Abstract
Germline KLLN promoter hypermethylation was recently identified as a potential genetic etiology of the cancer predisposition syndrome, Cowden syndrome (CS), when no causal PTEN gene mutation was found. We screened for KLLN promoter methylation in a large prospective series of CS patients and determined the risk of benign and malignant CS features in patients with increased methylation both with and without a PTEN mutation/variant of unknown significance. In all, 1012 CS patients meeting relaxed International Cowden Consortium criteria including 261 PTEN mutation-positive CS patients, 187 PTEN variant-positive CS patients and 564 PTEN mutation-negative CS patients, as well as 111 population controls were assessed for germline KLLN promoter methylation by MassARRAY EpiTYPER analysis. KLLN promoter methylation was analyzed both as a continuous and a dichotomous variable in the calculation of phenotypic risks by stepwise logistic regression and Kaplan–Meier/standardized incidence ratio methods, respectively. Significantly increased KLLN promoter methylation was seen in CS individuals with and without a PTEN mutation/VUS compared with controls (P<0.001). Patients with high KLLN promoter methylation have increased risks of all CS-associated malignancies compared with the general population. Interestingly, KLLN-associated risk of thyroid cancer appears to be gender and PTEN status dependent. KLLN promoter methylation associated with different benign phenotypes dependent on PTEN status. Furthermore, increasing KLLN promoter methylation is associated with a greater phenotype burden in mutation-negative CS patients. Germline promoter hypermethylation of KLLN is associated with particular malignant and benign CS features, which is dependent on the PTEN mutation status.
Introduction
Cowden syndrome (CS) is a dominantly inherited cancer predisposition syndrome with protean phenotypes including both malignant and benign features.1, 2 A large prospective study of CS patients revealed PTEN mutation-associated lifetime risks of 85% for breast cancer, 35% for thyroid cancer, 28% for endometrial cancer, 33% for renal cancer, 9% for colorectal cancer, and 6% for melanoma.3 Similar risks have been replicated in independent studies.4, 5 These studies have helped to revise appropriately diagnostic and screening guidelines for patients with PTEN mutations.
Because CS is difficult to recognize, our lab developed a weighted regression-based system, resulting in a PTEN Cleveland Clinic (CC) score. A higher CC score reflects a higher phenotypic burden, occurring at younger ages, and hence, denotes a higher pretest probability of carrying a predisposing mutation.6 Although the CC score was based on the study of adult patients, Tan et al.6 also developed pediatric criteria for consideration of testing for PTEN mutations. Approximately 25% of classic CS cases carry pathogenic PTEN mutations when accrued prospectively from the community.6 Patients who do not meet the full diagnostic criteria for CS but have combinations of CS-associated features are referred to as CS-like (CSL). Only ~5% of CSL patients harbor a PTEN mutation, making molecular diagnosis, predictive testing of family members, and preventative screening difficult.7 To estimate neoplasia risk in PTEN mutation-negative patients, we need to identify as many etiologic factors as possible. To this end, we previously identified germline hypermethylation of the shared KLLN/PTEN promoter, which decreased KLLN expression by 250-fold, and in contrast, slightly increased PTEN expression in 37% of 123 PTEN mutation-negative CS and CSL patients.8 KLLN is a p53-regulated tumor suppressor gene.9 When KLLN methylation was reversed with demethylating agents in patient lymphoblasts, there was a restoration of KLLN expression.10 Thus, KLLN promoter methylation was identified as potentially significant for the development of CS.
Our initial studies linking KLLN promoter methylation to CS/CSL were performed with a small series of selected PTEN mutation-negative patients using a binary technique to determine methylation (COBRA). To fully characterize the broad spectrum of clinical features potentially associated with KLLN promoter methylation, we sought to semiquantitatively analyze KLLN promoter methylation in a large independent series of PTEN mutation-negative adult and pediatric CS/CSL patients and characterize the clinical spectrum and lifetime cancer risks of patients with germline KLLN promoter methylation. We also sought to address the hypothesis that KLLN promoter methylation and a PTEN mutation are not mutually exclusive in CS/CSL patients and that KLLN promoter methylation modifies the PTEN phenotype.
