The present study establishes poly(ADP-ribose)polymerase's (PARP's) role in chronic asthma, demonstrates that it is activated in human asthma, increases the clinical relevance of targeting PARP for blocking or preventing chronic asthma in humans and presents olaparib as a likely candidate drug.
Keywords: eosinophilia, house dust mite, poly(adenosine 5′-diphosphate-ribose)polymerase (PARP) inhibition, T helper 2 (Th2) cytokines, olaparib (AZD2281)
Abstract
Our laboratory established a role for poly(ADP-ribose)polymerase (PARP) in asthma. To increase the clinical significance of our studies, it is imperative to demonstrate that PARP is actually activated in human asthma, to examine whether a PARP inhibitor approved for human testing such as olaparib blocks already-established chronic asthma traits in response to house dust mite (HDM), a true human allergen, in mice and to examine whether the drug modulates human cluster of differentiation type 4 (CD4+) T-cell function. To conduct the study, human lung specimens and peripheral blood mononuclear cells (PBMCs) and a HDM-based mouse asthma model were used. Our results show that PARP is activated in PBMCs and lung tissues of asthmatics. PARP inhibition by olaparib or gene knockout blocked established asthma-like traits in mice chronically exposed to HDM including airway eosinophilia and hyper-responsiveness. These effects were linked to a marked reduction in T helper 2 (Th2) cytokine production without a prominent effect on interferon (IFN)-γ or interleukin (IL)-10. PARP inhibition prevented HDM-induced increase in overall cellularity, weight and CD4+ T-cell population in spleens of treated mice whereas it increased the T-regulatory cell population. In CD3/CD28-stimulated human CD4 +T-cells, olaparib treatment reduced Th2 cytokine production potentially by modulating GATA binding protein-3 (gata-3)/IL-4 expression while moderately affecting T-cell proliferation. PARP inhibition inconsistently increased IL-17 in HDM-exposed mice and CD3/CD28-stimulated CD4+ T cells without a concomitant increase in factors that can be influenced by IL-17. In the present study, we provide evidence for the first time that PARP-1 is activated in human asthma and that its inhibition is effective in blocking established asthma in mice.
CLINICAL PERSPECTIVES
-
•
Our laboratory pioneered the studies that described the role of PARP in asthma and examined the underlying mechanisms by which PARP participates in the diseases using OVA-based animal models. To increase the clinical significance of our findings, it was imperative to demonstrate that PARP is activated in asthma and that its inhibition by a clinically relevant drug prevents or modulates chronic asthma traits using an animal model that resembles the human condition.
-
•
In the present study, we show that PARP-1 is activated in human asthma and that PARP inhibition genetically or by olaparib blocks chronic asthma traits in mice that were chronically exposed to HDM extract. Such effect was related to a modulation of CD4+ T-cell function and an increase in T-reg cells.
-
•
Our results support the potential of PARP inhibition as a novel therapeutic strategy in blocking chronic asthma in humans with olaparib as a likely candidate drug.
INTRODUCTION
Through a series of studies, our laboratory has established the role of poly(ADP-ribose)polymerase (PARP) in asthma pathogenesis using an ovalbumin (OVA)-based animal model of the disease [1–5]. These observations have been confirmed by several independent reports [6–9]. PARP-1 constitutes a prominent member of the PARP protein family and is classically characterized as a DNA repair enzyme. We have shown that PARP-1 regulates the expression of many genes whose products are crucial for asthma pathogenesis by controlling the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) nuclear trafficking and the fate of signal transducer and activator of transcription-6 (STAT-6) upon interleukin (IL)-4 or allergen exposure [5,10–12]. PARP-1 is not only involved in allergen-induced inflammation but also in airway hyper-responsiveness (AHR) [3,6]. Furthermore, PARP-1 appears to contribute to airway remodelling upon chronic exposure to allergens potentially through its regulation of inducible nitric oxide synthase (iNOS) and transforming growth factor (TGF)-β [6,13]. Collectively, these findings suggest that the protein may represent a viable therapeutic target to prevent or dampen onset of asthma symptoms upon allergen exposure. However, to increase the viability of PARP-1 as a therapeutic target it becomes imperative for us to demonstrate that PARP-1 is activated in human asthma, to validate the findings using an allergen that is more relevant to the human disease and to further examine the efficacy of a PARP inhibitor in asthma.
House dust mites (HDMs), the most prominent sources of indoor allergens, constitute a major health concern worldwide as it is one of the principal factors causing allergic asthma and rhinitis [14], with up to 50% of asthmatics being sensitized to the allergen [14,15]. Accruing evidence from animal and human studies has identified T helper 2 (Th2) cells and associated processes as major contributors to HDM-induced allergy and asthma [14,15]. The combined adaptive and innate immune responses are the key in rendering HDM such a potent allergen [14]. The mechanisms by which the body responds to HDM are complex and remain poorly understood. It is therefore very critical to decipher these mechanisms and unravel the players that govern the response to this allergen. The identification of these mediators may lead to new therapeutic targets and the development of drugs that may interfere with the ability of this allergen to cause asthma manifestations.
In the present study, we examined whether PARP is activated in peripheral blood mononuclear cells (PBMCs) and lung tissues of human asthmatics and used olaparib (AZD-2281), a clinically tested PARP-1 and PARP-2 inhibitor, to conduct a pre-clinical study to determine the efficacy of this drug in blocking established asthma. We purposely used an experimental model based on chronically-inhaled HDM given the fact that it closely resembles human asthma. This model involves mucosal sensitization within the lungs with a true human allergen to ultimately induce Th2-driven inflammation and AHR [16]. Lastly, we examined the effect of olaparib on the expression of Th2 cytokines and associated transcription factors upon T-cell receptor stimulation in human cluster of differentiation type 4 (CD4+) T-cells isolated from healthy volunteers.
MATERIALS AND METHODS
Animals, HDM challenge and airway hyper-responsiveness
Six eight-week-old C57BL/6J male mice were purchased from Jackson Laboratories. C57BL/6 PARP-1−/− mice were bred at the Louisiana State University Health Sciences Center (LSUHSC) vivarium and allowed unlimited access to sterilized chow and water. Husbandry, experimental protocols and procedures were all approved by the LSUHSC Animal Care & Use Committee. Mice were anaesthetized by isoflurane and challenged intranasally with 25 μl of saline or 1 mg/ml whole HDM (Dermatophagoides pteronyssinus) extract (Greer Labs), three times per week for 5 weeks. Intraperitoneal (i.p.) administration of olaparib (5 mg/kg) or vehicle (saline) was conducted 30 min after HDM challenge. Whereas one group received the drug only once (single administration protocol), another group received the drug once daily for 3 days (multiple administration protocol). Mice were killed 48 h later for bronchoalveolar lavage (BAL) or lung fixation and processing. Some mice were subjected to AHR measurements 24 h after the last challenge as recently described [17]. AHR to inhaled methacholine was assessed in unrestrained, conscious mice by recording ‘enhanced pause’ (Penh) using whole-body barometric plethysmography (EMKA Systems).
Organ recovery, cytokine assessments and FACS analysis
Lungs from killed mice were subjected to BAL; the BAL fluids (BALF) were assessed for inflammatory cells and cytokine production as described [2,17]. Measurement of spleen weight, total splenic cell count and FACS analysis for CD4+ T-cells, B-cells and T-regulatory (T-reg) cells were done as previously described [17]. Assessment of cytokines and chemokines in BALF, sera or cell culture media were conducted using the Bio-Rad Bioplex or the Millipore MILLIPLEX MAP human cytokine/chemokine system for mouse or human, according to the manufacturers' instructions. The multiplex assay and FACS analysis were conducted at the LSUHSC Comprehensive Alcohol Research Center Core.
