | — | — | 49.1 (1952) | 51.2 (1899) | 50.1 (48.5–51.7) | 53.1 (1780) | 50.1 (1845) | 51.6 (50.0–53.2) |
| 745 | 46.5 (5449) | 48.0 (5195) | 47.2 (46.3–48.2) | — | — | — | — | — | — | — | — | — | 49.2 (1127) | 51.4 (1940) | 50.6 (48.8–52.4) | 45.4 (1714) | 50.2 (3998) | 48.7 (47.4–50.0) |
| 777 | 93.9 (1871) | 95.0 (3330) | 94.6a (94.0–95.2) | 48.2 (1929) | 48.6 (1023) | 48.3 (46.5–50.1) | — | — | — | — | — | — | 45.3 (1742) | 50.6 (2925) | 48.6 (47.2–50.0) | 49.2 (1121) | 48.7 (2865) | 48.8 (47.3–50.4) |
Abbreviations: CI, confidence interval; WM, weighted mean. Variant frequencies, expressed in percentages, represent the weighted mean values calculated on the number of forward and reverse reads. Each amplicon has been sequenced twice (1, 2). Highlighted in dark grey, the weighted mean values calculated on the total of reads number of the two experiments. Highlighted in light grey, mosaic cases.
The fraction of normal allele detected in tumor probably depends on contamination with normal cells during scraping.
Discussion
In disorders such as RB, associated with a high rate of de novo mutations, mosaicism is not expected as a rare event.23 Accordingly, since the late seventies, an increased number of patients with mosaicism of the RB1 gene has been reported.10, 11, 12, 13 The reported incidence of mosaicism in RB has become increasingly accurate with the advancement of technologies employed for RB1 variant analysis. In particular, next generation sequencing technology, based on single-molecule counting, allows to detect even mosaic variants with low frequencies.16 In the present study, we re-tested by deep sequencing 40 sporadic RB cases (11 bilateral and 29 unilateral) in order to investigate the presence of mosaicism. In these samples, traditional methods (Sanger sequencing and MLPA) had previously identified 10 RB1 sequence variants (9 bilateral and 1 unilateral cases). Next generation sequencing was performed in the blood and in the additional available tissues (ocular tissues, urine and/or oral mucosa).
The present study detected a significant percentage (10%) of patients with sporadic RB and no apparent germline mutations in RB1 that are actually cases of mosaicism showing low variant frequency (8–24%) in the blood. These results are in accordance with a previous study reporting that the lowest proportion of the RB1 mutant alleles detectable unambiguously by Sanger sequencing ranges between 15 and 30%, depending on the specific change and sequence context.13 Unfortunately, among unilateral samples, we had the availability of only five tumor tissues for mutation analysis. This is a limit compared with the previous study by Chen et al,13 but it can represent a not infrequent situation in a diagnostic setting. The limited availability of tumor tissues was partially due to the fact that enucleation has decreased in favor of conservative treatments, including the recent superselective ophthalmic artery infusion chemotherapy promoted by Italian radiologists and ophthalmologists.24 The added value of deep sequencing with respect to Sanger analysis is that it allows for more sensitive and accurate detection of mosaic variants even without hints from the tumor. In our series, thanks to the employment of next generation sequencing, detection rate changed from 3 to 10% in unilateral cases and from 82 to 91% in bilateral cases.
The failure to identify RB1 mutations in the other samples, in particular the remaining bilateral case, could be due to the presence of mosaic sequence variants present at lower frequencies with respect to the LODs of our assay (Supplementary Figure S1) or to the presence of variants limited to tissues that were not tested in the present study.13, 25 Alternatively, deep intronic splice mutations in regions not included in the amplicon library might be responsible for failed identification.26, 27 Finally, mosaic large rearrangements of RB1 were not investigated by our approach.
