Abstract
Centrioles are evolutionarily conserved cylindrical cell organelles with characteristic radial symmetry. Despite their considerable size (400 nm × 200 nm, in humans), genetic studies suggest that relatively few protein components are involved in their assembly. We recently characterized the molecular architecture of the centrosomal P4.1-associated protein (CPAP), which is crucial for controlling the centriolar cylinder length. Here, we review the remarkable architecture of the C-terminal domain of CPAP, termed the G-box, which comprises a single, entirely solvent exposed, antiparallel β-sheet. Molecular dynamics simulations support the stability of the G-box domain even in the face of truncations or amino acid substitutions. The similarity of the G-box domain to amyloids (or amyloid precursors) is strengthened by its oligomeric arrangement to form continuous fibrils. G-box fibrils were observed in crystals as well as in solution and are also supported by simulations. We conclude that the G-box domain may well represent the best analogue currently available for studies of exposed β-sheets, unencumbered by additional structural elements or severe aggregations problems.
Keywords: CPAP, centriole, fibril, structure, b-sheet
Introduction
Centrioles are large cylindrical organelles, ubiquitous in eukaryotic cells, with a microtubule-based outer wall and characteristic 9-fold radial symmetry. We have limited structural information on most proteins found in centrioles but, so far, the few components resolved yielded exciting structures with the propensity to form large assemblies through oligomerization [1–4]. Recently, we and others, determined the structure of the C-terminal domain of the centriolar protein CPAP, termed the G-box [5–7]. The G-box domain adopts a unique conformation comprising a single continuous antiparallel β-sheet of approximately 7.5 nm length. Further, this domain oligomerizes in vitro in a concentration-depended manner and, in crystals, it self-associates through a head-to-tail interaction to form continuous β-sheets reminiscent of amyloid fibrils [5–7].
Functionally, CPAP is one of the five proteins, excluding microtubules, necessary for centriole formation. CPAP is a positive regulator of centriole length and, together with the centrosomal protein 120kDa (Cep120), Cep135 and the spindle and centriole associated protein 1 (SPICE1), it controls the centriole elongation process [8–11]. In centrioles, CPAP localizes close to the outer microtubule wall [12], where the α/β tubulin units impose a spatial periodicity of ∼8 nm [13]. Inspired by the similar length of the G-box, we proposed a model [5] in which G-box fibrils act as a molecular ruler aligning the centriolar core structure [14,15] with outer microtubules. Our model provides a mechanistic explanation for the abnormally long centrioles observed upon CPAP overexpression [11,16,17] and it crucially relies on the G-box domain to provide a platform for protein interactions important for centriole assembly. The importance of this domain to centriole assembly and consequently to human health, is demonstrated by a mis-sense mutation therein, E1235V that negatively affects centriole formation [6] and leads to primary microcephaly [18].
The G-box structure is the sole example from a unique protein fold. Although all-β proteins are common, they typically have a hydrophobic core formed by interactions between β-sheets as seen in the β-sandwich, β-helix or β-barrel folds. In these folds, open (non-hydrogen bonded) edges of β-strands are protected against interacting with other open β-strand edges, for example through steric hindrance by nearby loops or helices [19]. Single β-sheet proteins, in contrast, tend to form large aggregates through β-strand edge-to-edge interactions as well as exposed hydrophobic patches; such associations are well documented in amyloid fibrils where long parallel sheets, formed by β-strand interactions, sandwich hydrophobic surfaces [20–22]. As a result, biophysical studies of single β-sheets in isolation have primarily relied on protein analogues [23] and thus far only one short three-stranded β-sheet has been engineered [24]. To our knowledge, naturally occurring single-layered β-sheet proteins are restricted to the Borrelia burgdorferi outer surface protein A (OspA) [25] and the surface protein G (SasG) identified in Staphylococcus aureus [26]. However, OspA features globular domains capping its exposed single-layered central β-sheet, whereas the SasG topology does not define a continuous β-sheet. Hence, the G-box domain is, to our knowledge, the only available example of a repetitive class III single-antiparallel β-structure protein [27] and it can thus provide valuable information about the stability, flexibility and properties of such assemblies.