Human subjects, cell culture, immunoblot, immunofluorescence, proliferation measurement and RT-PCR analysis
Four healthy and six asthmatic individuals were recruited under a protocol (#8450) approved by the LSUHSC institutional review board. Subjects were included if they were ≥18 years of age with a physician diagnosis of asthma. Exclusion criteria were a diagnosis of another lung disease other than asthma, active malignancy or inflammatory condition or ≥10 pack-years of smoking. Human PBMCs isolated from peripheral blood of healthy donors or asthmatic individuals were subjected to protein extractions followed by immunoblot analysis with antibodies to the poly (ADP-ribose) moiety (PAR; Enzo Life Sciences). The blots were subjected to a Ponceau staining prior to incubation with the antibodies. Two de-identified lung specimens from individuals who died from severe asthma and normal lungs from two individuals who died from asthma-unrelated conditions (verified by a pathologist from the Stanley S. Scott Cancer Center (SSSCC) Molecular Histopathology and Analytical Microscopy Core) were acquired from the LSUHSC Pathology Department (kindly provided by Dr C. Espinoza). Sections from different experimental groups were subjected to Hematoxylin and eosin (H&E) or PAS (periodic acid—Schiff) staining or subjected to immunofluorescence labelling using antibodies to PAR followed by FITC-labelled secondary antibodies. Pictures were captured using a Leica DMRA2 fluorescence microscope (Leica). Human and mouse CD4+-enriched T-cells were purified using EasySep™ Human/Mouse CD4+ T Cell Isolation Kit following the vendor's recommendations. Cells were then stimulated in the presence or absence of 1 or 5 μM olaparib with immuno-immobilized anti-CD3 plus anti-CD28 antibodies, as described [17]. T-cell proliferation was assessed by 5,6-carboxyfluorescein diacetate succinimidyl ester (CFSE) staining, as described [17]. A subset of CD4+-enriched T-cells were further sorted for IL-4, interferon (IFN)-γ or forkhead box P3 (FoxP3)-producing cells using flow cytometry. All the antibodies used were from BD biosciences. Human or mouse CD4+-enriched T-cells were also stimulated with anti-CD3 and anti-CD28 antibodies for different time periods followed by assessment of cytokines in culture media or subjected to RNA extraction as described [17]. The extracted RNA was reverse transcribed into cDNA using reverse transcriptase III (Invitrogen), and the resulting cDNA was subjected to conventional or quantitative PCR using primer sets (IDT) specific for human GATA binding protein-3 (gata-3), T-box transcription factor (t-bet) or gapdh (glyceraldehyde-3-phosphate dehydrogenase) as described [17] or mouse il17 (forward: 5′-GGT CAA CCT CAA AGT CTT TAA CTC-3′; reverse: 5′-TTA AAA ATG CAA GTA AGT TTG CTG-3′) or mouse β-actin (forward: 5′-CGGTTCCGATGCCCTGAGGCTCTT-3′; reverse: 5′-CGTCACACTTCATGATGGAATTGA-3′).
Data analysis
Experiments are repeated at least two times. All data are expressed as means ± S.E.M. of values from multiple replicates per group. PRISM software (GraphPad) was used to analyse the differences between experimental groups by one-way ANOVA followed by Tukey's multiple comparison test.
RESULTS
PARP is activated in PBMCs and lung tissues of asthmatic individuals
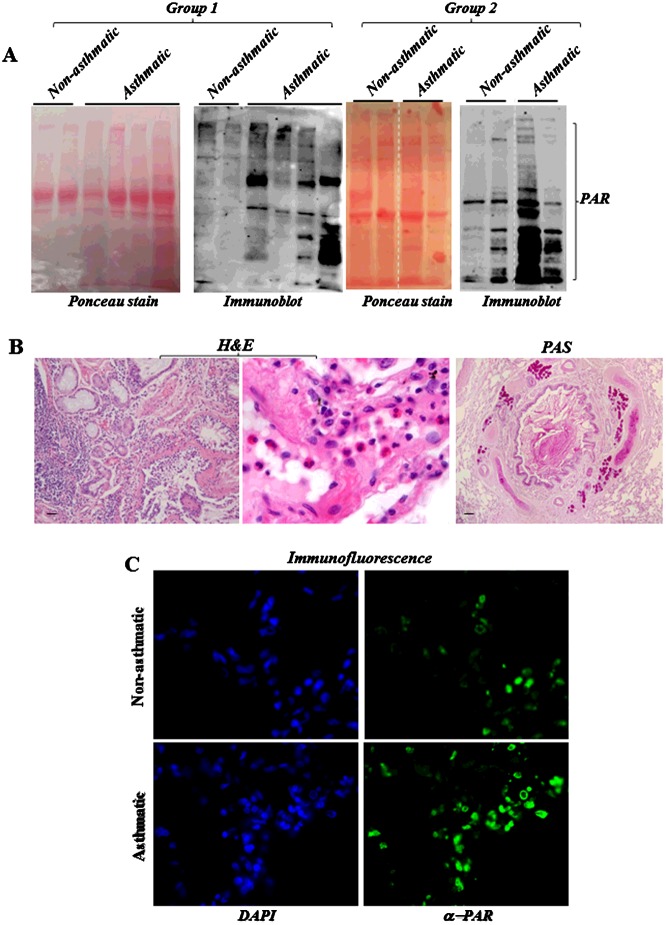
PBMCs collected from asthmatics or healthy volunteers were subjected to immunoblot analysis with antibodies to the PAR of PARP-modified proteins to determine whether PARP is activated in these cells. Figure 1 (A) shows a series of bands with PAR-immunoreactivity representing poly(ADP-ribosyl)ated proteins in PBMCs of asthmatics, which were largely absent from extracts of PBMCs derived from healthy individuals. We next examined whether PARP is also activated in lung tissue of two individuals who died from asthma and the lack thereof in tissue from an individual who died from an asthma-unrelated cause. Figure 1 (B) shows the typical eosinophilic inflammation and extensive mucus production in the lung of the asthmatic individual as assessed by H&E and PAS staining respectively. Figure 1 (C) shows a marked PARP activation in lung tissue of the asthmatic but not in the non-asthmatic individual as assessed by immunofluorescence with antibodies to PAR. These results demonstrate qualitatively for the first time that PARP is activated in human asthma.
Figure 1. PARP is activated in PBMCs and lung tissues of asthmatics.
(A) PBMCs collected from asthmatics or healthy volunteers were subjected to protein extraction followed by immunoblot analysis with antibodies to PAR. Following protein transfer, the blots were subjected to Ponceau staining. The dashed line indicates that the lane was cut from the same blot. The two groups of samples were done separately. (B) H&E and PAS staining of lung tissue of an individual who died from asthma. (C) Lung sections were subjected to immunofluorescence with antibodies to PAR.