Materials and Methods
Patients
From October 2005 to April 2013, 4120 CS/CSL patients were enrolled into CC IRB protocol no. 8458. Research participants were enrolled based on the full NCCN 2006 criteria for CS (Supplementary Table 1) or relaxed criteria for CSL. Patients under the age of 18 years met the pediatric criteria adapted from Tan et al.6 (Supplementary Table 2). Both affected and unaffected family members of probands were also enrolled when possible. Of the 4120 prospectively enrolled CS/CSL patients, 1012 with complete methylation data were analyzed in this study, including 261 PTEN mutation-positive, 187 PTEN variant-positive and 564 PTEN mutation-negative CS patients. One hundred and eleven controls from the region with no known cancer history were recruited through CC IRB protocol no. 06-716. All patients provided written informed consent. Patients were selected based on PTEN status, CC score, and age. Inclusion criteria included individuals with a germline pathogenic PTEN mutation, PTEN mutation-negative patients with a CC score above 6 or high clinical suspicion, or all available minors meeting described pediatric criteria. Pathological and imaging reports were used to establish the phenotype when necessary and mandatorily since 1 July 2009. Twelve unaffected family members belonging to 11 probands were available for analysis. Because 37% of the pilot series of PTEN mutation-negative CS/CSL patients were previously found to harbor germline KLLN promoter methylation, this study should be powered to detect methylation in CS/CSL patients and more precisely delineate the phenotype of CS/CSL patients associated with KLLN promoter methylation.8
Variant analysis
Germline genomic DNA was extracted from peripheral blood leukocytes by the Genomic Medicine Biorepository (GMB) (GMB protocols available at http://www.lerner.ccf.org/gmi/gmb/methods.php). The human reference genome used for mutation annotation was build 36 version 2, NC_000010.9. Mutation analysis encompassed all nine exons of PTEN and flanking intronic regions, using a high-resolution, melt-curve assay (Idaho Technology, Salt Lake City, UT, USA), followed by direct Sanger sequencing (ABI 3730xl; Applied Biosystems, Foster City, CA, USA) as described previously.6 The PTEN minimal promoter region was directly sequenced. Large deletion/rearrangement analysis was performed using multiplex ligation-dependent probe amplification (MLPA) following the P225 MLPA kit protocol (MRC-Holland, Amsterdam, The Netherlands). Of the 1012, 261 CS/CSL patients with available germline DNA and a known pathogenic germline PTEN mutation, encompassing large deletion, missense, indel, frameshift, truncation, and splice site mutations were included in this study. All mutations, including missense ones, within the coding region of PTEN and flanking intronic regions (±1 and 2) are pathogenic, and typically show decreased PTEN protein levels, increased P-AKT, and/or P-ERK1/2.6 A further 187 CS/CSL patients have a PTEN variant including variants in the minimal promoter region, synonymous changes, large intronic deletions detected by MLPA, and non-splice site intronic variants within −10 bases from exon/intron boundary. If a patient has both a known pathogenic PTEN mutation and a variant, they are only included in the group with known pathogenic mutations. The remaining 564 CS/CSL patients were PTEN mutation negative.
Methylation analysis
Methylation analysis was performed using 750 ng of germline genomic DNA extracted from peripheral blood lymphocytes. To ensure that the methylation assay was specific to PTEN and not the pseudogene, the restriction site PvuII (New England BioLabs, Ipswich, MA, USA) unique to the PTEN pseudogene was used before bisulfite treatment. Three independent digestions with PvuII to remove the pseudogene followed by bisulfite sequencing showed 2/28 (7.14%), 4/15 (26.7%), and 3/15 (20.0%) of clones to be PTEN pseudogene. These percentages of PTEN pseudogene were determined to have a negligible effect on further MassARRAY EpiTYPER analysis. Digested DNA was bisulfite treated according to the EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA) as per the manufacturer's instructions.
Targeted methylation analysis of the KLLN promoter (−1579 to −1908, KLLN ATG +1) was analyzed for methylation with the MassARRAY EpiTYPER (Sequenom, San Diego, CA, USA) system. Briefly, bisulfite-treated DNA was PCR amplified, followed by in vitro transcription with T7 polymerase, base-specific digestion, and data acquisition. Each 384-well plate of samples was run with a range of methylation controls to ensure between-plate variation data correction. Of the 33 CpGs in the amplicon, 11 CpGs had informative methylation percentages using the EpiTYPER analyses workflow. The average percent methylation for all 11 CpGs for each sample was used for further analysis.
For bisulfite sequencing, bisulfite-treated DNA was PCR amplified and the product gel purified using the GeneJET Gel Extraction Kit (Thermo Scientific, Pittsburg, PA, USA). Purified product was cloned according to the StrataClone PCR Cloning Kit (Agilent Technologies, Santa Clara, CA, USA) and plated on ampicillin resistance plates. Colonies were picked for each sample and PCR amplified using T7/T3 primers, followed by Exonuclease 1 (New England Biolabs) digestion and direct Sanger sequencing (ABI 3730xl; Applied Biosystems)
Statistical analysis
Analyses were performed using SPSS statistical software (IBM, Armonk, NY, USA). Methylation was analyzed as a continuous variable using the Kruskal–Wallis rank-sum test. For each phenotype analyzed, incidence was assumed to be 0 if the data was not provided. For analysis of benign phenotypes, a stepwise multivariate linear regression was used to calculate coefficients and 95% confidence intervals. One PTEN mutation-positive CS patient was not included in the phenotype analysis because no phenotype data was provided.