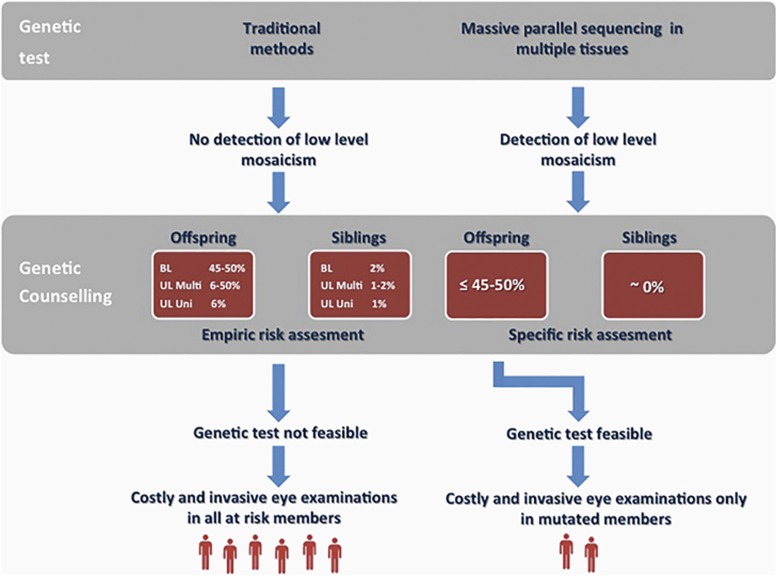
When the tumor tissue is not available, the identification of low-frequency mosaic variants in DNA isolated from the blood of patients is an important finding for genetic counseling. The failure to identify the low-level mosaic mutations by traditional methods would have led to give empirical risks based on tumor presentation (ie, unifocal or multifocal) to the family (Figure 1, left). The identification of the mutation instead is sufficient to propose testing in future offspring and make early identification of tumors without costly and invasive (under general anesthesia) eye examinations in those at-risk family members who have not inherited the mutation (Figure 1, right). Concerning the recurrence risk of the couple with the affected child, it is close to zero and thus lower than that attributable to a de novo germline mutation (Figure 1).
Figure 1.
Implications of detection of low-frequency mosaic variants in sporadic retinoblastoma patients. Screening of the RB1 gene performed by next generation sequencing in multiple tissues allows the detection of low-frequency mosaic variants (right), while this is not possible by traditional sequencing (left). This has important implications in genetic counseling. When an RB1 mutation is not detected in DNA from leukocytes of the proband, empirical risks are given for siblings and offspring depending on tumor presentation (bilateral, unilateral multifocal, unilateral unifocal). Genetic test is not feasible and frequent eye examination under anesthesia are performed in all at-risk members. When a low-frequency mosaic variant is detected, specific recurrence risks are established for siblings and offspring. Genetic test is performed only in offspring and frequent eye examination are performed only in members who have inherited mutation. BL, bilateral; UL, unilateral; Multi, multifocal; Uni, unifocal.
Present results show for the first time that the use of next generation sequencing combined with analysis in multiple tissues can reveal a significant fraction (3/7; 43%) of patients with high-level mosaicism in the blood who have previously been interpreted as cases with constitutional germline mutations. In fact, using deep sequencing, we were able to demonstrate that three cases (samples 768, 762 and 704) interpreted on the basis of Sanger electropherograms as heterozygotes were indeed mosaic cases (read frequencies of 36.6, 37.6 and 45.6%). This observation was confirmed by the analysis in other available tissues in which deep sequencing detected the same variants at variable percentages (Table 5). These results are in accordance with a recent paper showing that amplicon deep sequencing is more accurate for the quantification of mosaic variants and that Sanger sequencing gives estimations of higher percentages of mutated alleles.25 We therefore concluded that all these cases derived from deleterious postzygotic events instead of germline mutations. Depending on the time of occurrence during embryonic development, mosaicism is expressed at variable percentages in different tissues. When blood – that is, the tissue that is generally collected for DNA analysis – is the tissue that eventually shows a variant frequency close to 50%, misinterpretation with a constitutional variant may easily occur.
