Abstract
The tribbles protein family, an evolutionarily conserved group of pseudokinases, have been shown to regulate multiple cellular events including those involved in normal and malignant haematopoiesis. The three mammalian Tribbles homologues, Trib1, Trib2 and Trib3 are characterized by conserved motifs, including a pseudokinase domain and a C-terminal E3 ligase-binding domain. In this review, we focus on the role of Trib (mammalian Tribbles homologues) proteins in mammalian haematopoiesis and leukaemia. The Trib proteins show divergent expression in haematopoietic cells, probably indicating cell-specific functions. The roles of the Trib proteins in oncogenesis are also varied and appear to be tissue-specific. Finally, we discuss the potential mechanisms by which the Trib proteins preferentially regulate these processes in multiple cell types.
Keywords: haematopoiesis, leukaemia, tribbles
Tribbles
Tribbles proteins belong to an evolutionarily conserved protein family that is implicated in diverse cellular events including proliferation, metabolism and oncogenic transformation. Tribbles was first characterized in Drosophila as an important cell cycle regulator [1,2]. The three mammalian Tribbles homologues, Trib1, Trib2 and Trib3 [3] are characterized by a conserved pseudokinase domain [4] and a C-terminal E3 ligase-binding domain [5]. Tribbles proteins function as scaffolding molecules that facilitate the degradation of proteins via a proteasome-dependent mechanism. In Drosophila, tribbles mediates the degradation of the cell division cycle 25 homolog A (CDC25a) homologue string, which regulates cell cycle progression [1,6] and the CCAAT/enhancer-binding protein (C/EBP) homologue slow border cells (slbo) which functions in oocyte migration [7]. In mammals, Trib1 and Trib2 promote C/EBPα and C/EBPβ degradation by recruiting the E3 ligase, Caspase recruitment domain-containing protein 16 (CARD16, also known and defined in this article as COP1) [8,9]. Similarly, Trib3 promotes COP1-dependent degradation of acetyl CoA carboxylase (ACC), an enzyme involved in fatty acid synthesis [5]. TRIB proteins also modulate signalling pathways such as protein kinase B (PKB, also known and defined in this article as AKT) [9,10] and Mitogen-activated protein kinase (MAPK) [11] (Figure 1). However, it is unclear if these functions are related to protein degradation or alternative TRIB functions.
Figure 1. The Trib family members regulate protein function.
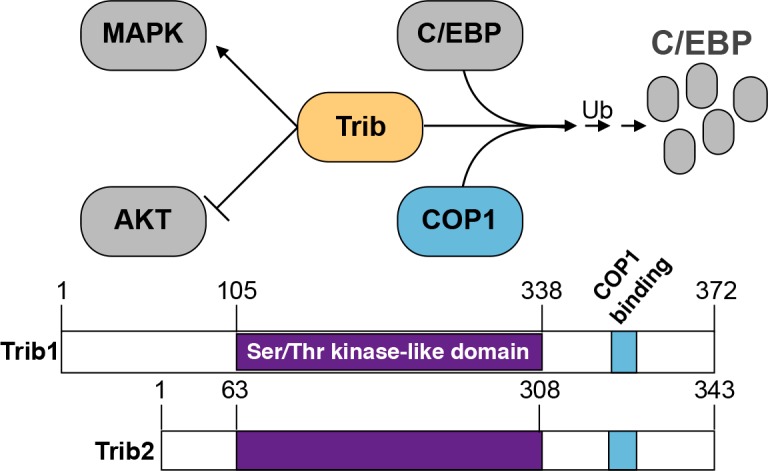
Trib proteins facilitate the degradation of C/EBP in a COP1-dependent manner. Additionally, Trib inhibits AKT phosphorylation and activates MAPK signalling.
Tribbles in normal haematopoiesis
The expression of the Trib genes in mammals is widely divergent, suggesting the compartmentalization of their function. Trib3 is largely expressed by non-haematopoietic tissues and therefore will not be discussed in this review. In contrast, both Trib1 and Trib2 are expressed in haematopoietic lineage cells; in mice, Trib1 is predominantly expressed in myeloid lineage cells, whereas Trib2 has a lymphoid bias [12]. This separation is not absolute as Trib2 is moderately expressed in the common myeloid progenitor (CMP) and megakaryocyte/erythroid progenitor (MEP; Figure 2). It is important to note that these data are derived from mRNA expression as antibodies capable of detecting endogenous expression are still being developed.