To analyze methylation as a dichotomous variable, samples were assigned high versus low methylation status based on a cutoff of 32% of regional-CpG methylation; 32% reflects the boundary for outlier values, which have methylation percentages above 1.5 times the interquartile range from the 75th percentile for the entire CS/CSL cohort (Supplementary Figure 1). This cutoff is higher and more rigorously defined than the commonly used methylation cutoff of 25%.11, 12, 13 Kaplan–Meier curves were constructed for each major CS/CSL cancer phenotype for cumulative risk assessment, with significance determined by a generalized Wilcoxon's test. To analyze thyroid histology likelihood ratio, χ2 was used to determine differences between groups.
The standardized incidence ratio (SIR) is a comparison of the observed number of cases of malignancy for those with KLLN promoter hypermethylation with the expected number in the general population. The expected number of cases for each cancer phenotype was calculated using the Surveillance Epidemiology and End Results age-adjusted incidence rates from the National Cancer Institute from 2003 to 2007.12 Gender-specific incidence rates are used for breast, endometrial, and thyroid cancer. The number of observed person-years is calculated starting at birth until the age of cancer diagnosis or age at consent. The expected incidence is calculated using the person-years of observation multiplied by age-specific incidence. The SIR and two-sided mid-P exact test with 95% confidence intervals was calculated using the OpenEpi software program (http://www.openepi.com).
There were 51 families comprising 102 affected and 12 unaffected family members. Concordance of methylation between family members with CS was calculated with Kendall's coefficient of concordance by pairwise analysis between proband and first- and second-degree relatives. Similarly, concordance of methylation was examined between CS/CSL-affected proband and first- and second-degree relatives who were unaffected. All P-values were two sided.
Results
KLLN promoter methylation in PTEN mutation-positive, mutation-negative, and VUS-positive CS/CSL patients compared with controls
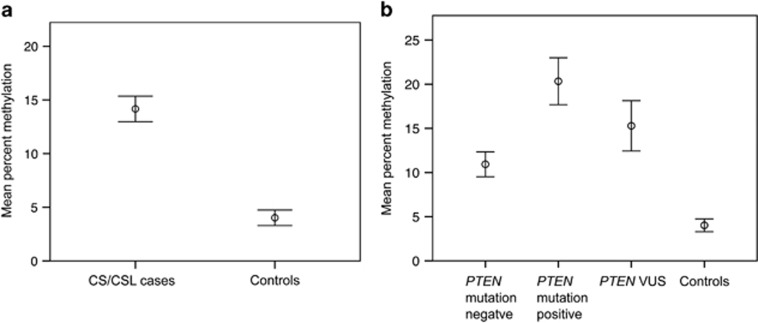
MassARRAY EpiTYPER analysis of the KLLN promoter for methylation shows significantly increased methylation in 1012 CS/CSL patients with a mean methylation of 14.3% compared with 111 population controls with a mean methylation of 4.03% (P<0.001, Kruskal–Wallis rank-sum test; Figure 1a). KLLN promoter methylation was significantly increased above controls for all three CS/CSL subgroups defined by PTEN mutation status: mutation-negative (n=564, P<0.001), mutation-positive (n=261, P<0.001), and VUS-positive (n=187, P<0.001; Figure 1b). Interestingly, KLLN promoter methylation was strongly associated with PTEN mutation status: 20.6% mean methylation for mutation-positive, 15.6% PTEN VUS, and 11.0% PTEN mutation-negative patients (P<0.001, Kruskal–Wallis rank-sum test; Figure 1b and Table 1).
Figure 1.
KLLN promoter methylation was analyzed in CS/CSL patients and controls. (a) The mean methylation percentage is increased in 1012 CS/CSL cases compared with 111 controls. Error bars represent 95% confidence intervals. (b) Mean percent methylation is increased in 564 PTEN mutation-negative patients, 261 PTEN mutation-positive patients, and 187 PTEN VUS-positive patients compared with 111 controls. Error bars represent 95% confidence intervals.
