Abstract
Nrf2 (nuclear factor erytheroid-derived-2-like 2) transcriptional programmes are activated by a variety of cellular stress conditions to maintain cellular homoeostasis. Under non-stress conditions, Nrf2 is under tight regulation by the ubiquitin proteasome system (UPS). Detailed mechanistic investigations have shown the Kelch-like ECH-associated protein 1 (Keap1)–cullin3 (Cul3)–ring-box1 (Rbx1) E3-ligase to be the primary Nrf2 regulatory system. Recently, both beta-transducin repeat-containing E3 ubiquitin protein ligase (β-TrCP) and E3 ubiquitin-protein ligase synoviolin (Hrd1) have been identified as novel E3 ubiquitin ligases that negatively regulate Nrf2 through Keap1-independent mechanisms. In addition to UPS-mediated regulation of Nrf2, investigations have revealed a cross-talk between Nrf2 and the autophagic pathway resulting in activation of Nrf2 in a non-canonical manner. In addition to regulation at the protein level, Nrf2 was recently shown to be regulated at the transcriptional level by oncogenic K-rat sarcoma (Ras). A consequence of these differential regulatory mechanisms is the dual role of Nrf2 in cancer: the canonical, protective role and the non-canonical ‘dark-side’ of Nrf2. Based on the protective role of Nrf2, a vast effort has been dedicated towards identifying novel chemical inducers of Nrf2 for the purpose of chemoprevention. On the other hand, upon malignant transformation, some cancer cells have a constitutively high level of Nrf2 offering a growth advantage, as well as rendering cancer cells resistant to chemotherapeutics. This discovery has led to a new paradigm in cancer treatment; the initially counterintuitive use of Nrf2 inhibitors as adjuvants in chemotherapy. Herein, we will discuss the mechanisms of Nrf2 regulation and how this detailed molecular understanding can be leveraged to develop Nrf2 modulators to prevent diseases, mitigate disease progression or overcome chemoresistance.
Keywords: chemoprevention/chemoresistance, Hrd1, Kelch-like ECH-associated protein 1 (Keap1), nuclear factor erytheroid-derived-2-like 2 (Nrf2), reactive oxygen species (ROS), ubiquitin proteasome system (UPS)
Introduction
Nuclear factor erytheroid-derived-2-like 2 (Nrf2) is a member of the ‘cap-n-collar’ class of basic leucine zipper transcription factors. Under basal conditions, Nrf2 is tightly regulated by several ubiquitin proteasome systems (UPSs)-mediated mechanisms. Upon activation, the levels of Nrf2 rise and nuclear Nrf2 heterodimerizes with one of the small Maf proteins. These Nrf2–Maf heterodimers recognize antioxidant response elements (AREs), 11- (or 16) bp enhancer sequences in the regulatory region of Nrf2 target genes, thereby allowing the recruitment of key factors for transcript synthesis [1,2]. Typical ARE-harbouring genes include redox balancing factors, detoxifying enzymes, transporters, stress response proteins and metabolic enzymes [2–4]. Up-regulation of this series of Nrf2-target genes helps the cell to combat harmful stressors such as reactive oxygen species (ROS) and electrophilic xenobiotics, effectively providing a cellular survival mechanism. This cytoprotective activity of Nrf2 has been implicated in disease prevention, including cancer [5–8]. In the case of cancer, controlled Nrf2 up-regulation has shown to protect against the initiation of many types of cancer [9,10]. Intriguingly, whereas it has been demonstrated that Nrf2 activation is effective in preventing oxidation-related disease pathogenesis, its role in disease progression once onset has occurred remains controversial [6]. In addition, the ‘dark-side’ of Nrf2 has recently been revealed. In this context, uncontrolled Nrf2 expression facilitates tumour growth and causes chemoresistance [11–14]. In the present review, we will discuss the mechanisms of Nrf2 regulation (both canonical and non-canonical) and chemical modulation to activate or inhibit the Nrf2 pathway as a chemopreventive or therapeutic strategy respectively.