Figure 2. Variable expressions of Trib1 and Trib2 mRNA in haematopoietic cells.
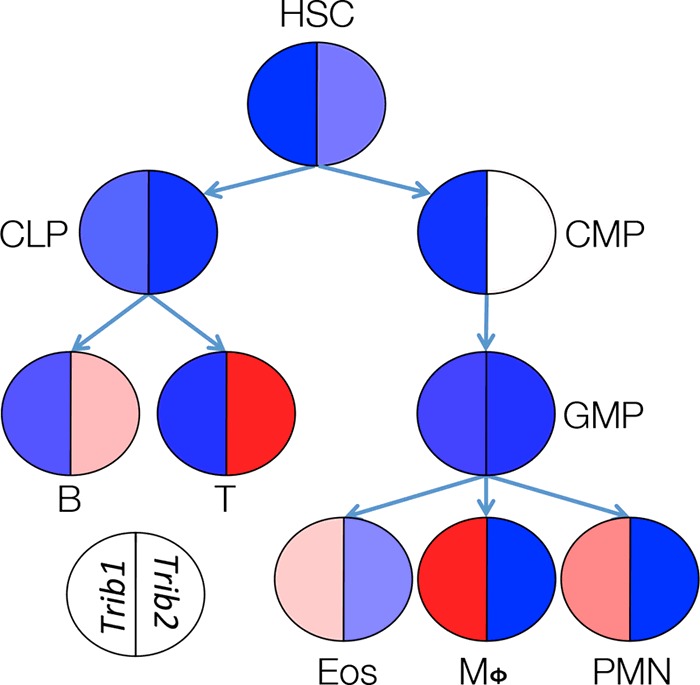
The expression data were obtained from the Immgen database [12]. Red denotes high expression, blue denotes low expression.
Although the expression of Trib1 and Trib2 is largely segregated between the myeloid and lymphoid lineages, there is evidence that both affect myeloid development. It was initially noted that retroviral expression of Trib2 in murine bone marrow progenitors reduced the percentage of granulocytes and concomitantly increased the number of monocytes prior to the development of acute myeloid leukemia (AML). This phenomenon correlated with decreased C/EPBα p42 protein expression with minimal impact on C/EPBα p30 [8]. Although increased Trib2 expression can affect myelopoiesis, the physiological relevance of this finding is uncertain as myeloid defects have not been described in Trib2 knockout mice [13]. Furthermore, given the absence of Trib2 expression in the majority of myeloid progenitors and mature cells, it is unclear what affect this protein has on normal myelopoiesis.
Trib1 is expressed in myeloid cells and probably regulates development of this lineage. A recent study by Satoh et al. [13] examined the role of Trib1 in myeloid development in a knockout mouse model. The results showed that Trib1 loss is associated with a decrease in splenic macrophage populations, including ‘M2’ biased cells [13]. In addition, these mice also exhibited an expanded peripheral neutrophil population and absence of eosinophils. These phenotypic changes correlated with increased C/EBPα protein expression, highlighting the potential importance of the interaction between Trib1 and C/EBP family members in myelopoiesis. Furthermore, these alterations promoted the development of lipodystrophy and metabolic disturbance, which could be rescued by transferring wild-type macrophages. The data, to date, suggest a role for Trib1 in physiologic myelopoiesis; however, overexpressed Trib2 may affect myelopoiesis in pathologic conditions [8]. C/EBPα appears to be connected to these phenotypes; however, this does not rule out the possibility that additional Trib-regulated signalling pathways are involved. For example, Trib proteins in mice and humans interact with the MAPK pathway [11,14], suggesting a potential role in cell proliferation. Further, Trib2 can inhibit AKT in a model of adipocyte differentiation [9] and Trib3 is also known to inhibit AKT activation (Figure 1) [10].
Tribbles in leukaemia
Acute myeloid leukemia
Despite the role of Drosophila tribbles as a negative regulator of the cell cycle, which suggested that it functioned as a tumour suppressor, exogenous expression of murine Trib1 or Trib2 is sufficient to induce AML in vivo, a process that requires COP1-mediated proteasomal degradation of C/EBPα [15,16]. Deletion of C/EBPα resulting in its complete absence in vivo leads to the development of a myeloproliferative disorder [17], whereas expression of the p30 C/EBPα isoform drives AML [18]. Therefore, an important function of Trib proteins in AML pathogenesis is the degradation or modification of C/EBPα that results in an altered ratio of the p42 and p30 C/EBPα isoforms. These findings are consistent with studies showing that TRIB1 and TRIB2 are highly expressed in molecularly-defined sub-types of human AML. However, distinct differences between TRIB1 and TRIB2 are apparent in AML pathogenesis. Mice transplanted with haematopoietic progenitors expressing Trib1 develop leukaemia more quickly (116±28 days) than those that receive cells expressing Trib2 (174±21 days) [19]. Additionally, increased TRIB2 expression is limited to a distinct subset of AML patients in which C/EBPα is not mutated, but displays decreased function [8]. In contrast, TRIB1 is highly expressed among multiple sub-types of AML that are not grouped by C/EBPα status including those driven by inv(16) or t(16;16) as well as M4 and M5 as compared with healthy controls [20,21].