Table 1. Clinical and demographic features in CS/CSL patients and KLLN promoter methylation.
| Characteristic | No. of patients | % Patients | Mean methylation % (SE) |
|---|---|---|---|
| Gender | |||
| Female | 779 | 77.0 | 13.9 (0.69) |
| Male | 233 | 23.0 | 15.7 (1.32) |
| P-value | 0.14 | ||
| Race/ethnicity | |||
| Caucasian | 594 | 58.7 | 13.6 (0.77) |
| African American | 29 | 2.87 | 15.8 (3.88) |
| Hispanic | 31 | 3.06 | 9.13 (2.46) |
| Other | 97 | 9.58 | 13.3 (1.90) |
| Unknown | 261 | 25.8 | 16.8 (1.34) |
| P-value | 0.12 | ||
| Age (years) | |||
| <18 | 160 | 15.8 | 13.7 (1.46) |
| 18–50 | 451 | 44.6 | 15.0 (0.97) |
| >50 | 401 | 39.6 | 13.7 (0.94) |
| P-value | 0.60 | ||
| CC score | |||
| ≤15 | 729 | 72.0 | 11.6 (0.65) |
| >15 | 282 | 27.9 | 21.3 (1.33) |
| P-value | <0.001 | ||
| PTEN status | |||
| Mut. negative | 564 | 55.7 | 11.0 (0.72) |
| Pathogenic mut. | 261 | 25.8 | 20.5 (1.36) |
| VUS | 187 | 18.5 | 15.6 (1.49) |
| P-value | <0.001 | ||
Note: Kruskal–Wallis rank-sum test was used to determine significance.
Nine CS/CSL cases representative of low or high KLLN promoter methylation and four controls were subjected to bisulfite sequencing to further validate the MassARRAY EpiTYPER results, with good concordance (Supplementary Figure 2). All four controls show a lack of methylation in the bisulfite sequencing results (Supplementary Figure 3).
KLLN promoter methylation and clinical and demographic features in CS/CSL patients
For the entire CS/CSL cohort, no significant association was observed between KLLN promoter methylation and patient demographic features, including gender, race/ethnicity, and age (Table 1). Notably, KLLN promoter methylation was associated with CC scores >15, denoting a greater phenotypic burden (P<0.001; Table 1). In planned subgroup analysis, the association between hypermethylation and CC score was seen in the PTEN-mutation-negative group alone (P<0.001).
SIR for CS-related malignancies in CS patients with high germline KLLN promoter methylation
We calculated the SIR for each component cancer in CS/CSL patients with high KLLN promoter methylation (>32%CpG island methylated, see Materials and methods). The SIR for each component cancer was significantly elevated (Table 2). The most significantly increased risk was for male thyroid cancer with an SIR of 176.8 (95% CI: 86.2–324.5).
Table 2. SIR for CS-related malignancies in patients with high KLLN promoter methylation.
| Cancer type | Observed | Expected | SIR | 95% CI | P-value |
|---|---|---|---|---|---|
| Female breast | 72 | 2.82 | 25.53 | 20.12–31.97 | <0.001 |
| Female thyroid | 36 | 0.831 | 43.32 | 30.8–59.33 | <0.001 |
| Male thyroid | 9 | 0.0509 | 176.8 | 86.23–324.5 | <0.001 |
| All thyroid | 45 | 0.645 | 69.77 | 51.5–92.53 | <0.001 |
| Endometrial | 20 | 0.643 | 31.1 | 19.53–47.19 | <0.001 |
| Renal | 12 | 0.485 | 24.77 | 13.41–42.06 | <0.001 |
| Colorectal | 4 | 0.721 | 5.55 | 1.76–13.38 | 0.007 |
| Melanoma | 5 | 0.829 | 6.03 | 2.21–13.37 | 0.002 |
Relationship between KLLN promoter methylation and PTEN mutation/VUS with respect to risk of CS-related malignancies
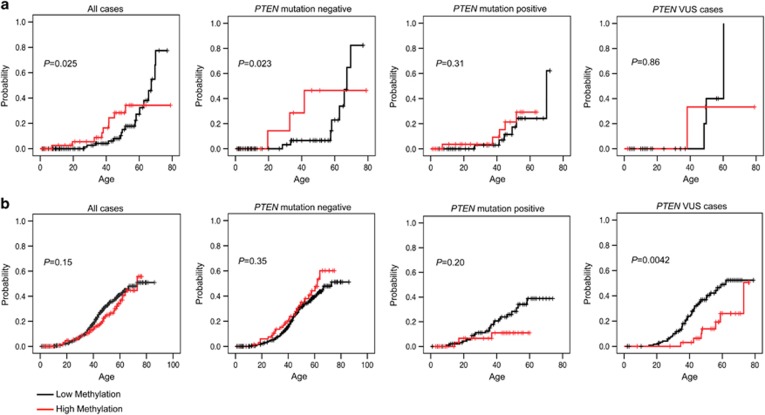
To analyze the relationship among KLLN promoter methylation status, PTEN mutation status, and age-adjusted risk of developing the most common CS-associated malignancies, specifically, breast, thyroid, endometrial, and renal carcinomas, KLLN promoter methylation was analyzed as a dichotomous variable (>32% methylated versus <32%). Of 1012 total patients, 197 (19.5%) have high KLLN promoter methylation. This is broken down as 76 of 564 (13.5%) PTEN mutation-negative, 81 of 260 (31.2%) PTEN mutation-positive, and 40 of 187 (21.4%) PTEN VUS cases. Only thyroid cancer showed significant differences in cumulative risk between CS/CSL patients with high KLLN promoter methylation and those with low KLLN promoter methylation (P=0.025; Figure 2 and Supplementary Figure 4). The association remained significant in the PTEN mutation-negative subgroup (P=0.023, Figure 