PARP inhibition by olaparib or gene knockout blocks asthma-like manifestation in a chronic HDM asthma model
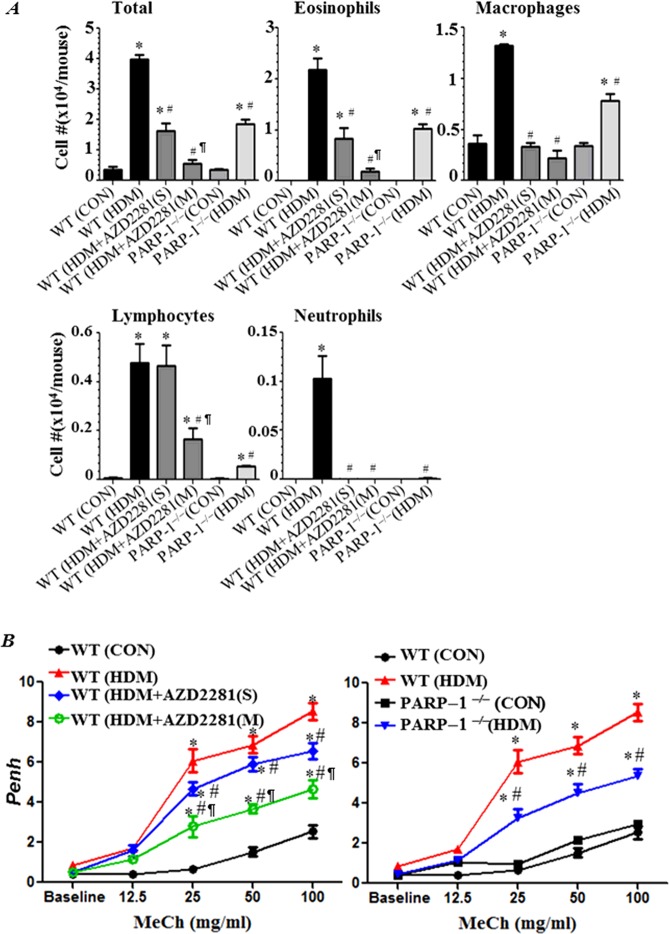
We next examined whether PARP inhibition pharmacologically by olaparib or genetically by gene knockout blocks asthma-like manifestation upon intraneural (i.n.) administration of HDM. Figure 2 (A) shows that a single administration of olaparib at the end of the HDM exposure protocol was highly effective in decreasing recruitment of eosinophils and macrophages as well as overall cellularity in the lungs. However, the increase in the number of lymphocytes was not affected. A remarkable protection was achieved upon two additional administrations of the drug including a reduction in the number of lymphocytes. Similar results were observed in HDM-exposed PARP-1−/− mice, which provide evidence for the specificity of such protective effects. Interestingly, repeated administration of olaparib provided significantly better reduction in recruitment of the total number of inflammatory cells, eosinophils and macrophages, than that provided by PARP-1 gene deletion.
Figure 2. PARP inhibition by olaparib or gene knockout blocks asthma-like traits in chronically HDM-exposed mice.
C57BL/6J WT or PARP-1−/− mice were subjected to HDM challenge or left untreated. HDM-challenged WT mice received 5 mg/kg of olaparib or saline once (S) 30 min after the last HDM challenge or once daily for 3 days (M). All mice were killed 48h later and BALF and organs were collected. (A) Cells of BALF were differentially stained and total eosinophils, macrophages, lymphocytes and neutrophils were counted. Data are expressed as total number of cells per mouse. (B) WT mice were subjected to HDM challenge followed by an i.p. injection of saline (▲), single (◆) or multiple administrations of 5 mg/kg olaparib (○). Control mice were not sensitized or challenged (●). PARP-1−/− mice were also subjected to HDM challenge (▼) and control mice were left unchallenged (■). Penh was recorded 24 h later using a whole body plethysmograph system before and after the indicated concentrations of aerosolized methacholine (MeCh). Results are plotted as maximal fold increase in Penh relative to baseline and expressed as mean ± S.E.M., where n=6 mice per group. *Difference from control WT mice, P<0.01; #Difference from HDM-challenged WT mice, P<0.01; ¶Difference from HDM-challenged WT mice that received a single dose of olaparib, P<0.01.
The manifestation of AHR upon chronic HDM exposure was modestly affected by a single administration of olaparib; a more pronounced reduction in AHR required two additional administrations of the drug (Figure 2B). PARP-1 gene deletion and repeated olaparib administration provided a similar protection against AHR (Figure 2B).
PARP inhibition by olaparib or gene knockout reduces Th2 cytokine production without a prominent effect on IFN-γ or IL-10
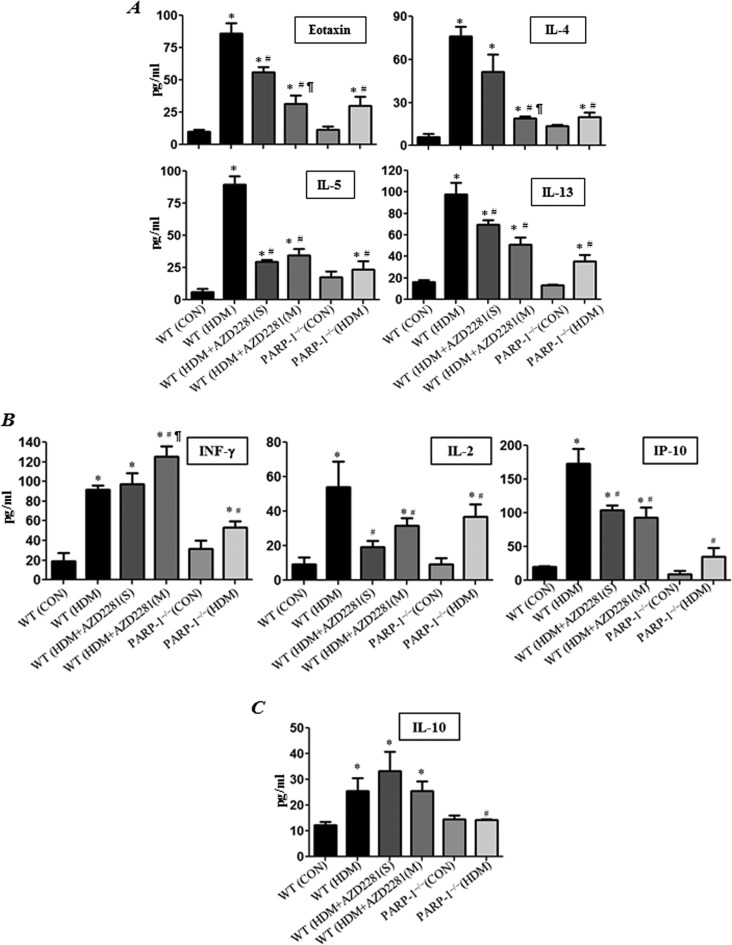
Figure 3 (A) shows that HDM-induced lung eosinophilia was accompanied with an increase in production of a number of Th2 cytokines in BALF collected from the treated animals, such as eotaxin, IL-4, IL-5 and IL-13. These cytokines were markedly reduced in BALF of mice that received a single or triple administration of olaparib. Similar reduction was observed in HDM-exposed PARP-1−/− mice. Although PARP inhibition pharmacologically or by gene knockout reduced production of the Th1 cytokines IL-2 and interferon gamma-induced protein-10 (IP-10) in HDM-treated mice, the IFN-γ levels either slightly increased or remained unaffected by PARP inhibition (Figure 3B). Interestingly, although the levels of the anti-inflammatory cytokine IL-10 were not affected by olaparib treatment, the levels of the cytokine in HDM-exposed PARP-1−/− mice remained lower than those detected in BALF of HDM-exposed wild-type (WT) mice.
Figure 3. PARP inhibition by olaparib reduces Th2 cytokine production without a prominent effect on IFN-γ or IL-10.
WT and PARP-1−/− mice were subjected to HDM challenge or left untreated. HDM challenged WT mice received a single dose of 5 mg/kg of olaparib or saline 30 min after the last challenge or were given multiple administrations of olaparib every 24 h for a total of three times. All mice were killed 48 h later and lungs were subjected to BAL. Assessment of BALF from different groups for Th2 cytokines: eotaxin, IL-4, IL-5 or IL-13 (A), INF-γ, IL-2 or IP-10 (B) and IL-10 (C). Data are means ± S.D. of values from at least six mice per group. *Difference from control mice, P<0.01; #difference from HDM challenged WT mice; P<0.01; ¶difference from HDM-challenged WT mice that received a single dose of olaparib, P<0.01.