Materials and Methods
Details of the molecular dynamics (MD) simulations and small angle X-ray scattering (SAXS) methods are available as supporting information.
The β-sheet fold of the G-box domain
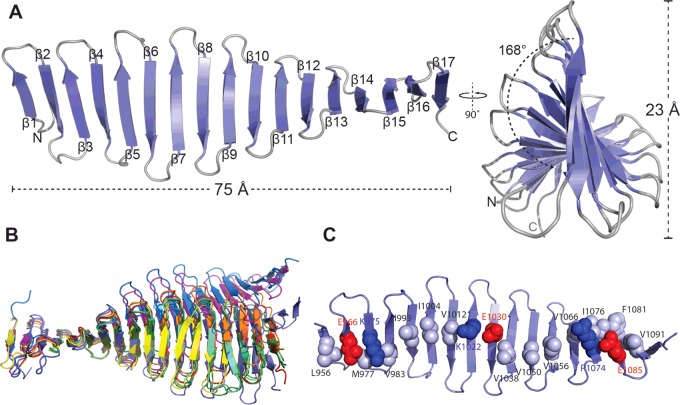
Our structure of the G-box domain from Danio rerio CPAP (4LD1, 4LD3 and 4LZF) [5] consists of 17 antiparallel β-strands that form a 75 Å (1 Å=0.1 nm) long and 23 Å wide β-sheet, with 168° of twist across its length (Figure 1A). Remarkably, all residues of this structure are solvent exposed, thus the G-box domain lacks a well-defined hydrophobic core. However, despite adopting a unique protein fold, all metrics of β-sheet geometry in the G-box domain match those observed in other systems. The mean twist per strand is 18°, which is comparable with that of globular proteins (30±30°) [28,29] and the overall β-sheet twist is left handed, consistent with reports that this is the most energetically favourable case [30]. Further, studies on the dependence of protein stability to β-strand lengths, using synthetic peptides forming antiparallel β-hairpins, found that seven residues per strand were optimal [31], this is also the length of most β-strands in the G-box domain.
Figure 1. Structural analysis of the CPAP G-box domain.
(A) Schematic representation of G-box in two perpendicular views. The secondary structure elements, protein dimensions and the β-sheet twist are shown. (B) Superposition of the first five β-strands of all available G-box structures. 4LD1 in slate, 4LZ in red, 4LD3 in orange, 4MPZ in yellow, 4BY2_A in light green, 4BY2_B n dark green, 4BY2_C in lime green, 4BXR_A in marine, 4BXR_B in magenta and 4BXP in blue. (C) Representation of electrostatic and hydrophobic interactions on the surface of the G-box β-sheet. Hydrophobic residues are shown in grey in space-filling representation, whereas charged residues are in blue (for positive charge) and red (for negative charge).
In addition to our model, further structures of the D. rerio CPAP G-box are now resolved (4BXP and 4BXR [6]), as well as structures from the Drosophila melanogaster CPAP G-box homologue (4BY2 and 4MPZ [6,7]). Although all structures show the same basic fold they differ in the number of β-strands formed, with the D. rerio G-box comprising 17–20 strands depending on the crystallographic model and the D. melanogaster domain having 14–15 strands. Superposition of the first five β-strands of these models shows that they differ in overall β-sheet geometry (Figure 1B), however no single point of large deviation could be identified. Rather, minor differences in inter-strand angles, amplified across the 14–20-strand long sheet, result in different β-sheet orientations. Further, β-sheet geometry is likely constrained and influenced by crystal packing forces, which naturally vary in these models due to different crystal contacts.
The G-box domain lacks a canonical, well-defined hydrophobic core as all its amino acid side chains are solvent exposed, yet it is stable in solution as shown by thermal denaturation experiments [5]. The structure is stabilized through a network of hydrophobic contacts and salt bridges that form between adjacent β-strands (Figure 1C). On one face of the G-box β-sheet strings of hydrophobic residues, such as Val1038, Val1050 and Ile1056, mediate contacts between adjacent strands. Any gaps in the continuity of the hydrophobic network are complemented with salt bridges, such as between Lys1022 and Glu1030. The second face of the G-box β-sheet has similar motifs of stabilization and, in addition, contains multiple aromatic and arginine residues involved in π-stacking interactions.