2a). Additionally, there was a significant difference in the thyroid histologies of CS/CSL male patients with high KLLN promoter methylation compared with those with low methylation (P=0.039; Supplementary Figure 5). Of 17 male patients with thyroid cancer and low KLLN promoter methylation, 9 (53%) had papillary thyroid cancer compared with only 1 (5.9%) with follicular histology. In contrast, among the nine male patients with high KLLN promoter methylation and thyroid cancer, two (22%) had papillary thyroid cancer and three (33%) follicular thyroid cancer. For male patients with thyroid cancer, PTEN mutation status was not significantly different in those with high versus low KLLN promoter methylation.
Figure 2.
Kaplan–Meier curves for cumulative risk of thyroid cancer between patients with low compared with high KLLN promoter methylation. (a) An increased risk of thyroid cancer was observed in male patients with high KLLN promoter methylation for all cases and for PTEN mutation-negative cases. No significant differences in risk of thyroid cancer in males were observed for PTEN mutation-positive or PTEN VUS cases. (b) For females, no significant differences in thyroid cancer risk based on KLLN promoter methylation were seen for all cases, PTEN mutation-negative cases, or PTEN mutation-positive cases. A trend can be observed for increased risk for thyroid cancer with low KLLN methylation for all cases. For the PTEN VUS cases, there was a significant increase in thyroid cancer risk for females with low KLLN promoter methylation compared with high.
For all CS/CSL female patients with thyroid cancer, the data suggest a reverse trend of increased cumulative incidence of thyroid cancer cases associating with low KLLN promoter methylation compared with those with high methylation (P=0.15; Figure 2b). This difference became significant in the PTEN VUS subgroup (P=0.004; Figure 2b). No differences in thyroid cancer histology are seen in female CS/CSL patients with high versus low KLLN promoter methylation.
Multivariate analysis of benign CS features and KLLN promoter methylation
Stepwise multivariate linear regression was performed to determine whether KLLN promoter methylation, as a continuous variable, is associated with benign CS features (Supplementary Table 3) for the overall cohort and the three PTEN status subgroups. For the entire CS/CSL cohort and specifically the PTEN mutation-negative group, KLLN promoter methylation was positively associated with three benign CS features, trichilemmomas, lipomas, and vascular growths (Table 3). Remarkably, trichilemmomas, which are a pathognomonic feature of CS, are associated with increasing KLLN methylation for the entire cohort as well as the PTEN mutation-negative and PTEN mutation-positive subgroups (Table 3, Supplementary Table 4 and Supplementary Figure 6). Autism and acral keratoses are inversely associated with KLLN methylation for the PTEN mutation-negative subgroup. Interestingly, decreased prevalence of goiters/thyroid nodules correlated with increasing KLLN methylation in the PTEN mutation-positive subgroup (Table 3).
Table 3. Benign CS features associated with KLLN promoter methylation.
| Coefficient | 95% CI | P-value | |
|---|---|---|---|
| All CS/CSL cases, n=1011 | |||
| Trichilemmoma | 10.3 | 6.18 to 14.4 | <0.001 |
| Lipoma | 4.17 | 1.45 to 6.89 | 0.0027 |
| Hemangioma/AVM/vasc. tumor | 4.09 | 1.10 to 7.08 | 0.0073 |
| PTEN mutation-negative cases, n=564 | |||
| Trichilemmoma | 9.68 | 4.29 to 15.1 | <0.001 |
| Lipoma | 4.56 | 1.26 to 7.87 | 0.0068 |
| Autism | −5.54 | −10.2 to −0.90 | 0.019 |
| Hemangioma/AVM/vasc. tumor | 4.57 | 0.53 to 8.61 | 0.027 |
| Acral keratoses | −5.46 | −10.4 to −0.51 | 0.031 |
| PTEN mutation-positive cases, n=260 | |||
| Trichilemmoma | 14.8 | 7.89 to 21.8 | <0.001 |
| Goiter/nodule | −5.92 | −11.2 to −0.60 | 0.029 |
| PTEN VUS cases, n=187 | |||
| Fibroids | 7.74 | 1.63 to 13.8 | 0.013 |
Note: Stepwise multivariate linear regression was used to determine significance.