The mechanistic role of Trib1 in AML is unclear. Trib1 is known to collaborate with HoxA7 and HoxA9 to decrease AML latency in mouse models [22], raising the possibility that TRIB1 and HOXA7/HOXA9 co-operate in human AML. Additionally, Trib1 and MEK1 interact, leading to the phosphorylation of ERK and the degradation of C/EBPα [23]. A TRIB1 gain of function mutation, R107L, was identified in human acute megakaryocytic leukaemia, the overexpression of which enhances Erk phosphorylation and accelerates the onset of AML in vivo [24]. These data suggest that the leukaemogenic functions of TRIB1 and TRIB2 are not identical.
T-cell acute lymphoblastic leukemia
Trib2 was first identified as a Neurogenic Locus Notch Homolog Protein homologue 1 (NOTCH1) target in murine T-ALL [8,25] and subsequent gene expression studies show that TRIB2 is highly expressed in T-ALL [21]. High TRIB2 expression was identified in T-ALL and correlates with NOTCH1/FBXW7 (F-box/WD repeat-containing protein 7, which is involved in Notch turnover) mutations in paediatric cases [26]. Additionally, studies showed that Trib2 is also a direct transcriptional target of TAL-1 and is required for the growth and survival of human T-ALL cell lines. The studies of Sanda et al. [27] show that TRIB2 is oppositely regulated by the TAL1 and immunoglobulin enhancer-binding factors E12/E47 (E2A) and Transcription Factor 12 (TCF12/HEB) transcription factors, where the former activates transcription and the latter represses. Their data showing that inhibiting TRIB2 leads to apoptosis and growth arrest of multiple human T-ALL cell lines suggest that TRIB2 may be a therapeutic target in T-ALL.
Perspectives on the oncogenic mechanisms of tribbles proteins
An oncogenic role for Trib proteins was first uncovered in a mouse model of AML. Recipients of haematopoietic progenitors overexpressing Trib2 developed AML with a median latency of 179 days [8]. Subsequent studies showed that Trib1 acts co-operatively with HoxA9 and Meis1 in a murine model of AML [22] and that the overexpression of Trib1, but not Trib3, in haematopoietic progenitors also induces AML in vivo [19]. Mechanistically, the Trib proteins promote leukaemogenesis by facilitating the degradation of C/EBPα [8] through interaction with the E3 ubiquitin ligase COP1 [15,16,19]. Additionally, Trib1 interacts with the MAPK pathway member MEK1 to drive disease [23].
The Trib proteins are also implicated as oncogenic factors in solid tumours, though the role of these proteins appears to be tumour dependent. TRIB1 was shown to be essential in the growth and survival of prostate cancer cells [28,29]. Trib2 is highly expressed in melanoma cells and promotes oncogenesis by inhibiting FOXO proteins [30] and was identified as a potential biomarker of melanoma progression [31]. Additionally, Trib2 was identified as a potential driver of lung tumorigenesis through C/EBPα inhibition [32]. Trib3 is highly expressed in human lung, colon and breast tumours [33]. Additionally, high Trib3 expression is associated with poor prognosis in breast cancer patients [34].
Despite having a potential oncogenic role in leukaemia and solid tumours, Trib proteins are also implicated as tumour suppressors. A recent study by Salazar et al. [35] showed that the genetic deletion of Trib3 accelerated progression mouse skin papillomas, resulting in a more aggressive phenotype. The mechanism of enhanced tumorigenesis was largely dependent on the de-regulated phosphorylation of AKT and the subsequent inactivation of the FOXO3 transcription factor [35]. Another study showed that TRIB1 interacts with p53 to suppress its transcriptional activity by enhancing deacetylation and decreasing DNA binding, resulting in increased cell viability [36].