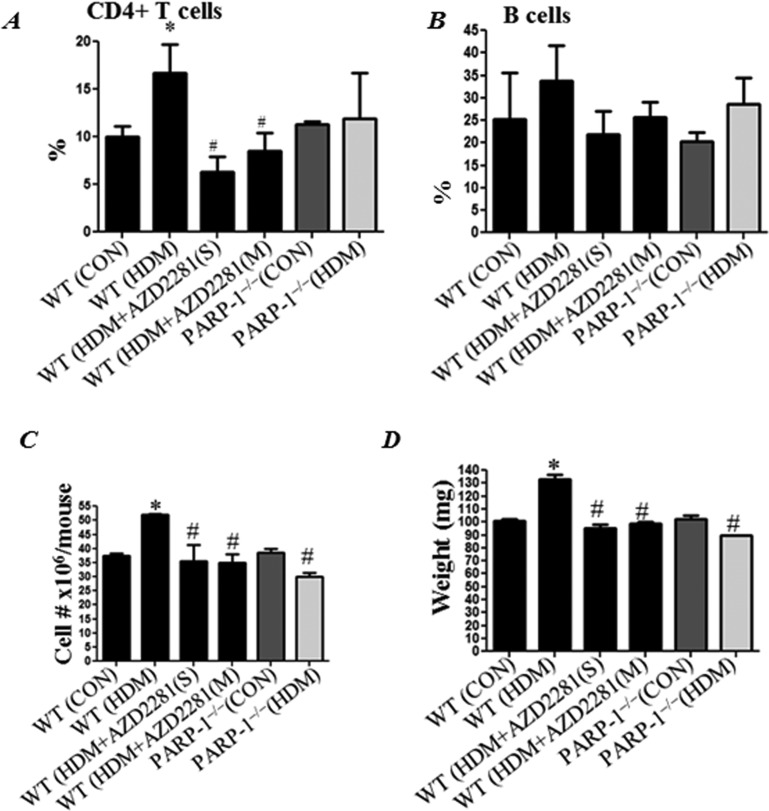
PARP inhibition prevents HDM-induced increase in overall cellularity, weight and CD4+ T-cell population in spleens of treated mice
HDM exposure induced a moderate but significant increase in the percentage of CD4+ T-cell population in spleens of treated mice; this increase was blunted by either a single or repeated administration of olaparib (Figure 4A). Although the percentage of CD4+ T populations trended lower in HDM-exposed PARP-1−/− mice, the difference was not statistically significant. HDM exposure did not change the percentage of the B-cell population in WT mice and PARP inhibition (pharmacologically or by gene knockout) did not cause any change either (Figure 4B). As expected, chronic exposure to HDM significantly increased the overall numbers of cells in spleens of treated animals (Figure 4C); this increase was prevented by PARP inhibition either by olaparib or by gene knockout. PARP inhibition also prevented the consequent increase in spleen weight (Figure 4D) in HDM-exposed WT mice.
Figure 4. PARP inhibition prevents HDM-induced increase in overall cellularity, weight and CD4+ T-cell population in spleens of treated mice.
WT or PARP-1−/− mice were subjected to HDM challenge or left untreated. HDM-challenged WT mice received a single dose or multiple doses of olaparib (5 mg/kg) or saline as explained in the previous figure legends. Spleens from different groups were used to prepare single cell suspensions followed by FACS analysis for (A) CD3+/CD4+ and (B) B220+/CD19+. Total cell counts (C) and spleen weights (D) from different groups were assessed.*Difference from control WT mice, P<0.01; #difference from HDM-challenged WT mice, P<0.01.
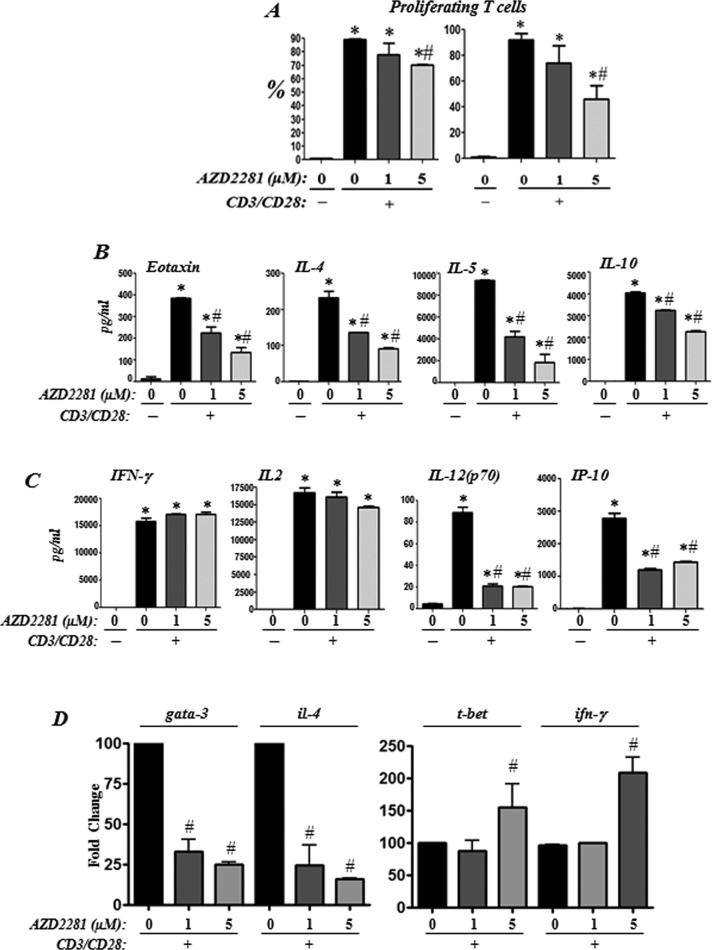
Olaparib treatment reduces Th2 cytokine production upon T-cell receptor (TCR) stimulation in human CD4+ T-cells with a moderate effect on overall T-cell proliferation
We first examined the effect of a single treatment with olaparib on overall proliferation of CD4+-enriched T-cells isolated from PBMCs of healthy human subjects. Figure 5 (A) shows that olaparib treatment did not cause a substantial inhibition of CD4+-enriched T-cells upon TCR activation. Upon addition of fresh inhibitor every day, a significant decrease in growth was observed at the 5 μM of the drug. Figure 5 (B) shows that olaparib treatment markedly reduced the ability of CD4+-enriched T-cells to produce the Th2 cytokines eotaxin, IL-4, IL-5 and IL-10 even at the 1 μM concentration of the drug. Figure 5 (C) shows that olaparib treatment differentially affected production of the Th1 cytokines as it exerted no effect on IFN-γ or IL-2 production but significantly reduced IL-12 and IP-10 production upon CD3/CD28 stimulation (Figure 5C).
Figure 5. Effects of PARP inhibition by olaparib on human CD4+ T-cell proliferation, cytokine production and expression of Th1/Th2 regulators upon TCR stimulation.
Negatively selected human CD4+-enriched T-cells from healthy donors were stimulated in triplicates with antibodies to CD3/CD28 in the absence or presence of 1 or 5 μM olaparib. (A) Cell proliferation was assessed by CFSE staining. (B and C) Culture supernatant was collected 24 h after TCR stimulation and cytokines were assessed. Data are given as means ± S.E.M. of values obtained from duplicates of the triplicates. (D) CD3/CD28-stimulated and control CD4+ T were subjected to RNA extraction after an incubation of 12 h. RNA was then reverse-transcribed and the resulting cDNA was subjected to PCR with primer sets specific to human gata-3, il-4, t-bet, ifn-γ or gapdh; *Difference from non-stimulated cells, P<0.01; #difference from CD3/CD28-stimulated cells, P<0.01.