Family pattern of KLLN methylation status
Pairwise analysis revealed concordance of KLLN methylation between 41 unrelated CS/CSL probands and 51 of their first- and second-degree relatives who also have CS/CSL (Supplementary Figure 7; P=0.39, Kendall's coefficient of concordance). There are 11 probands with 12 unaffected first-degree relatives (Supplementary Figure 7). In these 11 families, the methylation status was discordant between affected proband and the unaffected relative (P=0.035, Kendall's coefficient of concordance). All unaffected relatives have low/no KLLN methylation, and this holds true even when the affected proband has high KLLN methylation.
Discussion
Through this large case series of 1012 CS/CSL patients, we were able to show significantly increased germline KLLN promoter methylation in CS/CSL patients (mean methylation 14.29%) compared with controls (4.03%, P<0.001). Increasing methylation correlates with increasing PTEN CC score, and hence the phenotypic load, specifically in the PTEN mutation-negative subgroup. Furthermore, KLLN methylation status was overall concordant for 41 affected families, corroborating KLLN as a CS/CSL predisposition gene, which acts by epigenetic regulation. Notably, this study demonstrates for the first time that KLLN promoter hypermethylation and PTEN variation are not mutually exclusive, suggesting the potential for KLLN methylation to modify PTEN-related phenotypes.
KLLN promoter methylation was significantly associated with PTEN mutation status with the greatest mean methylation of 20.6% for the PTEN mutation-positive CS/CSL patients (Table 1). The relationship between PTEN variation and KLLN promoter methylation may be understood by the effect of mutations on decreasing transcriptional activity, which in turn alters methylation.14 It is possible that CS/CSL patients with KLLN hypermethylation but without a detectable PTEN mutation may carry cryptic alterations such as deep intronic mutations. It is known that genes with decreased expression are more likely to be methylated.14 PTEN variations can lead to decreased PTEN transcription through effects on transcription factor binding and decreased p53 stability.15 Because PTEN and KLLN share a bidirectional promoter, the potential effect of PTEN mutations on gene expression could explain the increase in KLLN promoter methylation with a PTEN mutation.14 The possibility of a direct relationship between KLLN promoter methylation and PTEN variation requires further investigation.
Our large prospective series of patients revealed specific KLLN epigenotype–malignant phenotype association. As expected for a CS predisposition gene, increased incidence of all CS-related cancers was evident in those with high KLLN promoter methylation compared with the general population. KLLN-related SIRs for CS-component cancers were similarly high as previously reported for patients with PTEN mutations.3 Interestingly, the highest KLLN-related SIR occurred for male thyroid cancer. In our original pilot study, there was elevation of male thyroid cancer SIR with KLLN hypermethylation compared with females, but the sample size in that study was too small to make definitive conclusions about gender differences.16
For thyroid cancer, we were able to further tease apart the role of KLLN promoter methylation in modifying gender-dependent risk and association with PTEN mutation/VUS. High KLLN promoter methylation was associated with an increased age-adjusted cumulative risk of thyroid cancer in CS/CSL males, whereas the association appears reversed in women, where KLLN promoter methylation trended with decreased cumulative risk of thyroid cancer. Interestingly, the lowest risk for thyroid cancer was for female patients with high KLLN promoter methylation and a PTEN VUS, which tend to be in the promoter region and confer a high risk of breast cancer.3 The gender paradox is tantalizing in the context that thyroid cancer is more prevalent among women than men in the general population.17, 18 Early reports link differences in androgen receptor (AR) expression in male versus female thyroid cancers to the gender bias in thyroid cancer risk in the general population.17 Of relevance, bidirectional AR-binding motifs lie between KLLN and PTEN.19 In prostate cancer, KLLN suppresses AR activity, and AR itself decreases KLLN and PTEN transcription; whereas in female breast cancer, AR was found to activate KLLN and PTEN expression.20, 21 It would be plausible to postulate that increasing KLLN promoter methylation, further transcriptionally suppressing KLLN, is dose additive in the male context, thus raising the likelihood of developing thyroid cancer. Understanding KLLN and AR expression in thyroid cancer may further elucidate the gender-specific risks of thyroid cancer in the context of KLLN promoter methylation.