Following these results, genetic test in siblings is not proposed. Moreover, the recurrence risk to offspring can be <45–50% depending on the level of germline involvement in mosaicism. Quantitative analysis in sperm may be helpful.12 However, as the germline cells may not be stable over time, the best approach would be to perform the test in close proximity to child conception.11, 28, 29, 30
Reaching the average depth (~2500 × ) required for the identification of low-level mosaicism leads to an increase in costs (~800 Euros/sample) that become comparable to the costs of traditional analysis. However, deep sequencing significantly reduces working time and molecular diagnosis is achieved in 5 days instead of ~30-40 days necessary for conventional analysis in our country.
In conclusion, we identified a significant fraction (6/40; 15%) of mosaic patients among RB sporadic cases. Concerning clinical presentation, mosaic cases included the same number of unilateral and bilateral cases. These findings have to be taken into serious consideration in genetic counseling, where mosaicism has important implications for both recurrence risk assessment and family care. Moreover, this study confirms the higher sensitivity of deep sequencing technology in comparison with traditional methods for the detection and quantification of low-frequency mosaic variants. Finally, this paper highlights the importance of collecting other tissues in addition to blood, because they can be relevant to confirm somatic mosacism suspected by next generation sequencing.
Acknowledgments
We thank all the families who participated in this study. We also thank Professor Lucio Luzzatto for critical reading of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- 1Gallie BL, Campbell C, Devlin H, Duckett A, Squire JA: Developmental basis of retinal-specific induction of cancer by RB mutation. Cancer Res 1999; 59: 1731s–1735s. [PubMed] [Google Scholar]
- 2Donaldson SS, Smith LM: Retinoblastoma: biology, presentation, and current management. Oncology 1989; 3: 45–51, discussion 51–2. [PubMed] [Google Scholar]
- 3Knudson AG Jr.: Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA 1971; 68: 820–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Dimaras H, Khetan V, Halliday W et al: Loss of RB1 induces non-proliferative retinoma: increasing genomic instability correlates with progression to retinoblastoma. Hum Mol Genet 2008; 17: 1363–1372. [DOI] [PubMed] [Google Scholar]
- 5Sampieri K, Amenduni M, Papa FT et al: Array comparative genomic hybridization in retinoma and retinoblastoma tissues. Cancer Sci 2009; 100: 465–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Vogel F: Genetics of retinoblastoma. Hum Genet 1979; 52: 1–54. [DOI] [PubMed] [Google Scholar]
- 7Dimaras H, Kimani K, Dimba EA et al: Retinoblastoma. Lancet 2012; 379: 1436–1446. [DOI] [PubMed] [Google Scholar]
- 8Eng C, Li FP, Abramson DH et al: Mortality from second tumors among long-term survivors of retinoblastoma. J Natl Cancer Inst 1993; 85: 1121–1128. [DOI] [PubMed] [Google Scholar]
- 9Dryja TP, Morrow JF, Rapaport JM: Quantification of the paternal allele bias for new germline mutations in the retinoblastoma gene. Hum Genet 1997; 100: 446–449. [DOI] [PubMed] [Google Scholar]
- 10Carlson EA, Desnick RJ: Mutational mosaicism and genetic counseling in retinoblastoma. Am J Med Genet 1979; 4: 365–381. [DOI] [PubMed] [Google Scholar]
- 11Sippel KC, Fraioli RE, Smith GD et al: Frequency of somatic and germ-line mosaicism in retinoblastoma: implications for genetic counseling. Am J Hum Genet 1998; 62: 610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Rushlow D, Piovesan B, Zhang K et al: Detection of mosaic RB1 mutations in families with retinoblastoma. Hum Mutat 2009; 30: 842–851. [DOI] [PubMed] [Google Scholar]