We next examined whether the reduction in the anti-CD3/CD28-stimulated Th2 cytokines by olaparib treatment was associated with a reduction in the expression of the transcription factor GATA-3, the master regulator of the Il4/Il5/Il13 cytokine locus [18]. Figure 5 (D) shows that olaparib treatment was very effective in reducing gata-3 expression in human CD4+ T-cells upon TCR stimulation with a concomitant decrease in IL-4 mRNA as assessed by real-time PCR. Conversely, olaparib treatment did not affect the expression of t-bet in TCR-stimulated cells at the 1 μM concentration; however, a slight increase in the levels of the transcription factor was observed at the 5 μM concentration of the drug (Figure 5D). The effect of olaparib treatment on IFN-γ mRNA levels mirrored that of t-bet.
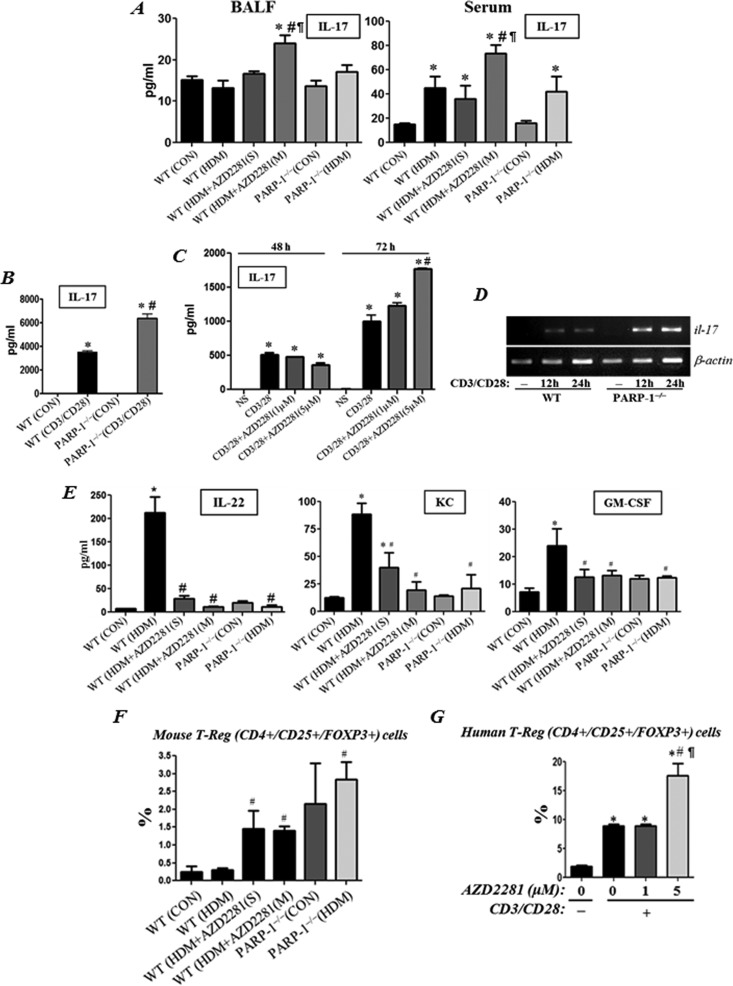
PARP inhibition inconsistently increases IL-17 production without a concomitant increase in IL-17-associated factors but with an increase in the percentage of T-reg cells in vitro and in HDM-exposed mice
Several recent studies have reported conflicting findings on the relationship between PARP and IL-17. Whereas some reports show an increase in IL-17 production [19], others showed either no change [20] or a decrease in the cytokine upon PARP inhibition [21–23]. It therefore became imperative to examine whether olaparib or PARP-1 gene deletion affects the levels of the cytokine in our experimental models. Figure 6 (A) shows chronic HDM exposure induced a slight increase in IL-17 only in sera but not in BALF of treated mice at the end of the exposure protocol. In BALF of HDM-exposed mice, the level of the cytokine modestly increased upon repeated, but not upon a single, administration of olaparib. IL-17 did not increase in BALF of HDM-exposed PARP-1−/− mice. In sera of HDM-exposed mice, the level of IL-17 also increased upon repeated, but not upon a single, administration of olaparib. Unlike in BALF, IL-17 increased in sera of HDM-exposed PARP-1−/− mice. The increase in IL-17 upon PARP inhibition was confirmed in CD4+ T-cells derived from PARP-1−/− mice that were activated with antibodies to CD3/CD28 (Figure 6B). In CD3/CD28-activated human CD4+ T-cells, olaparib treatment only increased IL-17 production at the 5 μM concentration and only after an extended time of treatment (Figure 6C). Figure 6 (D) shows that the increase in IL-17 in CD3/CD28-stimulated PARP-1−/− CD4+ T-cells occurred at the mRNA level. Surprisingly, the PARP inhibition-associated increase in IL-17 was accompanied with a decrease, rather than an increase, in factors that can be influenced by IL-17 such as IL-22, keratinocyte chemoattractant (KC) and granulocyte macrophage colony-stimulating factor (GM-CSF) in HDM-treated mice (Figure 6D). Furthermore, such increase in IL-17 but decrease in IL-22 coincided with a significant increase in the percentage of CD4+/CD25+/Foxp3+ T-reg cell population in spleens of HDM-exposed animals (Figure 6F). HDM exposure did not increase the percentage of T-reg cells in spleens of WT mice (Figure 6F), which is consistent with the report by Kim et al. [24]. The T-reg cell population was also increased in PARP-1−/− mice without any HDM exposure. Consistent with these results, olaparib treatment also promoted a significant increase in the T-reg cell population in CD3/CD28-activated human CD4+ T-cells but only in response to the 5 μM concentration of the drug (Figure 6G).
Figure 6. Effect of PARP inhibition on IL-17 production and T-reg cell population upon CD3/CD28 stimulation in vitro and in HDM-exposed mice.
WT or PARP-1−/− mice were subjected to HDM challenge or left untreated. HDM-challenged WT mice received a single dose or multiple doses of olaparib (5 mg/kg) or saline as described above. Mice were killed 48 h later. (A) BALF and sera of the killed mice were assessed for IL-17. *Difference from control WT mice, P<0.05; #difference from HDM-challenged WT mice, P<0.05; ¶, difference from HDM-challenged WT mice subjected a single olaparib administration, P<0.05. (B) Splenic CD4+ T-cells isolated from naive WT or PARP-1 −/− mice were activated with antibodies to CD3/CD28. After an incubation of 96 h, culture supernatants were assessed for IL-17. *Difference from non-stimulated cells, P<0.01; #difference from anti-CD3/CD28-stimulated cells; P<0.01. (C) Negatively selected human CD4+ T-enriched cells from healthy donors were stimulated with antibodies to CD3/CD28 in the absence or presence of 1 or 5 μM of olaparib. Cell supernatant was tested for IL-17 production. *Difference from non-stimulated cells, P<0.01; #difference from anti-CD3/CD28-stimulated cells; P<0.01. (D) mRNA was isolated from WT or PARP-1−/− splenic CD4+ T-enriched cells that were activated with anti-CD3/CD28 antibodies for 12 or 24 h. mRNA was reverse-transcribed and the resulting cDNA was subjected to conventional PCR with primer sets specific to mouse il-17 or β-actin. (E) BALF from the different experimental groups described in (A) were tested for IL-22, KC and GM-CSF production. *Difference from control WT mice, P<0.01; #difference from HDM-challenged WT mice, P<0.01. (F) Spleens from the different experimental groups described in (A) were processed to generate single cell suspensions, which were analysed by FACS for CD4+/CD25+/Foxp3+ cells. #difference from HDM-challenged WT mice, P<0.05; ¶Difference from HDM-challenged WT mice subjected a single olaparib administration, P<0.05. (G) Negatively selected human CD4+ T-enriched cells from healthy donors were stimulated with antibodies to CD3/CD28 for 72 h in the absence or presence of 1 or 5 μM of olaparib. Cells were analysed by FACS for CD4+/CD25+/Foxp3+ cells. *Difference from non-stimulated cells, P<0.01; #difference from anti-CD3/CD28-stimulated cells; P<0.01; ¶difference from anti CD3/CD28-stimulated cells in presence of 1 μM of olaparib.