Nrf2 regulation at the protein level by the ubiquitin proteasome system
Keap1–Cul3–Rbx1 E3 ubiquitin ligase (the canonical mechanism of Nrf2 regulation)
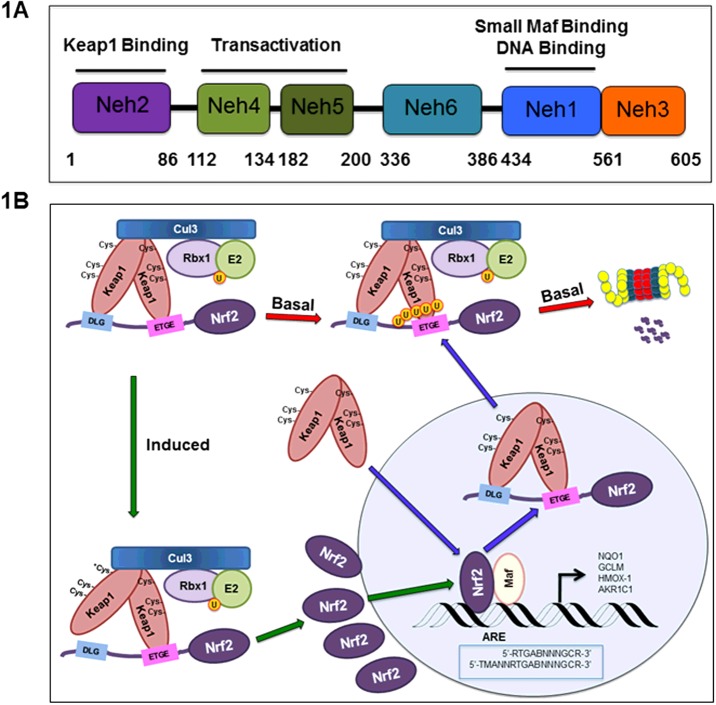
In an unstressed state, the Nrf2 protective response is not needed. In fact, genetic ablation of Nrf2 is tolerated in mice, although they are highly sensitive to xenobiotic stress [15]. Under basal conditions, cells mediate the constant degradation of Nrf2 through the UPS, keeping Nrf2 protein levels low and preventing transcription of un-needed genes. This regulation occurs through Kelch-like ECH-associated protein 1 (Keap1), an adaptor protein of a cullin3 (Cul3)–ring-box 1 (Rbx1) containing E3 ubiquitin ligase complex [16,17]. Dimeric Keap1 is responsible for recognition of Nrf2 through two key motifs in the Neh2 domain of Nrf2 located in its N-terminus (Figures 1A and 2A). The Kelch domain of each Keap1 binds to the ‘DLG’ and ‘ETGE’ motifs, recognized as the low affinity and high-affinity-binding sites respectively [18,19]. Nrf2 is subsequently polyubiquitylated at seven key lysine residues within the Neh2 domain, condemning Nrf2 to proteasomal destruction [17] (Figure 1B).
Figure 1. The canonical Nrf2 regulatory pathway.
(A) The domain architecture of Nrf2 and known functions of the individual domains. (B) The canonical, Keap1–Cul3–Rbx1-mediated Nrf2 regulatory pathway. Upon cellular insult, cysteine residues in Keap1 are modified and the activity of the E3 ubiquitin ligase is suppressed. Nrf2 levels rise and Nrf2 enters the nucleus, where it dimerizes with Maf and turns on ARE-containing genes.
Figure 2. The three E3 ubiquitin ligases for Nrf2.
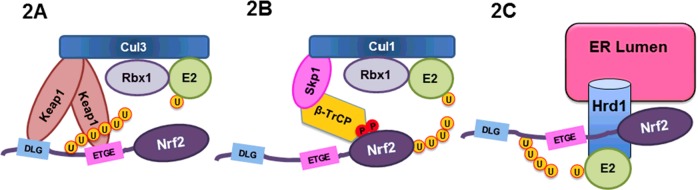
(A) Keap1–Cul3–Rbx1 E3 ubiquitin ligase. (B) β-TrCP–Skp1–Cul1–Rbx1 E3 ubiquitin ligase. (C) The Hrd1 E3 ubiquitin ligase.
Upon introduction of electrophiles or ROS, the Nrf2-mediated cytoprotective response is activated. Critical cysteine residues in Keap1, especially Cys151, act as sensors of these cellular insults and become covalently-modified by electrophilic species or ROS [20]. Additional cysteine residues in Keap1 may also be modified by electrophilic species [21,22]. Such modifications induce a conformational change in Keap1, probably by disrupting the low-affinity interaction between the Kelch domain and the DLG-motif, which leads to impaired ubiquitylation of Nrf2, blocking UPS-mediated degradation and thus increasing Nrf2 protein levels [23]. As newly synthesized Nrf2 accumulates, there are no longer sufficient Keap1 molecules available for Nrf2 binding. This cytosolic Nrf2 is then free to translocate into the nucleus and transcriptionally activate its target genes. After induction, when homoeostasis is restored, Keap1 translocates into the nucleus in a karyopherin alpha 6 (importin alpha 7) (KPNA6)-dependent manner to facilitate nuclear export of Nrf2 and rejoins the Keap1-mediated ubiquitylation and degradation machinery in the cytosol [24,25]. These events turn off the transcription of the Nrf2 target genes and restore the low basal level of Nrf2 to maintain cellular redox homoeostasis (Figure 1B).