We saw significant differences in thyroid cancer histology between male patients with high versus low KLLN methylation (P=0.039), with no significant differences in PTEN mutation status between these groups. Papillary thyroid cancer is the predominant histological subtype in the general population by about 14:1, whereas follicular thyroid cancer is predominant (PTC:FTC 3:1) among PTEN mutation-positive CS patients.16, 22, 23 The thyroid cancer histology for the high KLLN promoter methylation group was predominantly follicular, which is classic for CS, while the low KLLN promoter methylation group resembled the general population with the predominant histology being papillary. These data indicate that similar thyroid cancer surveillance to those with a PTEN mutation may be important for male patients with KLLN promoter methylation with specific attention paid to histology.
KLLN promoter methylation modified the risk of specific non-malignant CS phenotypes dependent on the PTEN mutation context. Specifically, trichilemmonas, which are hamartomas and a pathognomonic feature of CS, are associated with increased KLLN promoter methylation for patients both with and without a PTEN mutation. For PTEN mutation-negative cases, high KLLN promoter methylation was also associated with lipomas and vascular malformations, which are abnormal overgrowths, and for the latter, may be considered a hamartoma as well. This suggests a role for KLLN in maintaining proper cellular proliferation. While vascular malformations have been noted as part of the CS phenotype, they are not specifically included in the NCCN diagnostic criteria.6 Surveillance and management of vascular malformations may be particularly important for patients with KLLN promoter methylation. Additionally, there is a decreased risk of autism with KLLN promoter hypermethylation. These findings help to delineate gene-testing strategies based on phenotype and to construct gene-specific patient management for CS. It will be important to understand how KLLN promoter hypermethylation mediates risk of specific CS phenotypes, in the presence or absence of PTEN abnormalities to aid in the development of precise phenotype prediction at the individual level, beyond the current cohort level.
This study was able to identify specific CS phenotypes associated with KLLN promoter hypermethylation but would miss yet unrecognized CS features, or CS-unrelated clinical features, which were associated with KLLN hypermethylation. We have stratified our cohort based on PTEN mutation status. While we have analyzed patients for PTEN mutations, we may be missing mutations, which were not identified, in our mutation screening. Given our careful mutation screening technique, we believe this would be too small of a number of cases to influence the percentage methylation in each group. We have chosen to use MassARRAY EpiTYPER methylation analysis because it is a high-throughput method for candidate region analysis at a base-specific resolution that provides reliable quantifiable results.24 The threshold % methylation used here to assign high compared to low methylation is based on >1000 patients but with continuing patient analysis, the cutoff may need to be adjusted for future clinical utility.
Overall, our observations reveal that germline KLLN methylation is associated with CS/CSL phenotypes, with particular association with increasing phenotypic burden (CC score >15) in those without a PTEN mutation. Because KLLN methylation is associated with CC scores >15, after these findings have been repeated, KLLN methylation testing may be applicable for patients with CC scores over 15 and are PTEN mutation negative. The finding of trichilemmomas may be a significant indicator for KLLN promoter methylation testing. Furthermore, our data here allow us to counsel KLLN methylation-positive patients of the very elevated risks of CS-component cancers, with attendant high-risk medical management. Specifically, in patients who are PTEN mutation negative but found to have high KLLN promoter methylation, surveillance for both male thyroid cancer and vascular malformations may be important to potentially reduce morbidity and mortality. Our study clearly demonstrates the complex interplay among PTEN, KLLN and gender in neoplasia risks and suggests further study in this regard.