The G-box domain has a stable but dynamic structure
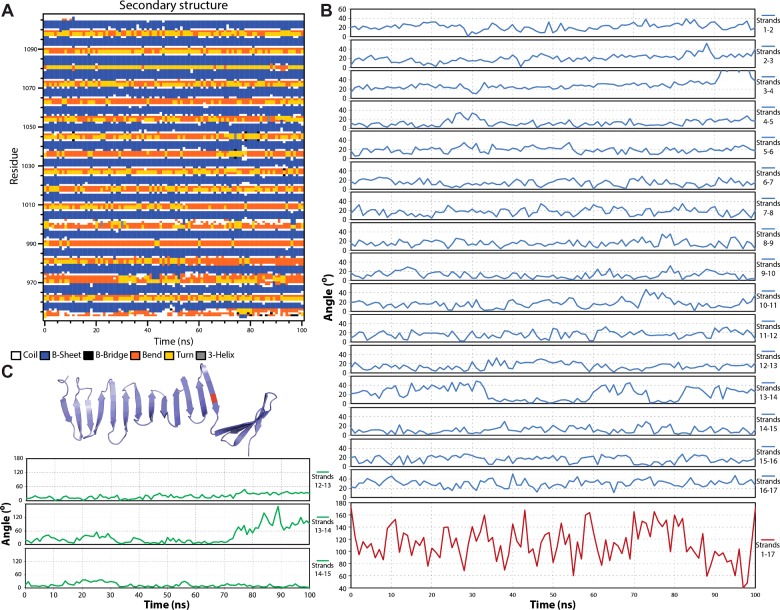
In order to further investigate the unique nature of this domain, we used atomistic MD simulations to gain insight in the stability and flexibility of the structure. Starting from the crystallographic model 4LD1 we performed 100 ns-long simulations [5], throughout which the G-box structure remains stable as it retains the main chain hydrogen bonding network and secondary structure elements (Figure 2A; Supplementary Table S1; Supplementary Movie S1). Minimal distortions are visible in the terminal β-strands where the protein N- and C-termini tend to fold back and interact with the β-sheet; these distortions are transient and manifest as slightly higher than average inter-strand twist angles at the terminal β-strands (Figure 2B). The overall G-box β-sheet twist remains left-handed throughout the MD simulations although it flattens somewhat to 130±30° from the 168° starting point. In addition, an average length of seven amino acids is maintained for the majority of β-strands throughout the simulations (Figure 2A). Hence, the G-box structure retains an energetically favourable conformation with respect to its twist [30] and typical strand length [31]. Further domain motions in the MD simulations result in slight β-sheet bending, such that the average end-to-end distance is 72 Å compared with 75 Å in the starting model and transient fanning of β-strand termini as single hydrogen bonds break and reform.
Figure 2. Analysis of the G-box structure throughout 100 ns of atomistic MD simulations.
(A) Secondary structure composition as function of amino acid residue and simulation time and (B) inter-strand twist angle as function of simulation time. The specific strand pairs are indicated. (C) One MD simulation of an I1067G substitution on β-strand 13 yielded large fluctuations in twist and eventually in breakage of the β-sheet. Shown here are (top) a snapshot of the G-box model at the end of this MD simulation and (bottom) inter-strand twist angles for the region surrounding the I1067G substitution.
The variable number of β-strands resolved in the different crystallographic models of the G-box domain, as well as biophysical data showing that G-box variants harbouring single amino acid substitutions remain folded [5–7], led us to study the effects of truncations or substitutions on the G-box structure in MD simulations. We constructed in silico C-terminally or N-terminally truncated models of the G-box domain comprising just eight out of 17 strands; however, 100 ns-long MD simulations of these models did not show significant structural perturbations (Supplementary Tables S2 and S3 respectively). G-box variants harbouring amino acid substitutions V1050A, V1050S, Y995A, F1024A, F1069A or V1050A/I1056A also did not yield significant or reproducible differences in simulations compared with the wild-type domain (result not shown). These results suggest that the G-box structure is resistant to truncations or most substitutions. However, we were able to perturb the G-box structure using a single I1067G substitution, located on β-strand 13. Glycine residues are known to destabilize β-sheets [32] and in one out of three MD simulations of the I1067G G-box variant we observed large fluctuations in the inter-strand twist angle (Figure 2C; Supplementary Table S4) and increased loss of hydrogen bonds near the substitution, which resulted in the complete breakage of the β-sheet (Figure 2C).