GSK-3β/β-TrCP–Skp1–Cul1–Rbx1 E3 ubiquitin ligase
Although Keap1 has been revealed as the primary redox-sensitive regulator of Nrf2 through reactive cysteine residues, a redox-insensitive degron within the Neh6 domain of Nrf2 was reported in 2004 [26] (Figure 1A). Subsequently, it was found that the Neh6 domain of mouse Nrf2 contains a group of serine residues that can be phosphorylated by glycogen synthase kinase 3 (GSK-3), a serine/threonine kinase. This phosphorylation event in the Neh6 domain creates a phosphorylated destruction motif (phosphodegron), which can then be recognized by the β-TrCP–Skp1–Cul1–Rbx1 E3 ubiquitin ligase complex [27] (Figure 2B). This E3 ligase complex ubiquitylates Nrf2 and sends it to the proteasome for destruction. Further characterization of the Neh6 domain found two distinct motifs recognized by β-TrCP, DSAPGS and DSGIS, the latter containing a GSK-3β phosphorylation site [28]. However, the conditions that favour the GSK-3β/β-TrCP E3 ubiquitin ligase over the Keap1–Cul3–Rbx1 E3 ligase in controlling Nrf2 remains to be determined.
Hrd1 E3 ubiquitin ligase
More recently, our laboratories discovered that the Nrf2-mediated protective response was suppressed during liver cirrhosis [29]. This was a surprising result because Keap1 should be inactivated by the high levels of ROS in cirrhotic livers, leading to Nrf2 signalling by the canonical mechanism. Liver cirrhosis is a pathogenic state typically caused by chronic alcohol consumption or viral hepatitis infection, resulting in a profound scarring of the liver. Endoplasmic reticulum (ER) stress has been implicated during the pathogenesis of liver cirrhosis. ER stress occurs when misfolded proteins accumulate and the unfolded protein response (UPR) is then initiated. Three sensors, inositol-requiring enzyme 1 (IRE1), protein kinase RNA-like ER kinase (PERK) and activating transcription factor 6 (ATF6), located on the ER membrane detect the accumulation of misfolded proteins and relays signalling cascades, resulting in induction of heat-shock proteins, autophagy factors, proteasomal subunits and apoptotic factors and a decrease in other secretory proteins until homoeostasis is reached [30]. If the UPR system fails to correct the insult, apoptotic programmes are activated.
To understand the decrease in Nrf2 in cirrhotic livers, we investigated the cross-talk between the ER stress pathways and the Nrf2-mediated antioxidant stress pathway. It was found that decreased Nrf2 levels correlated with activation of the IRE1 arm of the UPR. ER stress is known to release the association between IRE1 and 78 kDa glucose-regulated protein (GRP78) (a chaperone also known as BiP, part of the heat-shock protein 70 (HSP70) family), enabling free IRE1 to homodimerize and actively splice X-box-binding protein 1 (XBP1) mRNA into a mature mRNA encoding XBP1s, a transcription factor. Hrd1 is an XBP1s target gene that is up-regulated upon activation of the IRE1–XBP1 signalling pathway. Based on the fact that the protein level of Nrf2 was decreased in a Hrd1-dependent manner whenever the IRE1 arm is activated, we identified Hrd1 as a novel E3 ubiquitin ligase (Figure 2C) [29]. This discovery has important implications for the treatment and protection of cirrhotic livers.