Acknowledgments
This work was supported, in part, by the National Cancer Institute (P01CA124570 to CE) and the Breast Cancer Research Foundation (to CE). CE is the Sondra J and Stephen R Hardis Endowed Chair of Cancer Genomic Medicine at the Cleveland Clinic, and an ACS Clinical Research Professor. EN was supported, in part, by National Institute of General Medicine (T32 GM007250) and is supported by National Cancer Institute (F30CA168151).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- 1Eng C: Will the real Cowden syndrome please stand up: revised diagnostic criteria. J Med Genet 2000; 37: 828–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Hobert JA, Eng C: PTEN hamartoma tumor syndrome: an overview. Genet Med 2009; 11: 687–694. [DOI] [PubMed] [Google Scholar]
- 3Tan MH, Mester JL, Ngeow J, Rybicki LA, Orloff MS, Eng C: Lifetime cancer risks in individuals with germline PTEN mutations. Clin Cancer Res 2012; 18: 400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Bubien V, Bonnet F, Brouste V et al: High cumulative risks of cancer in patients with PTEN hamartoma tumour syndrome. J Med Genet 2013; 50: 255–263. [DOI] [PubMed] [Google Scholar]
- 5Nieuwenhuis MH, Kets CM, Murphy-Ryan M et al: Cancer risk and genotype–phenotype correlations in PTEN hamartoma tumor syndrome. Fam Cancer 2013; 13: 57–63. [DOI] [PubMed] [Google Scholar]
- 6Tan MH, Mester J, Peterson C et al: A clinical scoring system for selection of patients for PTEN mutation testing is proposed on the basis of a prospective study of 3042 probands. Am J Hum Genet 2011; 88: 42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Marsh DJ, Dahia PL, Caron S et al: Germline PTEN mutations in Cowden syndrome-like families. J Med Genet 1998; 35: 881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Bennett KL, Mester J, Eng C: Germline epigenetic regulation of KILLIN in Cowden and Cowden-like syndrome. JAMA 2010; 304: 2724–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Cho YJ, Liang P: Killin is a p53-regulated nuclear inhibitor of DNA synthesis. Proc Natl Acad Sci USA 2008; 105: 5396–5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Bennett KL, Campbell R, Ganapathi S et al: Germline and somatic DNA methylation and epigenetic regulation of KILLIN in renal cell carcinoma. Genes Chromosomes Cancer 2011; 50: 654–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Seisenberger S, Andrews S, Krueger F et al: The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell 2012; 48: 849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Haworth KE, Farrell WE, Emes RD et al: Combined influence of gene-specific cord blood methylation and maternal smoking habit on birth weight. Epigenomics 2013; 5: 37–49. [DOI] [PubMed] [Google Scholar]
- 13Alhejaily A, Day AG, Feilotter HE, Baetz T, Lebrun DP: Inactivation of the CDKN2A tumor-suppressor gene by deletion or methylation is common at diagnosis in follicular lymphoma and associated with poor clinical outcome. Clin Cancer Res 2014; 20: 1676–1686. [DOI] [PubMed] [Google Scholar]
- 14Jones PA: Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012; 13: 484–492. [DOI] [PubMed] [Google Scholar]
- 15Tang Y, Eng C: PTEN autoregulates its expression by stabilization of p53 in a phosphatase-independent manner. Cancer Res 2006; 66: 736–742. [DOI] [PubMed] [Google Scholar]
- 16Ngeow J, Mester J, Rybicki LA, Ni Y, Milas M, Eng C: Incidence and clinical characteristics of thyroid cancer in prospective series of individuals with Cowden and Cowden-like syndrome characterized by germline PTEN, SDH, or KLLN alterations. J Clin Endocrinol Metab 2011; 96: E2063–E2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Stanley JA, Aruldhas MM, Chandrasekaran M et al: Androgen receptor expression in human thyroid cancer tissues: a potential mechanism underlying the gender bias in the incidence of thyroid cancers. J Steroid Biochem Mol Biol 2012; 130: 105–124. [DOI] [PubMed] [Google Scholar]
- 18Siegel R, Naishadham D, Jemal A: Cancer statistics, 2013CA Cancer J Clin 2013; 63: 11–30. [DOI] [PubMed] [Google Scholar]
- 19Wang Y, Romigh T, He X et al: Differential regulation of PTEN expression by androgen receptor in prostate and breast can cers. Oncogene 2011; 30: 4327–4338. [DOI] [PubMed] [Google Scholar]
- 20Wang Y, He X, Yu Q, Eng C: Androgen receptor-induced tumor suppressor, KLLN, inhibits breast cancer growth and transcriptionally activates p53/p73-mediated apoptosis in breast carcinomas. Hum Mol Genet 2013; 22: 2263–2272. [DOI] [PubMed] [Google Scholar]
- 21Wang Y, Radhakrishnan D, He X, Peehl DM, Eng C: Transcription factor KLLN inhibits tumor growth by AR suppression, induces apoptosis by TP53/TP73 stimulation in prostate carcinomas, and correlates with cellular differentiation. J Clin Endocrinol Metab 2013; 98: E586–E594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Harach HR, Soubeyran I, Brown A, Bonneau D, Longy M: Thyroid pathologic findings in patients with Cowden disease. Ann Diagn Pathol 1999; 3: 331–340. [DOI] [PubMed] [Google Scholar]
- 23Laury AR, Bongiovanni M, Tille JC, Kozakewich H, Nose V: Thyroid pathology in PTEN-hamartoma tumor syndrome: characteristic findings of a distinct entity. Thyroid 2011; 21: 135–144. [DOI] [PubMed] [Google Scholar]
- 24Olkhov-Mitsel E, Bapat B: Strategies for discovery and validation of methylated and hydroxymethylated DNA biomarkers. Cancer Med 2012; 1: 237–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.