The G-box domain forms fibril
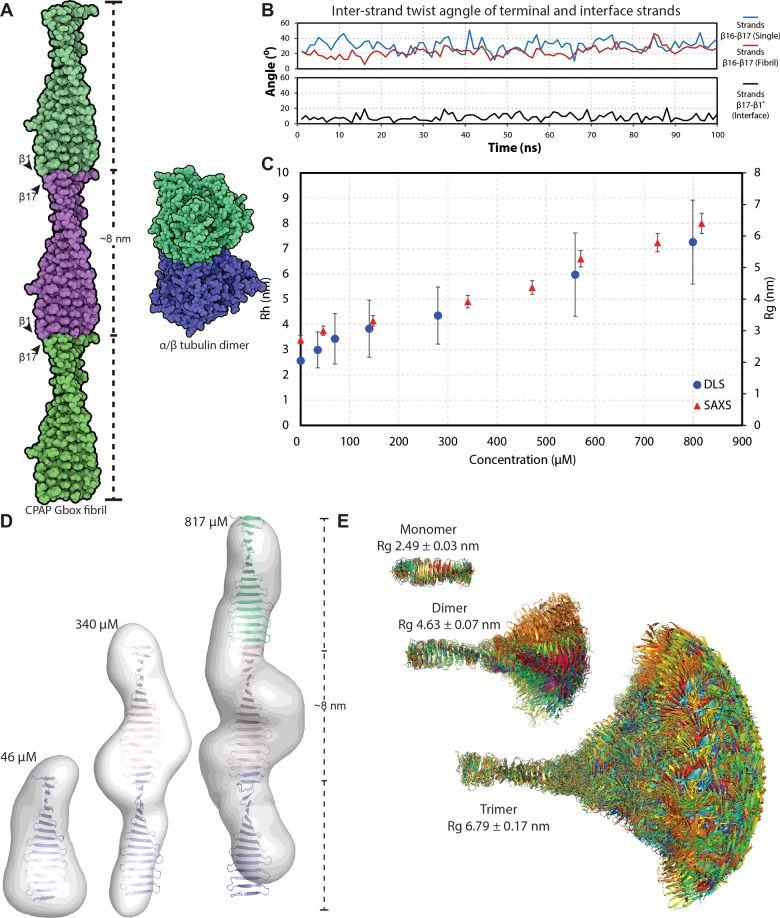
In several crystallographic structures (4LD1, 4LD3, 4LZF, 4BY2 and 4MPZ), G-box domains associate through a head-to-tail interaction along the open ends of the first and last β-strands [5–7]. This interaction is stabilized by the formation of five hydrogen bonds and the burial of ∼420 Å2 of accessible surface area. The resulting assembly is a continuous β-sheet with ∼8 nm periodicity, reminiscent of amyloid fibrils (Figure 3A). MD simulations of this interface show that it persists with no significant perturbations (Supplementary Movie S2) and that it has a lower twist angle as measured between strand 17 of one monomer and strand 1 of the next, compared with the average inter-strand twist of the G-box domain (∼8° compared with ∼18°, Figure 3B; Supplementary Table S5). Further, whereas MD simulations of the isolated G-box domain showed some fluctuations at the terminal strands, these were substantially reduced in the oligomer suggesting that oligomerization stabilizes the G-box conformation (Figure 3B).
Figure 3. The CPAP G-box domain forms elongated fibrils.