Nrf2 regulation at the protein level by (E/S)TGE-containing proteins (the non-canonical mechanism of Nrf2 regulation)
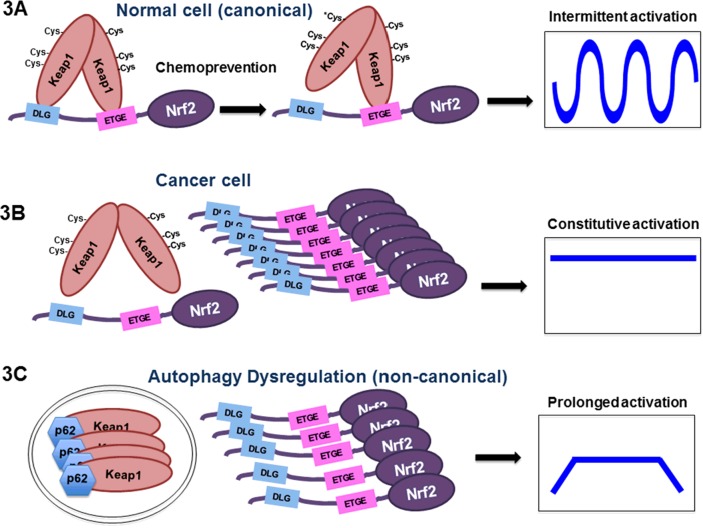
In addition to regulation through the UPS, Nrf2 is subjected to positive regulation by other proteins through disruption of the Nrf2–Keap1 interaction. A recent study identified numerous proteins with motifs identical (or similar) to the ETGE motif of Nrf2, which can compete with Nrf2 for Keap1 binding, thus stabilizing Nrf2 [31]. Some examples of (E/S)TGE containing proteins include dipeptidyl peptidase 3 (DPP3) and partner and localizer of BRCA2 (PALB2), but p62 (a protein containing the STGE motif) is perhaps the most recognized positive regulator of Nrf2. p62 is a scaffold protein recruited into autophagosomes to shuttle cargo proteins destined for degradation through autophagy-lysosome pathway, the cell's bulk degradation process. Autophagy dysregulation leads to accumulation of autophagosomes, where p62 captures Keap1, resulting in inactivation of Keap1-mediated Nrf2 ubiquitylation and thus activation of Nrf2 [32]. It is envisioned that activation of Nrf2 by this p62-dependent, but Keap1 cysteine-independent mechanism takes a much longer time to attenuate, which leads to heightened cell survival and potential cellular transformation (Figure 3C). Indeed, we demonstrated that the human carcinogen arsenic, widely distributed in the ground water presenting a global health issue that affects some 200 million people, activates Nrf2 through this non-canonical mechanism, resulting in prolonged Nrf2 activation and possibly explaining the carcinogenic nature of arsenic [33].
Figure 3. The three modes of Nrf2 activation in health and disease.
(A) The canonical mechanism of Nrf2 regulation in a normal cell. (B) Misregulated Nrf2 signalling in a cancer cell that leads to constitutive activation of Nrf2. The molecular events underlying this misregulation are discussed in the text. (C) Prolonged Nrf2 signalling from compromised autophagy. This is probably a major contributing factor in arsenic-mediated carcinogenesis.
Nrf2 regulation at the transcriptional level by oncogenes
Nrf2 was previously reported to be up-regulated at the transcriptional level by oncogenic activation of K-RasG12D, B-RafV619E and MycERT2 [34]. Our group demonstrated that activation of Nrf2 by oncogenic K-rat sarcoma (Ras) is facilitated through a TPA (12-O-Tetradecanoylphorbol-13-acetate)-responsive element (TRE) in the regulatory region of NRF2 [35]. This was the first demonstration of Nrf2 modulation at the transcriptional level. These studies also argued for the potential power of Nrf2 inhibitors, such as brusatol, in this context to facilitate chemotherapy. The precise mechanisms by which B-RafV619E and MycERT2 up-regulate the transcription of Nrf2 are currently unknown.
Chemical modulation targeting Nrf2 activation for disease prevention
The validity of the role of Nrf2 in cancer prevention is most drastically seen in the Nrf2 knockout (Nrf2−/−) mouse model. Despite their normal embryonic development and lifespan, Nrf2−/− mice are highly susceptible to carcinogenic species and readily develop tumours upon exposure to chemical carcinogens, compared with their wild-type littermates [36]. The most likely mechanism contributing to this Nrf2-mediated protection against cancer development is the cell's ability to detoxify carcinogens and to combat mutagenic ROS via the activation of the battery of Nrf2-responsive target genes. Whereas Nrf2 is maintained at low levels in normal cells, mounting evidence demonstrates a reduced tumour incidence upon co-administration of Nrf2 activators along with a carcinogen [37].
Canonical Keap1-dependent Nrf2 activators
The salubrious benefits of many foods and herbal medicines have been known for thousands of years. The mechanisms of many of these beneficial effects often remain without explanation, but recent research has revealed activation of Nrf2 by many foods and traditional medicines as an explanation of benefit [6]. As discussed, Keap1 is the primary regulator of Nrf2 and is subject to oxidation and electrophilic modification. Many foods (i.e., broccoli, grapes and cinnamon) may contain natural electrophiles that react with Keap1–Cys151 and increase Nrf2 levels via the canonical mechanism. Suforaphane and cinnamaldeyde, from broccoli or cinnamon respectively, are the two well-studied canonical Nrf2 activators [33].