In summary, the accumulating data support an important function for Trib proteins in oncogenesis. The precise function is unlikely to be stereotypical and will probably depend on context. For example, Trib proteins have the capacity to function as both dominant oncogenes and tumour suppressors. Although Trib inhibition of C/EBPα may be an important mechanism in some tumour types, there are data implicating other signalling pathways, such as MAPK and AKT. Clearly, further studies are needed to identify the downstream targets of oncogenic Trib signalling as well as targets during normal haematopoiesis, which may be distinct. We anticipate that ongoing studies will reveal both the mechanisms by which Trib functions in oncogenic transformation and in which tumours it may be therapeutic target.
Abbreviations
- AML
acute myeloid leukemia
- C/EBP
CCAAT/enhancer-binding protein
- MAPK
Mitogen-activated protein kinase
- T-ALL
T-cell acute lymphoblastic leukemia
- Trib
Tribbles mammalian homologue
Footnotes
Tribbles Pseudokinases on the Crossroads of Metabolism, Cancer, Immunity and Development: Held at The Aquincum Hotel, Budapest, 22–24 April 2015
Funding
This work was supported by the National Institute of Health [grant numbers T32 CA009140 (to S.J.S.), T32 HL0074 and T32 GM0071 (to E.A.M.) and F31CA189661 (K.S.R.)]; the Samuel Waxman Foundation for Cancer Research; the Alex's Lemonade Stand Foundation; the American Cancer Society [grant number PF-15-065-01-TBG (to (S.J.S.)]; the National Science Foundation [grant number DGE-1321851 (to K.S.R.)].
References
- 1.Grosshans J., Wieschaus E. A genetic link between morphogenesis and cell division during formation of the ventral furrow in Drosophila. Cell. 2000;101:523–531. doi: 10.1016/S0092-8674(00)80862-4. [DOI] [PubMed] [Google Scholar]
- 2.Seher T.C., Leptin M. Tribbles, a cell-cycle brake that coordinates proliferation and morphogenesis during Drosophila gastrulation. Curr. Biol. 2000;10:623–629. doi: 10.1016/S0960-9822(00)00502-9. [DOI] [PubMed] [Google Scholar]
- 3.Tang K., Finley R.L., Jr, Nie D., Honn K.V. Identification of 12-lipoxygenase interaction with cellular proteins by yeast two-hybrid screening. Biochemistry. 2000;39:3185–3191. doi: 10.1021/bi992664v. [DOI] [PubMed] [Google Scholar]
- 4.Hegedus Z., Czibula A., Kiss-Toth E. Tribbles: a family of kinase-like proteins with potent signalling regulatory function. Cell Signal. 2007;19:238–250. doi: 10.1016/j.cellsig.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 5.Qi L., Heredia J.E., Altarejos J.Y., Screaton R., Goebel N., Niessen S., Macleod I.X., Liew C.W., Kulkarni R.N., Bain J., et al. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science. 2006;312:1763–1766. doi: 10.1126/science.1123374. [DOI] [PubMed] [Google Scholar]
- 6.Mata J., Curado S., Ephrussi A., Rorth P. Tribbles coordinates mitosis and morphogenesis in Drosophila by regulating string/CDC25 proteolysis. Cell. 2000;101:511–522. doi: 10.1016/S0092-8674(00)80861-2. [DOI] [PubMed] [Google Scholar]
- 7.Rorth P., Szabo K., Texido G. The level of C/EBP protein is critical for cell migration during Drosophila oogenesis and is tightly controlled by regulated degradation. Mol. Cell. 2000;6:23–30. doi: 10.1016/S1097-2765(05)00008-0. [DOI] [PubMed] [Google Scholar]
- 8.Keeshan K., He Y., Wouters B.J., Shestova O., Xu L., Sai H., Rodriguez C.G., Maillard I., Tobias J.W., Valk P., et al. Tribbles homolog 2 inactivates C/EBPalpha and causes acute myelogenous leukemia. Cancer Cell. 2006;10:401–411. doi: 10.1016/j.ccr.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naiki T., Saijou E., Miyaoka Y., Sekine K., Miyajima A. TRB2, a mouse Tribbles ortholog, suppresses adipocyte differentiation by inhibiting AKT and C/EBPbeta. J. Biol. Chem. 2007;282:24075–24082. doi: 10.1074/jbc.M701409200. [DOI] [PubMed] [Google Scholar]