(A) Schematic representation of the G-box fibril in comparison with the α/β tubulin heterodimer. Three successive G-box domains are coloured green, magenta and green, with the β1 and β17 strands of each monomer and the fibril periodicity indicated. (B) Mean Rh and Rg of G-box oligomers in solution as a function of protein concentration, derived by DLS and SAXS respectively. (C) Analysis of the end-to-end interaction between G-box domains by MD simulations. The inter-strand twist angle is shown for the last G-box β-hairpin of the domain when simulated alone (in red) or as part of a fibril (in blue) and for the interface β-strands in black. (D) Ab-initio SAXS reconstruction of G-box oligomers at increasing protein concentration, with G-box monomeric and fibrillar structures fitted within. (E) Overlay of G-box models extracted from the MD simulations and used to build fibrillar oligomers (monomer to dimer to trimer). The calculated Rg for each oligomeric species is shown.
To establish whether the G-box domain forms similar fibrils in solution we performed dynamic light scattering (DLS) and SAXS. As shown in Figure 3(C), the DLS and SAXS measurements show that G-box forms oligomers of increasing hydrodynamic radius (Rh) and radius of gyration (Rg) respectively, as a function of protein concentration. The ab initio reconstructions derived from the SAXS data are in good agreement with the monomeric G-box domain for the lowest sample concentration (Figure 3D). As the protein concentration increases, the ab initio models derived from SAXS data have increasingly elongated shape and match the dimensions of G-box oligomers, composed of 2 or 3 domains arranged in elongated fibrils as observed in the crystals (Figure 3D). Based on these data, we conclude that the G-box domain is inherently able to form oligomers in solution, in a manner probably similar to the fibrils observed in the crystal.
However, it should be noted that the fit of the crystallographic fibrils to the elongated SAXS reconstructions is not ideal, probably due to sample polydispersity. We anticipate that the G-box domain in solution will form a mixture of oligomers with different lengths and, in addition that the domain flexibility observed in MD simulations (Supplementary Movie S1) will result in larger G-box oligomers having variable geometries. To better visualize such variable geometries we constructed in silico fibrils using 15 structurally diverse G-box models extracted from the MD simulations. As seen in Figure 3(E), fibrils of even just three G-box domains show a vast number of possible conformations.
Conclusion
The G-box structure has shown that proteins comprising isolated, single β-sheets are possible. Although the structure is unique, it obeys the same basic principles of favourable β-sheet formation, such as strand twist and length, as those noted for globular proteins. Conversely, lessons learned from studying the G-box domain may be more generally applicable. Thus, we believe that this domain constitutes an ideal system to study the structural, dynamic and thermodynamic properties of β-sheets, without contributions from additional structural elements. Furthermore, the G-box domain forms end-to-end fibrils that may be useful in studies of amyloidogenesis, yet be easier to characterise than true amyloids as G-box oligomers are soluble and reversible. More broadly, the CPAP G-box domain is just one example of structural ‘solutions’ that have evolved to underpin formation of centrioles, a remarkable protein assembly in eukaryotes. As we delve further into this system, it is intriguing to speculate what other structural ‘surprises’ we may uncover.
Acknowledgments
We thank the ESRF and the EMBL Hamburg DESY for provision of synchrotron radiation facilities.
Abbreviations
- CPAP
centrosome P4.1-associated protein
- DLS
dynamic light scattering
- MD
molecular dynamics
- OspA
outer surface protein A
- Rg
radius of gyration
- Rh
hydrodynamics radius
- SAXS
small angle X-ray scattering
Footnotes
Repetitive, Non-Globular Proteins: Nature to Nanotechnology: Held at the University of York, U.K., 30 March 2015–1 April 2015.
Funding
This work was supported by the Wellcome Trust [grant number 088497/Z/09/Z (to I.V.) and PhD studentship (to E.E.C.)]; and the Biotechnology and Biological Sciences Research Council [grant number BB/J008265/1 (to I.V.) and BB/1019855/1 (to P.J.S.)].