In addition to phytochemicals from foods and traditional medicines, two Nrf2 inducers; Bardoxolone-methyl (CDDO-Me) and dimethylfumarate (DMF) have advanced to clinical trials and the clinic respectively. Bardoxolone, a natural product derived triterpenoid, was reported to interact with the BTB domain of Keap1, disrupting the BTB/Cul3 interface, leading to Nrf2 activation [38]. Bardoxolone was entered into phase II clinical trials for the treatment of diabetic nephropathy and other complications. In this trial, an increased glomerular filtration rate in patients with chronic kidney disease was attributed to long-term bardoxolone treatment. However, upon matriculation to phase III, it was retracted due to safety issues [39]. Conversely, another compound capable of Keap1 adduction, DMF (also known as BG-12 and Tecfidera) was recently awarded Food and Drug Administration (FDA) approval for the treatment of multiple sclerosis (MS). Pre-clinically, DMF was shown to have significant neuroprotective effects in transgenic murine models of Huntington's disease as well as experimental models of demyelination and neurodegeneration [40,41].
Keap1-independent Nrf2 activators
Recently, Keap1-independent Nrf2 activators have emerged. For example, nordihydroguaiaretic acid (NDGA) was shown to increase Nrf2 and hemeoxygenase-1 (HO-1) levels through inhibition of phosphorylation of the Neh6 motif in Nrf2 by GSK-3β [42]. Additionally, we demonstrated that 4U8C and LS-102, an IRE1 inhibitor and a Hrd1 inhibitor respectively, were able to reactivate the Nrf2 protective response to improve liver function in the CCl4-induced liver cirrhosis model [29]. Therefore, it is essential to understand the physiological or pathological state under which a specific E3 has a dominant role. Only with this knowledge can the correct E3 ubiquitin ligase be targeted to effectively activate Nrf2 for disease prevention.
Nrf2 inhibition to overcome chemoresistance
Nrf2 undeniably plays a prominent role in cancer prevention; however, reports from our laboratory have revealed the ‘dark-side’ of Nrf2 [11]. Because Nrf2 improves cellular survival under cytotoxic challenges, cells heavily rely on Nrf2 activation to circumvent cell death. Unfortunately, upon malignant transformation, certain cancer cells have a constitutively high level of Nrf2 resulting in uncontrolled Nrf2 expression. Mounting evidence has indicated that elevated Nrf2 levels are associated with resistance to chemotherapeutic agents and a poor prognosis in many cancer types including non-small cell lung carcinoma, endometrial carcinoma and ovarian carcinoma [11,43,44]. In addition to protecting against cell death, many Nrf2 target genes are responsible for glucose metabolism, purine biogenesis and fatty acid oxidation, a necessity for rapid growth and proliferation of cancer cells [45,46]. More intriguingly, we have demonstrated that prolonged Nrf2 activation by chronic, low-level arsenic exposure through the non-canonical p62-dependent mechanism may be the underlying mechanism of arsenic-mediated carcinogenesis [33]. Figure 3 summarizes the modes of Nrf2 activation based on the canonical regulatory mechanism (Figure 3A), constitutive activation seen in cancer cells (Figure 3B) and prolonged activation due to autophagy dysregulation and accumulation of autophagosomes (Figure 3C).
Cancer cells can utilize several mechanisms to achieve prolonged or constitutive Nrf2 activation. Somatic mutations in KEAP1, NRF2 and CUL3 genes in cancer cells have been identified that disrupt the interaction between Nrf2 and Keap1, resulting in accumulation of Nrf2 (Figure 3B) [13,47,48]. It is certain that many somatic mutations in other genes encoding Nrf2-regulatory proteins will be identified in addition to these. Another mechanism for Nrf2 overexpression in cancer cells, which is consistent in brain, lung and prostate cancers, is the epigenetic silencing of the KEAP1 gene, leading to up-regulation of Nrf2 [49,50]. In these cases, the KEAP1 promoter region was found to be hypermethylated, preventing the expression of Keap1 mRNA.