- 10.Du K., Herzig S., Kulkarni R.N., Montminy M. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 2003;300:1574–1577. doi: 10.1126/science.1079817. [DOI] [PubMed] [Google Scholar]
- 11.Kiss-Toth E., Bagstaff S.M., Sung H.Y., Jozsa V., Dempsey C., Caunt J.C., Oxley K.M., Wyllie D.H., Polgar T., Harte M., et al. Human tribbles, a protein family controlling mitogen-activated protein kinase cascades. J. Biol. Chem. 2004;279:42703–42708. doi: 10.1074/jbc.M407732200. [DOI] [PubMed] [Google Scholar]
- 12.Heng T.S., Painter M.W. Immunological Genome Project, C. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 13.Satoh T., Kidoya H., Naito H., Yamamoto M., Takemura N., Nakagawa K., Yoshioka Y., Morii E., Takakura N., Takeuchi O., Akira S. Critical role of Trib1 in differentiation of tissue-resident M2-like macrophages. Nature. 2013;495:524–528. doi: 10.1038/nature11930. [DOI] [PubMed] [Google Scholar]
- 14.Eder K., Guan H., Sung H.Y., Ward J., Angyal A., Janas M., Sarmay G., Duda E., Turner M., Dower S.K., et al. Tribbles-2 is a novel regulator of inflammatory activation of monocytes. Int. Immunol. 2008;20:1543–1550. doi: 10.1093/intimm/dxn116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keeshan K., Bailis W., Dedhia P.H., Vega M.E., Shestova O., Xu L., Toscano K., Uljon S.N., Blacklow S.C., Pear W.S. Transformation by Tribbles homolog 2 (Trib2) requires both the Trib2 kinase domain and COP1 binding. Blood. 2010;116:4948–4957. doi: 10.1182/blood-2009-10-247361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida A., Kato J.Y., Nakamae I., Yoneda-Kato N. COP1 targets C/EBPalpha for degradation and induces acute myeloid leukemia via Trib1. Blood. 2013;122:1750–1760. doi: 10.1182/blood-2012-12-476101. [DOI] [PubMed] [Google Scholar]
- 17.Zhang D.E., Zhang P., Wang N.D., Hetherington C.J., Darlington G.J., Tenen D.G. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirstetter P., Schuster M.B., Bereshchenko O., Moore S., Dvinge H., Kurz E., Theilgaard-Monch K., Mansson R., Pedersen T.A., Pabst T., et al. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 2008;13:299–310. doi: 10.1016/j.ccr.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 19.Dedhia P.H., Keeshan K., Uljon S., Xu L., Vega M.E., Shestova O., Zaks-Zilberman M., Romany C., Blacklow S.C., Pear W.S. Differential ability of Tribbles family members to promote degradation of C/EBPalpha and induce acute myelogenous leukemia. Blood. 2010;116:1321–1328. doi: 10.1182/blood-2009-07-229450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rothlisberger B., Heizmann M., Bargetzi M.J., Huber A.R. TRIB1 overexpression in acute myeloid leukemia. Cancer Genet. Cytogenet. 2007;176:58–60. doi: 10.1016/j.cancergencyto.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 21.Liang K.L., Rishi L., Keeshan K. Tribbles in acute leukemia. Blood. 2013;121:4265–4270. doi: 10.1182/blood-2012-12-471300. [DOI] [PubMed] [Google Scholar]
- 22.Jin G., Yamazaki Y., Takuwa M., Takahara T., Kaneko K., Kuwata T., Miyata S., Nakamura T. Trib1 and Evi1 cooperate with Hoxa and Meis1 in myeloid leukemogenesis. Blood. 2007;109:3998–4005. doi: 10.1182/blood-2006-08-041202. [DOI] [PubMed] [Google Scholar]
- 23.Yokoyama T., Kanno Y., Yamazaki Y., Takahara T., Miyata S., Nakamura T. Trib1 links the MEK1/ERK pathway in myeloid leukemogenesis. Blood. 2010;116:2768–2775. doi: 10.1182/blood-2009-10-246264. [DOI] [PubMed] [Google Scholar]
- 24.Yokoyama T., Toki T., Aoki Y., Kanezaki R., Park M.J., Kanno Y., Takahara T., Yamazaki Y., Ito E., Hayashi Y., Nakamura T. Identification of TRIB1 R107L gain-of-function mutation in human acute megakaryocytic leukemia. Blood. 2012;119:2608–2611. doi: 10.1182/blood-2010-12-324806. [DOI] [PubMed] [Google Scholar]