References
- 1.Kitagawa D., Vakonakis I., Olieric N., Hilbert M., Keller D., Olieric V., Bortfeld M., Erat M.C.M.C., Flückiger I., Gönczy P., et al. Structural basis of the 9-fold symmetry of centrioles. Cell. 2011;144:364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Breugel M., Hirono M., Andreeva A., Yanagisawa H.A., Yamaguchi S., Nakazawa Y., Morgner N., Petrovich M., Ebong I.-O.O., Robinson C.V., et al. Structures of SAS-6 suggest its organization in centrioles. Science. 2011;331:1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- 3.Hilbert M., Erat M.C., Hachet V., Guichard P., Blank I.D., Flückiger I., Slater L., Lowe E.D., Hatzopoulos G.N., Steinmetz M.O., et al. Caenorhabditis elegans centriolar protein SAS-6 forms a spiral that is consistent with imparting a ninefold symmetry. Proc. Natl. Acad. Sci. U.S.A. 2013;110:11373–11378. doi: 10.1073/pnas.1302721110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Breugel M., Wilcken R., McLaughlin S.H., Rutherford T.J., Johnson C.M. Structure of the SAS-6 cartwheel hub from Leishmania major. Elife. 2014;3:e01812. doi: 10.7554/eLife.01812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hatzopoulos G.N., Erat M.C., Cutts E., Rogala K.B., Slater L.M., Stansfeld P.J., Vakonakis I. Structural analysis of the G-box domain of the microcephaly protein CPAP suggests a role in centriole architecture. Structure. 2013;21:2069–2077. doi: 10.1016/j.str.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cottee M.A., Muschalik N., Wong Y.L., Johnson C.M., Johnson S., Andreeva A., Oegema K., Lea S.M., Raff J.W., van Breugel M. Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly. Elife. 2013;2:e01071. doi: 10.7554/eLife.01071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng X., Gooi L.M., Wason A., Gabriel E., Mehrjardi N.Z., Yang Q., Zhang X., Debec A., Basiri M.L., Avidor-Reiss T., et al. Conserved TCP domain of Sas-4/CPAP is essential for pericentriolar material tethering during centrosome biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E354–E363. doi: 10.1073/pnas.1317535111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comartin D., Gupta G.D., Fussner E., Coyaud É., Hasegan M., Archinti M., Cheung S.W.T., Pinchev D., Lawo S., Raught B., et al. CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr. Biol. 2013;23:1360–1366. doi: 10.1016/j.cub.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Lin Y.-C., Chang C.-W., Hsu W.-B., Tang C.-J.C., Lin Y.-N., Chou E.-J., Wu C.-T., Tang T.K. Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. EMBO J. 2013;32:1141–1154. doi: 10.1038/emboj.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang C.C.J., Lin S.Y., Hsu W.B., Lin Y.Y.N.C., Wu C.T., Lin Y.Y.N.C., Chang C.W., Wu K.S., Tang T.K. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 2011;30:4790–4804. doi: 10.1038/emboj.2011.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang C.-J.C., Fu R.-H., Wu K.-S., Hsu W.-B., Tang T.K. CPAP is a cell-cycle regulated protein that controls centriole length. Nat. Cell Biol. 2009;11:825–831. doi: 10.1038/ncb1889. [DOI] [PubMed] [Google Scholar]
- 12.Sonnen K.F., Schermelleh L., Leonhardt H., Nigg E.A. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open. 2012;1:965–976. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prota A.E., Bargsten K., Zurwerra D., Field J.J., Díaz J.F., Altmann K.-H., Steinmetz M.O. Molecular mechanism of action of microtubule-stabilizing anticancer agents. Science. 2013;339:587–590. doi: 10.1126/science.1230582. [DOI] [PubMed] [Google Scholar]
- 14.Guichard P., Hachet V., Majubu N., Neves A., Demurtas D., Olieric N., Fluckiger I., Yamada A., Kihara K., Nishida Y., et al. Native architecture of the centriole proximal region reveals features underlying its 9-fold radial symmetry. Curr. Biol. 2013;23:1620–1628. doi: 10.1016/j.cub.2013.06.061. [DOI] [PubMed] [Google Scholar]