In order to combat this oncogenic function of Nrf2, we have focused on the discovery and development of inhibitors of the Nrf2 pathway. Our first success in this vain, brusatol, potently decreases Nrf2 protein levels and target gene expression at nanomolar concentrations in cancer cells with constitutively high Nrf2 expression, enhancing the cytotoxic effect of cisplatin and other chemotherapeutics [12]. As mentioned above, oncogenic K-Ras up-regulates Nrf2 mRNA levels, which offer a possible explanation for why oncogenic K-Ras mutations lead to chemoresistant tumours. This also provides further evidence for the ‘dark-side’ of Nrf2. Very recently, we demonstrated that co-administration of brusatol enhanced the efficacy of cisplatin and improved survival of mice with lung cancer using a K-RasG12D mouse model [35]. As a result, brusatol has illuminated the value of Nrf2 inhibitors as adjuvants to chemotherapeutic drugs that are typical first line regimens for cancers.
Conclusion
In this review, we have discussed the canonical and non-canonical mechanisms of Nrf2 regulation and how these relate to the dual role of Nrf2 in cancer. Understanding these mechanisms has revealed novel means to control Nrf2 expression to maintain cellular redox homoeostasis. It should also be highlighted that the mechanisms governing some of these Nrf2 regulatory pathways remain to be understood, but the knowledge of Nrf2 regulation will add to our understanding of Nrf2-mediated pathological states and how to exploit this critical protective pathway to prevent diseases or to treat diseases directly or in combination with other drugs. In addition, we have discussed the consequence of prolonged or chronic up-regulation of Nrf2 and how this can lead to cellular transformation and chemoresistance. Understanding this ‘dark-side’ of Nrf2 has fuelled the counterintuitive drive to develop Nrf2 inhibitors. The leader of this class, brusatol, has proven to be a powerful tool in understanding Nrf2 biology and has provided a strong incentive to develop the next generation of Nrf2 inhibitors.
Abbreviations
- ARE
antioxidant response element
- Cul3
cullin 3
- DMF
dimethylfumarate
- ER
endoplasmic reticulum
- GSK
glycogen synthase kinase
- Keap1
Kelch-like ECH-associated protein 1
- Nrf2
nuclear factor erytheroid-derived-2-like 2
- ROS
reactive oxygen species
- UPS
ubiquitin proteasome system
- UPR
unfolded protein response
Footnotes
The Keap1/Nrf2 Pathway in Health and Disease: Held at Robinson College, Cambridge, UK, 6–8 Jan 2015.
Funding
This work was supported by the National Institute of Environmental Health Sciences [grant numbers R01 ES015010 and R01 CA154377 (to D.D.Z), R01 ES023758 (to E.C. and D.D.Z.) and ES006694].
References
- 1.Wasserman W.W., Fahl W.E. Functional antioxidant responsive elements. Proc. Natl. Acad. Sci. U.S.A. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayes J.D., McMahon M., Chowdhry S., Dinkova-Kostova A.T. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid. Redox Signal. 2010;13:1713–1748. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- 3.Malhotra D., Portales-Casamar E., Singh A., Srivastava S., Arenillas D., Happel C., Shyr C., Wakabayashi N., Kensler T.W., Wasserman W.W., Biswal S. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kensler T.W., Wakabayashi N., Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 5.Kensler T.W., Wakabayashi N. Nrf2: friend or foe for chemoprevention? Carcinogenesis. 2010;31:90–99. doi: 10.1093/carcin/bgp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaramillo M.C., Zhang D.D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kundu J.K., Surh Y.J. Nrf2-Keap1 signaling as a potential target for chemoprevention of inflammation-associated carcinogenesis. Pharm. Res. 2010;27:999–1013. doi: 10.1007/s11095-010-0096-8. [DOI] [PubMed] [Google Scholar]
- 8.Cheung K.L., Khor T.O., Huang M.T., Kong A.N. Differential in vivo mechanism of chemoprevention of tumor formation in azoxymethane/dextran sodium sulfate mice by PEITC and DBM. Carcinogenesis. 2010;31:880–885. doi: 10.1093/carcin/bgp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.auf dem Keller U., Huber M., Beyer T.A., Kumin A., Siemes C., Braun S., Bugnon P., Mitropoulos V., Johnson D.A., Johnson J.A., et al. Nrf transcription factors in keratinocytes are essential for skin tumor prevention but not for wound healing. Mol. Cell Biol. 2006;26:3773–3784. doi: 10.1128/MCB.26.10.3773-3784.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thimmulappa R.K., Rangasamy T., Alam J., Biswal S. Dibenzoylmethane activates Nrf2-dependent detoxification pathway and inhibits benzo(a)pyrene induced DNA adducts in lungs. Med Chem. 2008;4:473–481. doi: 10.2174/157340608785700199. [DOI] [PubMed] [Google Scholar]