- 25.Wouters B.J., Jorda M.A., Keeshan K., Louwers I., Erpelinck-Verschueren C.A., Tielemans D., Langerak A.W., He Y., Yashiro-Ohtani Y., Zhang P., et al. Distinct gene expression profiles of acute myeloid/T-lymphoid leukemia with silenced CEBPA and mutations in NOTCH1. Blood. 2007;110:3706–3714. doi: 10.1182/blood-2007-02-073486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hannon M.M., Lohan F., Erbilgin Y., Sayitoglu M., O'Hagan K., Mills K., Ozbek U., Keeshan K. Elevated TRIB2 with NOTCH1 activation in paediatric/adult T-ALL. Br. J. Haematol. 2012;158:626–634. doi: 10.1111/j.1365-2141.2012.09222.x. [DOI] [PubMed] [Google Scholar]
- 27.Sanda T., Lawton L.N., Barrasa M.I., Fan Z.P., Kohlhammer H., Gutierrez A., Ma W., Tatarek J., Ahn Y., Kelliher M.A., et al. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell. 2012;22:209–221. doi: 10.1016/j.ccr.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mashima T., Soma-Nagae T., Migita T., Kinoshita R., Iwamoto A., Yuasa T., Yonese J., Ishikawa Y., Seimiya H. TRIB1 supports prostate tumorigenesis and tumor-propagating cell survival by regulation of endoplasmic reticulum chaperone expression. Cancer Res. 2014;74:4888–4897. doi: 10.1158/0008-5472.CAN-13-3718. [DOI] [PubMed] [Google Scholar]
- 29.Lin Z.Y., Huang Y.Q., Zhang Y.Q., Han Z.D., He H.C., Ling X.H., Fu X., Dai Q.S., Cai C., Chen J.H., et al. MicroRNA-224 inhibits progression of human prostate cancer by downregulating TRIB1. Int. J. Cancer. 2014;135:541–550. doi: 10.1002/ijc.28707. [DOI] [PubMed] [Google Scholar]
- 30.Zanella F., Renner O., Garcia B., Callejas S., Dopazo A., Peregrina S., Carnero A., Link W. Human TRIB2 is a repressor of FOXO that contributes to the malignant phenotype of melanoma cells. Oncogene. 2010;29:2973–2982. doi: 10.1038/onc.2010.58. [DOI] [PubMed] [Google Scholar]
- 31.Hill R., Kalathur R.K., Colaco L., Brandao R., Ugurel S., Futschik M., Link W. TRIB2 as a biomarker for diagnosis and progression of melanoma. Carcinogenesis. 2015;36:469–477. doi: 10.1093/carcin/bgv002. [DOI] [PubMed] [Google Scholar]
- 32.Grandinetti K.B., Stevens T.A., Ha S., Salamone R.J., Walker J.R., Zhang J., Agarwalla S., Tenen D.G., Peters E.C., Reddy V.A. Overexpression of TRIB2 in human lung cancers contributes to tumorigenesis through downregulation of C/EBPalpha. Oncogene. 2011;30:3328–3335. doi: 10.1038/onc.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowers A.J., Scully S., Boylan J.F. SKIP3, a novel Drosophila tribbles ortholog, is overexpressed in human tumors and is regulated by hypoxia. Oncogene. 2003;22:2823–2835. doi: 10.1038/sj.onc.1206367. [DOI] [PubMed] [Google Scholar]
- 34.Wennemers M., Bussink J., Scheijen B., Nagtegaal I.D., van Laarhoven H.W., Raleigh J.A., Varia M.A., Heuvel J.J., Rouschop K.M., Sweep F.C., Span P.N. Tribbles homolog 3 denotes a poor prognosis in breast cancer and is involved in hypoxia response. Breast Cancer Res. 2011;13:R82. doi: 10.1186/bcr2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salazar M., Lorente M., Garcia-Taboada E., Perez Gomez E., Davila D., Zuniga-Garcia P., Flores J., Rodriguez A., Hegedus Z., Mosen-Ansorena D., et al. Loss of Tribbles pseudokinase-3 promotes Akt-driven tumorigenesis via FOXO inactivation. Cell Death Differ, Maria. 2015;22:131–144. doi: 10.1038/cdd.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyajima C., Inoue Y., Hayashi H. Pseudokinase Tribbles 1 (TRB1) negatively regulates tumor-suppressor activity of p53 through p53 deacetylation. Biol. Pharm. Bull. 2015;38:618–624. doi: 10.1248/bpb.b15-00003. [DOI] [PubMed] [Google Scholar]