DISCUSSION
A great deal of effort has been made by our laboratory to establish a role for PARP-1 in asthma using OVA-based experimental models of the disease with the ultimate goal to translate our findings to the human condition [1,2,4,13,25]. However, the relevance and clinical significance of these studies were limited due to the lack of evidence that the enzyme is in fact activated in cells or tissues of human asthmatics. Results of the present study provide this critical evidence and demonstrate for the first time that PARP is indeed activated in PBMCs and lung tissue of asthmatics. Relevance to the human disease was bolstered by demonstrating the important role for PARP in a HDM-based mouse model of the disease that more closely reflects human asthma. More importantly, we provide convincing evidence that olaparib, a PARP inhibitor that is currently being tested in cancer clinical trials, efficiently blocked established asthma-like traits including production of Th2 cytokines and mucus and AHR. The drug was also efficient in reducing production of Th2 cytokines in response to TCR stimulation in human CD4+ T-cells. The anti-inflammatory effects conferred by PARP inhibition seemed to be aided by an increase in T-reg cells. Finally, we show that PARP inhibition inconsistently increased IL-17 production. Interestingly, the factors that may be influenced by IL-17 were decreased rather than increased upon PARP inhibition in both HDM mouse model and stimulated human CD4+-enriched T-cells. We thus propose PARP inhibition as a viable strategy in targeting at least some aspects of human asthma and that the therapeutic strategy merits consideration in clinical trials.
The modulatory effects of PARP inhibition by olaparib or by gene knockout on eosinophilia and IL-4 are indicative of a potential reduction in IgE. However, the levels of immunoglobulin in the HDM mouse model used in the present study did not reach detectable levels (result not shown), which is consistent with our recent report [17] and that of De Alba et al. [26]. Detection of HDM-specific IgE requires a stronger sensitization of animals to the allergen primarily through an i.p. administration often with an adjuvant such as aluminum hydroxide [26]. This mechanism of sensitization was purposely avoided to follow a model that closely resembles human asthma involving mucosal sensitization within the lungs [16]. It is clear, however that PARP inhibition pharmacologically or by gene knockout blocks IgE production in OVA-based mouse models [2,3]. In a concurrent study [27], we show that olaparib very efficiently reduces OVA-specific IgE levels in BALF and sera of challenged mice. However, PARP-1 does not seem to play a direct role in IgE production. Indeed, adoptive transfer of OVA-sensitive CD4+ T-cells into naive PARP-1−/− mice reversed most of the IgE production. Thus, PARP-1 appears to play a role in CD4+ T-cells rather than B-cells.
We expanded our studies on the role of PARP in the function of TCR-stimulated CD4+-enriched T-cells derived from human PBMCs. The effects of PARP inhibition by olaparib were associated with a reduction in the ability of Th2 cells to produce Th2 cytokines as well as expression of factors that drive their differentiation including GATA-3 and IL-4. Such effects occurred despite a marginal effect of PARP inhibition on T-cell proliferation. Our results on the effect of PARP inhibition on T-cell proliferation are consistent with those reported by Saenz et al. [28]. The effect of PARP inhibition of Th2 cytokine production upon TCR stimulation may be directly linked to a modulation of gata-3 expression. Currently, the mechanism by which PARP-1 regulates the expression of gata-3 is not clear. It is however established that gata-3 is regulated by NF-κB [29] and STAT-6 [30], both of which can be regulated by PARP-1 as shown in our previous studies [10,12]. Studies are underway to decipher the exact role of PARP-1 in the regulation of the NF-κB and STAT-6 pathways. The role of PARP in regulating the expression of Th1 cytokines is rather intriguing as inhibition of the enzyme reduced some but not all Th1 cytokines despite the lack of clear effect on expression of t-bet. For instance, PARP inhibition did not affect expression of IFN-γ or IL-2 but decreased expression of IL-12(p70) and IP-10 in anti-CD3/CD28-activated human CD4+ T-cells. The differential effects on IFN-γ and IP-10 in human CD4+ T-cells was consistent with those observed in HDM-based animal model of asthma. Obviously, more experimentation is necessary to clarify such differential effects and identify the exact factors that can be regulated by PARP. It is important to note that sometimes PARP inhibition by olaparib provided a more pronounced effect compared with that achieved by gene knockout. For instance, the effect of multiple administration of olaparib on lung eosinophilia and total BALF cell count at the 5 mg/kg dose was better than that observed in PARP-1−/− mice. The enhanced effect may be associated with the ability of olaparib to inhibit both PARP-1 and PARP-2.
The connection between PARP-1 and IL-17 is rather interesting. Several reports showed conflicting results on the ability of PARP inhibition to modulate IL-17 in animal models of inflammatory diseases and in in vitro systems. Indeed, a report by Nasta et al. [20] showed that PARP-1 deficiency does not affect production of IL-17 in response to multiple stimuli including CD3 and CD28. In contrast, PARP inhibition was shown to reduce IL-17 production in adjuvant-induced arthritis mouse model [22] and carrageenan-induced lung inflammation in mice [23]. However, recently, Zhang et al. [19] showed that in TGF-β and IL-6-treated CD4+ T-cells, PARP inhibition can in fact increase IL-17. Using both the chronic HDM exposure mouse model and the CD3/CD28-activated CD4+ T-cells, we showed that the association between PARP inhibition and modulation of IL-17 production is inconsistent. PARP appears to regulate IL-17 at the level of mRNA in activated CD4+ T-cells. Surprisingly, the increase in IL-17 in both the animal and the cell culture models was accompanied by a decrease rather than an increase in factors that can be influenced by IL-17, such as IL-22, KC and GM-CSF. It is interesting that despite the increase in IL-17 in our experimental systems, the net effect was anti- rather than pro-inflammatory. The reduction in KC and GM-CSF can be unrelated to the effect on IL-17, as PARP inhibition, pharmacologically or by gene knockout, reduces expression of the two inflammatory factors in response to LPS [11]. It is important to note that if IL-17 is increased by PARP inhibition and if this increase may exert some pathology, then using olaparib for the treatment of asthma in humans constitutes an important limitation of the strategy. IL-17 has been regarded as an important player in severe forms of asthma, which display neutrophilic inflammation [31]. Interestingly, in our HDM-based model, neutrophilia is completely abrogated upon PARP inhibition by olaparib or gene knockout. These results suggest the relationship between IL-17, its effects and PARP-1 is complex and requires further detailed investigations to decipher the exact nature of such a relationship.
An emerging role for PARP in inflammation is its potential influence on T-reg cells [20–22,32]. This role is associated with the stabilization of the transcription factor Foxp3 [21]. Our results show that PARP inhibition by olaparib or gene knockout increased the T-reg cell population. This appears to be independent of HDM exposure as naive PARP-1−/− mice displayed higher levels of T-reg cells compared with the WT counterparts. In human CD4+ T-cells, the increase in T-reg cell population was only observed upon an exposure to the high dose (5 μM) but not to the low dose (1 μM) of olaparib. These results and those of aforementioned published reports strongly suggest that the role of PARP-1 in T-reg differentiation and function is complex; additional studies are required to reach a better understanding of the relationship. Nevertheless, an increase in T-reg cell population may be an added protective trait achieved upon PARP inhibition in the control of asthma-associated inflammation.