- 13Chen Z, Moran K, Richards-Yutz J et al: Enhanced sensitivity for detection of low-level germline mosaic RB1 mutations in sporadic retinoblastoma cases using deep semiconductor sequencing. Hum Mutat 2014; 35: 384–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Biesecker LG, Spinner NB: A genomic view of mosaicism and human disease. Nat Rev Genet 2013; 14: 307–320. [DOI] [PubMed] [Google Scholar]
- 15Valverde JR, Alonso J, Palacios I, Pestana A: RB1 gene mutation up-date, a meta-analysis based on 932 reported mutations available in a searchable database. BMC Genetics 2005; 6: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Rohlin A, Wernersson J, Engwall Y, Wiklund L, Bjork J, Nordling M: Parallel sequencing used in detection of mosaic mutations: comparison with four diagnostic DNA screening techniques. Hum Mutat 2009; 30: 1012–1020. [DOI] [PubMed] [Google Scholar]
- 17Pagnamenta AT, Lise S, Harrison V et al: Exome sequencing can detect pathogenic mosaic mutations present at low allele frequencies. J Hum Genet 2012; 57: 70–72. [DOI] [PubMed] [Google Scholar]
- 18Beicht S, Strobl-Wildemann G, Rath S et al: Next generation sequencing as a useful tool in the diagnostics of mosaicism in Alport syndrome. Gene 2013; 526: 474–477. [DOI] [PubMed] [Google Scholar]
- 19Pritchard CC, Smith C, Marushchak T et al: A mosaic PTEN mutation causing Cowden syndrome identified by deep sequencing. Genet Med 2013; 15: 1004–1007. [DOI] [PubMed] [Google Scholar]
- 20Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G: Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res 2002; 30: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Nichols KE, Houseknecht MD, Godmilow L et al: Sensitive multistep clinical molecular screening of 180 unrelated individuals with retinoblastoma detects 36 novel mutations in the RB1 gene. Hum Mutat 2005; 25: 566–574. [DOI] [PubMed] [Google Scholar]
- 22Margulies M, Egholm M, Altman WE et al: Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005; 437: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Wijsman EM: Recurrence risk of a new dominant mutation in children of unaffected parents. Am J Hum Genet 1991; 48: 654–661. [PMC free article] [PubMed] [Google Scholar]
- 24Venturi C, Bracco S, Cerase A et al: Superselective ophthalmic artery infusion of melphalan for intraocular retinoblastoma: preliminary results from 140 treatments. Acta Ophthalmol 2013; 91: 335–342. [DOI] [PubMed] [Google Scholar]
- 25Nota B, Hamilton EM, Sie D et al: Novel cases of D-2-hydroxyglutaric aciduria with IDH1 or IDH2 mosaic mutations identified by amplicon deep sequencing. J Med Genet 2013; 50: 754–759. [DOI] [PubMed] [Google Scholar]
- 26Dehainault C, Michaux D, Pages-Berhouet S et al: A deep intronic mutation in the RB1 gene leads to intronic sequence exonisation. Eur J Hum Genet 2007; 15: 473–477. [DOI] [PubMed] [Google Scholar]
- 27Zhang K, Nowak I, Rushlow D, Gallie BL, Lohmann DR: Patterns of missplicing caused by RB1 gene mutations in patients with retinoblastoma and association with phenotypic expression. Hum Mutat 2008; 29: 475–484. [DOI] [PubMed] [Google Scholar]
- 28Orye E, Benoit Y, Coppieters R et al: A case of retinoblastoma, associated with histiocytosis-X and mosaicism of a deleted D-group chromosome (13q14 leads to q31). Clin Genet 1982; 22: 37–39. [PubMed] [Google Scholar]
- 29Motegi T, Minoda K: A decreasing tendency for cytogenetic abnormality in peripheral lymphocytes of retinoblastoma patients with 13q14 deletion mosaicism. Hum Genet 1984; 66: 186–189. [DOI] [PubMed] [Google Scholar]
- 30Ribeiro MC, Andrade JA, Erwenne CM, Brunoni D: Bilateral retinoblastoma associated with 13q-mosaicism. Possible manifestation of a germinal mutation. Cancer Genet Cytogenet 1988; 32: 169–175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.