- 11.Wang X.J., Sun Z., Villeneuve N.F., Zhang S., Zhao F., Li Y., Chen W., Yi X., Zheng W., Wondrak G.T., Wong P.K., Zhang D.D. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren D., Villeneuve N.F., Jiang T., Wu T., Lau A., Toppin H.A., Zhang D.D. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1433–1438. doi: 10.1073/pnas.1014275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh A., Misra V., Thimmulappa R.K., Lee H., Ames S., Hoque M.O., Herman J.G., Baylin S.B., Sidransky D., Gabrielson E., et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padmanabhan B., Tong K.I., Ohta T., Nakamura Y., Scharlock M., Ohtsuji M., Kang M.I., Kobayashi A., Yokoyama S., Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Aoki Y., Sato H., Nishimura N., Takahashi S., Itoh K., Yamamoto M. Accelerated DNA adduct formation in the lung of the Nrf2 knockout mouse exposed to diesel exhaust. Toxicol. Appl. Pharmacol. 2001;173:154–160. doi: 10.1006/taap.2001.9176. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi A., Kang M.I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang D.D., Lo S.C., Cross J.V., Templeton D.J., Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMahon M., Thomas N., Itoh K., Yamamoto M., Hayes J.D. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J. Biol. Chem. 2006;281:24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- 19.Tong K.I., Kobayashi A., Katsuoka F., Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol. Chem. 2006;387:1311–1320. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- 20.Zhang D.D., Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinkova-Kostova A.T., Holtzclaw W.D., Cole R.N., Itoh K., Wakabayashi N., Katoh Y., Yamamoto M., Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMahon M., Lamont D.J., Beattie K.A., Hayes J.D. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18838–18843. doi: 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baird L., Lleres D., Swift S., Dinkova-Kostova A.T. Regulatory flexibility in the Nrf2-mediated stress response is conferred by conformational cycling of the Keap1-Nrf2 protein complex. Proc. Natl. Acad. Sci. U.S.A. 2013;110:15259–15264. doi: 10.1073/pnas.1305687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Z., Wu T., Zhao F., Lau A., Birch C.M., Zhang D.D. KPNA6 (Importin {alpha}7)-mediated nuclear import of Keap1 represses the Nrf2-dependent antioxidant response. Mol. Cell Biol. 2011;31:1800–1811. doi: 10.1128/MCB.05036-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Z., Zhang S., Chan J.Y., Zhang D.D. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol. Cell Biol. 2007;27:6334–6349. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMahon M., Thomas N., Itoh K., Yamamoto M., Hayes J.D. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem. 2004;279:31556–31567. doi: 10.1074/jbc.M403061200. [DOI] [PubMed] [Google Scholar]
- 27.Rada P., Rojo A.I., Chowdhry S., McMahon M., Hayes J.D., Cuadrado A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell Biol. 2011;31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chowdhry S., Zhang Y., McMahon M., Sutherland C., Cuadrado A., Hayes J.D. Nrf2 is controlled by two distinct β-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene. 2013;32:3765–3381. doi: 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu T., Zhao F., Gao B., Tan C., Yagishita N., Nakajima T., Wong P.K., Chapman E., Fang D., Zhang D.D. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev. 2014;28:708–722. doi: 10.1101/gad.238246.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 31.Hast B.E., Goldfarb D., Mulvaney K.M., Hast M.A., Siesser P.F., Yan F., Hayes D.N., Major M.B. Proteomic analysis of ubiquitin ligase KEAP1 reveals associated proteins that inhibit NRF2 ubiquitination. Cancer Res. 2013;73:2199–2210. doi: 10.1158/0008-5472.CAN-12-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lau A., Wang X.J., Zhao F., Villeneuve N.F., Wu T., Jiang T., Sun Z., White E., Zhang D.D. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol. Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau A., Zheng Y., Tao S., Wang H., Whitman S.A., White E., Zhang D.D. Arsenic inhibits autophagic flux, activating the Nrf2-Keap1 pathway in a p62-dependent manner. Mol. Cell Biol. 2013;33:2436–2446. doi: 10.1128/MCB.01748-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeNicola G.M., Karreth F.A., Humpton T.J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K.H., Yeo C.J., Calhoun E.S., et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao S., Wang S., Moghaddam S.J., Ooi A., Chapman E., Wong P.K., Zhang D.D. Oncogenic KRAS confers chemoresistance by upregulating NRF2. Cancer Res. 2014;74:7430–7441. doi: 10.1158/0008-5472.CAN-14-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramos-Gomez M., Kwak M.K., Dolan P.M., Itoh K., Yamamoto M., Talalay P., Kensler T.W. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yates M.S., Kwak M.K., Egner P.A., Groopman J.D., Bodreddigari S., Sutter T.R., Baumgartner K.J., Roebuck B.D., Liby K.T., Yore M.M., et al. Potent protection against aflatoxin-induced tumorigenesis through induction of Nrf2-regulated pathways by the triterpenoid 1-[2-cyano-3-,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole. Cancer Res. 2006;66:2488–2494. doi: 10.1158/0008-5472.CAN-05-3823. [DOI] [PubMed] [Google Scholar]
- 38.Cleasby A., Yon J., Day P.J., Richardson C., Tickle I.J., Williams P.A., Callahan J.F., Carr R., Concha N., Kerns J.K., et al. Structure of the BTB domain of Keap1 and its interaction with the triterpenoid antagonist CDDO. PLoS One. 2014;9:e98896. doi: 10.1371/journal.pone.0098896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang D.D. Bardoxolone brings Nrf2-based therapies to light. Antioxid. Redox Signal. 2013;19:517–518. doi: 10.1089/ars.2012.5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ellrichmann G., Petrasch-Parwez E., Lee D.H., Reick C., Arning L., Saft C., Gold R., Linker R.A. Efficacy of fumaric acid esters in the R6/2 and YAC128 models of Huntington's disease. PLoS One. 2011;6:e16172. doi: 10.1371/journal.pone.0016172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bomprezzi R. Dimethyl fumarate in the treatment of relapsing-remitting multiple sclerosis: an overview. Ther. Adv. Neurol. Disord. 2015;8:20–30. doi: 10.1177/1756285614564152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rojo A.I., Medina-Campos O.N., Rada P., Zuniga-Toala A., Lopez-Gazcon A., Espada S., Pedraza-Chaverri J., Cuadrado A. Signaling pathways activated by the phytochemical nordihydroguaiaretic acid contribute to a Keap1-independent regulation of Nrf2 stability: Role of glycogen synthase kinase-3. Free Radic. Biol. Med. 2012;52:473–487. doi: 10.1016/j.freeradbiomed.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 43.Jiang T., Chen N., Zhao F., Wang X.J., Kong B., Zheng W., Zhang D.D. High levels of Nrf2 determine chemoresistance in type II endometrial cancer. Cancer Res. 2010;70:5486–5496. doi: 10.1158/0008-5472.CAN-10-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shim G.S., Manandhar S., Shin D.H., Kim T.H., Kwak M.K. Acquisition of doxorubicin resistance in ovarian carcinoma cells accompanies activation of the NRF2 pathway. Free Radic. Biol. Med. 2009;47:1619–1631. doi: 10.1016/j.freeradbiomed.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 45.Mitsuishi Y., Taguchi K., Kawatani Y., Shibata T., Nukiwa T., Aburatani H., Yamamoto M., Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 46.Ludtmann M.H., Angelova P.R., Zhang Y., Abramov A.Y., Dinkova-Kostova A.T. Nrf2 affects the efficiency of mitochondrial fatty acid oxidation. Biochem. J. 2014;457:415–424. doi: 10.1042/BJ20130863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ooi A., Wong J.C., Petillo D., Roossien D., Perrier-Trudova V., Whitten D., Min B.W., Tan M.H., Zhang Z., Yang X.J., et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell. 2011;20:511–523. doi: 10.1016/j.ccr.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 48.Tong K.I., Katoh Y., Kusunoki H., Itoh K., Tanaka T., Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol. Cell Biol. 2006;26:2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muscarella L.A., Parrella P., D’Alessandro V., la Torre A., Barbano R., Fontana A., Tancredi A., Guarnieri V., Balsamo T., Coco M., et al. Frequent epigenetics inactivation of KEAP1 gene in non-small cell lung cancer. Epigenetics. 2011;6:710–719. doi: 10.4161/epi.6.6.15773. [DOI] [PubMed] [Google Scholar]
- 50.Wang R., An J., Ji F., Jiao H., Sun H., Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem. Biophys. Res. Commun. 2008;373:151–154. doi: 10.1016/j.bbrc.2008.06.004. [DOI] [PubMed] [Google Scholar]