In conclusion, the present study provides critical information on the role of PARP in a chronic asthma model that utilizes a true human allergen and provides evidence for the first time that PARP-1 is activated in PBMCs and lung tissue of human asthmatics. More importantly, our results lend support to the notion that PARP can be targeted for the treatment of human asthma.
Acknowledgments
We would like to thank the clinical co-ordinators, Marie Sandi and Connie Romaine, for their effort in collecting peripheral blood from the human subjects. We would like to thank the Pulmonary and Critical Care Section at LSUHSC for giving us access to the whole body plethysmograph system. We would also like to thank Mrs Constance Porretta from the LSUHSC Comprehensive Alcohol Research Center Core (CARC) for her assistance and data analysis with the FACS assays, as well as Dr J Weiss and the department of Ophthalmology for giving us access to their microtome.
Abbreviations
- AHR
airway hyper-responsiveness
- BAL
bronchoalveolar lavage
- BALF
BAL fluids
- CD4+
cluster of differentiation type 4; CFSE, 5,6-carboxyfluorescein diacetate succinimidyl ester
- FoxP3
forkhead box P3
- gapdh
glyceraldehyde-3-phosphate dehydrogenase
- gata-3
GATA binding protein-3
- GM-CSF
granulocyte macrophage colony-stimulating factor
- HDM
house dust mite
- H&E
Hematoxylin and eosin; i.p., intraperitoneal
- IFN
interferon
- IL
interleukin
- IP-10
interferon gamma-induced protein-10
- KC
keratinocyte chemoattractant
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells; OVA, ovalbumin
- PAR
poly (ADP-ribose) moiety
- PARP-1
poly(ADP-ribose)polymerase-1
- PAS
periodic acid–Schiff
- PBMC
peripheral blood mononuclear cell
- Penh
enhanced pause
- STAT-6
signal transducer and activator of transcription-6
- t-bet
T-box transcription factor
- TCR
T-cell receptor
- T-reg
CD4+ T-regulatory cell
- TGF
transforming growth factor
- Th2
T helper cells 2
- WT
wild-type
AUTHOR CONTRIBUTION
Mohamed Ghonim led the study, conducted the majority of the experiments and the statistical analyses. Kusma Pyakurel assisted in the animal studies, significantly contributed to the in vitro experiments and helped with statistical analyses. Salome Ibba conducted part of the experiments using human samples, RT-PCR and the statistical analyses. Jeffrey Wang and Christian Davis assisted with the animal studies. Paulo Rodriguez conducted some of the in vitro experiments and helped in the troubleshooting. Amir Al-Khami contributed to some of the ex vivo experiments/analysis. Matthew Lammi contributed to the human subjects' referral, assisted in the acquisition of human PBMCs, edited the manuscript and contributed to discussion. Hogyoung Kim conducted some of the IL-17 RT-PCR experiments. Arnold Zea provided PBMCs from healthy subjects. Samuel Okpechi provided technical assistance. Moselhy Mansy contributed to the training of the first author. Augusto Ochoa provided reagents and consultation. Amarjit Naura contributed to the training of several members of the laboratory, experimental design and troubleshooting. Hamid Boulares contributed to the design of the experiment and the training of researchers as well as provided the financial support for the conducted work.
FUNDING
This work was supported by the National Institute of Health (NIH) [grant number HL072889 (to A.H.B.)]; the Louisiana Cancer Research Center (to A.H.B.); the National Institute of General Medical Sciences of the National Institutes of Health, which funds the Louisiana Clinical and Translational Science Center [grant number 1 U54 GM104940 (A.A.A. and M.R.L.)]; the Egyptian Cultural and Educational Bureau [grant number JS-2634 (to M.A.G.)]; the American Heart Association [grant number 14PRE19630012 (to K.P.)]; the Department of Biotechnology, Government of India [grant number BT/RLF/Re-entry/36/2012 (to A.S.N.)]; and theNational Institute of General Medical Sciences [grant number COBRE 5P30GM106392].
References
- 1.Boulares A.H., Zoltoski A.J., Sherif Z.A., Jolly P., Massaro D., Smulson M.E. Gene knockout or pharmacological inhibition of poly(ADP-ribose) polymerase-1 prevents lung inflammation in a murine model of asthma. Am. J. Respir. Cell Mol. Biol. 2003;28:322–329. doi: 10.1165/rcmb.2001-0015OC. [DOI] [PubMed] [Google Scholar]
- 2.Oumouna M., Datta R., Oumouna-Benachour K., Suzuki Y., Hans C., Matthews K., Fallon K., Boulares H. Poly(ADP-ribose) polymerase-1 inhibition prevents eosinophil recruitment by modulating Th2 cytokines in a murine model of allergic airway inflammation: a potential specific effect on IL-5. J. Immunol. 2006;177:6489–6496. doi: 10.4049/jimmunol.177.9.6489. [DOI] [PubMed] [Google Scholar]
- 3.Naura A.S., Hans C.P., Zerfaoui M., You D., Cormier S.A., Oumouna M., Boulares A.H. Post-allergen challenge inhibition of poly(ADP-ribose) polymerase harbors therapeutic potential for treatment of allergic airway inflammation. Clin. Exp. Allergy. 2008;38:839–846. doi: 10.1111/j.1365-2222.2008.02943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naura A.S., Datta R., Hans C.P., Zerfaoui M., Rezk B.M., Errami Y., Oumouna M., Matrougui K., Boulares A.H. Reciprocal regulation of iNOS and PARP-1 during allergen-induced eosinophilia. Eur. Respir. J. 2009;33:252–262. doi: 10.1183/09031936.00089008. [DOI] [PubMed] [Google Scholar]
- 5.Datta R., Naura A.S., Zerfaoui M., Errami Y., Oumouna M., Kim H., Ju J., Ronchi V.P., Haas A.L., Boulares A.H. PARP-1 deficiency blocks IL-5 expression through calpain-dependent degradation of STAT-6 in a murine asthma model. Allergy. 2011;66:853–861. doi: 10.1111/j.1398-9995.2011.02549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lucarini L., Pini A., Gerace E., Pellicciari R., Masini E., Moroni F. Poly(ADP-ribose) polymerase inhibition with HYDAMTIQ reduces allergen-induced asthma-like reaction, bronchial hyper-reactivity and airway remodelling. J. Cell Mol. Med. 2014;18:468–479. doi: 10.1111/jcmm.12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki Y., Masini E., Mazzocca C., Cuzzocrea S., Ciampa A., Suzuki H., Bani D. Inhibition of poly(ADP-ribose) polymerase prevents allergen-induced asthma-like reaction in sensitized Guinea pigs. J. Pharmacol. Exp. Ther. 2004;311:1241–1248. doi: 10.1124/jpet.104.072546. [DOI] [PubMed] [Google Scholar]
- 8.Virag L. Poly(ADP-ribosyl)ation in asthma and other lung diseases. Pharmacol. Res. 2005;52:83–92. doi: 10.1016/j.phrs.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 9.Virag L., Bai P., Bak I., Pacher P., Mabley J.G., Liaudet L., Bakondi E., Gergely P., Kollai M., Szabo C. Effects of poly(ADP-ribose) polymerase inhibition on inflammatory cell migration in a murine model of asthma. Med. Sci. Monit. 2004;10:BR77–BR83. [PubMed] [Google Scholar]