- 15.Guichard P., Desfosses A., Maheshwari A., Hachet V., Dietrich C., Brune A., Ishikawa T., Sachse C., Gönczy P., Gonczy P., Gönczy P. Cartwheel architecture of Trichonympha basal body. Science. 2012;337:553. doi: 10.1126/science.1222789. [DOI] [PubMed] [Google Scholar]
- 16.Kohlmaier G., Lončarek J., Meng X., McEwen B.F., Mogensen M.M., Spektor A., Dynlacht B.D., Khodjakov A., Gönczy P., Loncarek J., et al. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr. Biol. 2009;19:1012–1018. doi: 10.1016/j.cub.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt T.I., Kleylein-Sohn J., Westendorf J., Le Clech M., Lavoie S.B., Stierhof Y.-D.D., Nigg E.A. Control of centriole length by CPAP and CP110. Curr. Biol. 2009;19:1005–1011. doi: 10.1016/j.cub.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 18.Thornton G.K., Woods C.G. Primary microcephaly: do all roads lead to Rome? Trends Genet. 2009;25:501–510. doi: 10.1016/j.tig.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richardson J.S., Richardson D.C. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esposito L., Pedone C., Vitagliano L. Molecular dynamics analyses of cross-beta-spine steric zipper models: beta-sheet twisting and aggregation. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11533–11538. doi: 10.1073/pnas.0602345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson R., Sawaya M.R., Balbirnie M., Madsen A.Ø., Riekel C., Grothe R., Eisenberg D. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makin O.S., Atkins E., Sikorski P., Johansson J., Serpell L.C. Molecular basis for amyloid fibril formation and stability. Proc. Natl. Acad. Sci. U.S.A. 2005;102:315–320. doi: 10.1073/pnas.0406847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makabe K., McElheny D., Tereshko V., Hilyard A., Gawlak G., Yan S., Koide A., Koide S. Atomic structures of peptide self-assembly mimics. Proc. Natl. Acad. Sci. U.S.A. 2006;103:17753–17758. doi: 10.1073/pnas.0606690103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kortemme T. Design of a 20-amino acid, three-stranded -sheet protein. Science. 1998;281:253–256. doi: 10.1126/science.281.5374.253. [DOI] [PubMed] [Google Scholar]
- 25.Li H., Dunn J.J., Luft B.J., Lawson C.L. Crystal structure of Lyme disease antigen outer surface protein A complexed with an Fab. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3584–3589. doi: 10.1073/pnas.94.8.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruszka D.T., Wojdyla J.A., Bingham R.J., Turkenburg J.P., Manfield I.W., Steward A., Leech A.P., Geoghegan J.A., Foster T.J., Clarke J., Potts J.R. Staphylococcal biofilm-forming protein has a contiguous rod-like structure. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E1011–E1018. doi: 10.1073/pnas.1119456109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kajava A.V. Tandem repeats in proteins: from sequence to structure. J. Struct. Biol. 2012;179:279–288. doi: 10.1016/j.jsb.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Nagarajaram H.A., Reddy B.V.B., Blundell T.L. Analysis and prediction of inter-strand packing distances between β-sheets of globular proteins. Protein Eng. 1999;12:1055–1062. doi: 10.1093/protein/12.12.1055. [DOI] [PubMed] [Google Scholar]
- 29.Ho B.K., Curmi P.M.G. Twist and shear in beta-sheets and beta-ribbons. J. Mol. Biol. 2002;317:291–308. doi: 10.1006/jmbi.2001.5385. [DOI] [PubMed] [Google Scholar]
- 30.Chothia C. Conformation of twisted beta-pleated sheets in proteins. J. Mol. Biol. 1973;75:295–302. doi: 10.1016/0022-2836(73)90022-3. [DOI] [PubMed] [Google Scholar]
- 31.Stanger H.E., Syud F.A., Espinosa J.F., Giriat I., Muir T., Gellman S.H. Length-dependent stability and strand length limits in antiparallel beta -sheet secondary structure. Proc. Natl. Acad. Sci. U.S.A. 2001;98:12015–12020. doi: 10.1073/pnas.211536998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith C.K., Withka J.M., Regan L. A thermodynamic scale for the beta-sheet forming tendencies of the amino acids. Biochemistry. 1994;33:5510–5517. doi: 10.1021/bi00184a020. [DOI] [PubMed] [Google Scholar]