- 10.Zerfaoui M., Suzuki Y., Naura A.S., Hans C.P., Nichols C., Boulares A.H. Nuclear translocation of p65 NF-kappaB is sufficient for VCAM-1, but not ICAM-1, expression in TNF-stimulated smooth muscle cells: Differential requirement for PARP-1 expression and interaction. Cell Signal. 2008;20:186–194. doi: 10.1016/j.cellsig.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zerfaoui M., Naura A.S., Errami Y., Hans C.P., Rezk B.M., Park J., Elsegeiny W., Kim H., Lord K., Kim J.G., Boulares A.H. Effects of PARP-1 deficiency on airway inflammatory cell recruitment in response to LPS or TNF: differential effects on CXCR2 ligands and duffy antigen receptor for chemokines. J. Leukoc. Biol. 2009;86:1385–1392. doi: 10.1189/jlb.0309183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zerfaoui M., Errami Y., Naura A.S., Suzuki Y., Kim H., Ju J., Liu T., Hans C.P., Kim J.G., Abd Elmageed Z.Y., et al. Poly(ADP-ribose) polymerase-1 is a determining factor in Crm1-mediated nuclear export and retention of p65 NF-kappa B upon TLR4 stimulation. J. Immunol. 2010;185:1894–1902. doi: 10.4049/jimmunol.1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naura A.S., Zerfaoui M., Kim H., Abd Elmageed Z.Y., Rodriguez P.C., Hans C.P., Ju J., Errami Y., Park J., Ochoa A.C., Boulares A.H. Requirement for inducible nitric oxide synthase in chronic allergen exposure-induced pulmonary fibrosis but not inflammation. J. Immunol. 2010;185:3076–3085. doi: 10.4049/jimmunol.0904214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calderon M.A., Linneberg A., Kleine-Tebbe J., De Blay F., Hernandez Fernandez de Rojas D., Virchow J.C., Demoly P. Respiratory allergy caused by house dust mites: what do we really know? J. Allergy Clin. Immunol. 2015;136:38–48. doi: 10.1016/j.jaci.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 15.Jacquet A. The role of the house dust mite-induced innate immunity in development of allergic response. Int. Arch. Allergy Immunol. 2011;155:95–105. doi: 10.1159/000320375. [DOI] [PubMed] [Google Scholar]
- 16.Stevenson C.S., Birrell M.A. Moving towards a new generation of animal models for asthma and COPD with improved clinical relevance. Pharmacol. Ther. 2011;130:93–105. doi: 10.1016/j.pharmthera.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Ghonim M.A., Pyakurel K., Ju J., Rodriguez P.C., Lammi M.R., Davis C., Abughazleh M.Q., Mansy M.S., Naura A.S., Boulares A.H. DNA-dependent protein kinase inhibition blocks asthma in mice and modulates human endothelial and CD4 T-cell function without causing severe combined immunodeficiency. J. Allergy Clin. Immunol. 2015;135:425–440. doi: 10.1016/j.jaci.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng W., Flavell R.A. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/S0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 19.Zhang P., Nakatsukasa H., Tu E., Kasagi S., Cui K., Ishikawa M., Konkel J.E., Maruyama T., Wei G., Abbatiello B., et al. PARP-1 regulates expression of TGF-beta receptors in T cells. Blood. 2013;122:2224–2232. doi: 10.1182/blood-2013-05-503250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nasta F., Laudisi F., Sambucci M., Rosado M.M., Pioli C. Increased Foxp3+ regulatory T cells in poly(ADP-Ribose) polymerase-1 deficiency. J. Immunol. 2010;184:3470–3477. doi: 10.4049/jimmunol.0901568. [DOI] [PubMed] [Google Scholar]
- 21.Zhang P., Maruyama T., Konkel J.E., Abbatiello B., Zamarron B., Wang Z.Q., Chen W. PARP-1 controls immunosuppressive function of regulatory T cells by destabilizing Foxp3. PLoS One. 2013;8:e71590. doi: 10.1371/journal.pone.0071590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmad S.F., Zoheir K.M., Bakheet S.A., Ashour A.E., Attia S.M. Poly(ADP-ribose) polymerase-1 inhibitor modulates T regulatory and IL-17 cells in the prevention of adjuvant induced arthritis in mice model. Cytokine. 2014;68:76–85. doi: 10.1016/j.cyto.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Ahmad S.F., Zoheir K.M., Ansari M.A., Korashy H.M., Bakheet S.A., Ashour A.E., Al-Shabanah O.A., Al-Harbi M.M., Attia S.M. The role of poly(ADP-ribose) polymerase-1 inhibitor in carrageenan-induced lung inflammation in mice. Mol. Immunol. 2015;63:394–405. doi: 10.1016/j.molimm.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 24.Kim B.Y., Park H.R., Shin J.H., Kim S.W., Cho J.H., Park Y.J., Kim S.W. The serine protease inhibitor, 4-(2-aminoethyl) benzene sulfonyl fluoride hydrochloride, reduces allergic inflammation in a house dust mite allergic rhinitis mouse model. Allergy Asthma Immunol. Res. 2014;6:558–566. doi: 10.4168/aair.2014.6.6.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naura A.S., Kim H., Ju J., Rodriguez P.C., Jordan J., Catling A.D., Rezk B.M., Abd Elmageed Z.Y., Pyakurel K., Tarhuni A.F., et al. Minocycline blocks asthma-associated inflammation in part by interfering with the T cell receptor-NF-kB-GATA-3-IL-4 axis without a prominent effect on PARP. J. Biol. Chem. 2013;288:1458–1468. doi: 10.1074/jbc.M112.419580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Alba J., Raemdonck K., Dekkak A., Collins M., Wong S., Nials A.T., Knowles R.G., Belvisi M.G., Birrell M.A. HDM induces direct airway inflammation in vivo: implications for future disease therapy? Eur. Respir. J. 2009;35:1377–1387. doi: 10.1183/09031936.00022908. [DOI] [PubMed] [Google Scholar]
- 27.Ghonim M.A., Pyakurel K., Ibba V.S., Al-Khami A.A., Wang J., Rodriguez P.C., Rady H.F., El-Bahrawy A.H., Lammi M.R., Mansy M.S., et al. PARP inhibition by olaparib or gene knockout blocks asthma-like manifestation in mice by modulating CD4+ T cell function J. Transl. Med. 2015;13:225. doi: 10.1186/s12967-015-0583-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saenz L., Lozano J.J., Valdor R., Baroja-Mazo A., Ramirez P., Parrilla P., Aparicio P., Sumoy L., Yelamos J. Transcriptional regulation by poly(ADP-ribose) polymerase-1 during T cell activation. BMC Genomics. 2008;9:171. doi: 10.1186/1471-2164-9-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das J., Chen C.H., Yang L., Cohn L., Ray P., Ray A. A critical role for NF-kappa B in GATA3 expression and TH2 differentiation in allergic airway inflammation. Nat. Immunol. 2001;2:45–50. doi: 10.1038/83158. [DOI] [PubMed] [Google Scholar]
- 30.Maier E., Duschl A., Horejs-Hoeck J. STAT6-dependent and -independent mechanisms in Th2 polarization. Eur. J. Immunol. 2012;42:2827–2833. doi: 10.1002/eji.201242433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lambrecht B.N., Hammad H. The immunology of asthma. Nat. Immunol. 2015;16:45–56. doi: 10.1038/ni.3049. [DOI] [PubMed] [Google Scholar]
- 32.Rosado M.M., Bennici E., Novelli F., Pioli C. Beyond DNA repair, the immunological role of PARP-1 and its siblings. Immunology. 2013;139:428–437. doi: 10.1111/imm.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]