It is unclear whether both CD1 and CD2 of A3B can deaminate C and mC. We showed that only CD2 is active, and has weak mC deaminating activity. We successfully engineered A3BCD2 to gain over 100-fold higher mC deamination activity.
Keywords: cytosine/methylcytosine (C/mC) selectivity, enzyme engineering, multiple determinants for mC specificity, substrate specificity alteration, zinc (Zn) deaminase
Abstract
APOBEC (apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like) is a family of enzymes that deaminates cytosine (C) to uracil (U) on nucleic acid. APOBEC3B (A3B) functions in innate immunity against intrinsic and invading retroelements and viruses. A3B can also induce genomic DNA mutations to cause cancer. A3B contains two cytosine deaminase domains (CD1, CD2), and there are conflicting reports about whether both domains are active. Here we demonstrate that only CD2 of A3B (A3BCD2) has C deamination activity. We also reveal that both A3B and A3BCD2 can deaminate methylcytosine (mC). Guided by structural and functional analysis, we successfully engineered A3BCD2 to gain over two orders of magnitude higher activity for mC deamination. Important determinants that contribute to the activity and selectivity for mC deamination have been identified, which reveals that multiple elements, rather than single ones, contribute to the mC deamination activity and selectivity in A3BCD2 and possibly other APOBECs.
INTRODUCTION
In the human genome, there are 11 members in the family of apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like (APOBEC) deaminases, including activation-induced cytidine deaminase (AID), APOBEC1, APOBEC2, APOBEC3A (A3A) to APOBEC3 H and APOBEC4 [1,2]. Among them, APOBEC3 members and AID are well known for their roles in innate and acquired immunity [3,4]. The major function of the APOBEC3s is to inhibit replication of retroelements or infectious retroviruses such as HIV-1 [5–15].
APOBEC3B (A3B) is a member of the APOBEC3 group. It has two cytosine deaminase domains (CDs): CD1 at the N-terminus and CD2 at the C-terminus. A3B displays anti-retroviral activity and inhibits retrotransposon replication [16–19], and has been shown to impair infection by the DNA virus hepatitis B virus (HBV) [15]. A3B expression is up-regulated in HBV- and human papillomavirus (HPV)-infected patients [20,21]. A3B is shown to deaminate HBV covalently closed circular DNA (cccDNA) to induce not only mutations of the viral genome [21,22] but also the degradation of the viral cccDNA [23]. However, uncontrolled deamination activity by A3B on genomic DNA can have a detrimental effect, as A3B has been reported to be an enzymatic source of mutation in multiple cancers including breast, lung and cervical cancers [24–34]. Despite the importance of A3B function and its involvement in cancer, there has been no comprehensive report yet on the in vitro biochemical characterization of A3B.
Cytosine methylation is a common modification of genomic DNA in epigenetic regulation of gene expression. So far, AID and A3A, both of which are single-domain deaminases, are the only APOBECs reported to be capable of deaminating methylcytosine (mC), with A3A being much more active than other APOBECs in catalysing this reaction in vitro [35–40]. The mC deamination activity associated with AID has been proposed as an alternative way, in addition to the ten-eleven translocation (TET) pathway [41], to contribute to DNA demethylation for regulating the methylation pattern in mouse germ cells [42] and for cell reprogramming in inducing pluripotent stem cells [43]. For A3A, its mC deamination is currently proposed to be involved in clearing foreign infectious DNA or degraded self-DNA from apoptotic cells [5,35].
The catalytic CD2 of APOBEC3G [A3G (A3GCD2)] shares significant sequence homology with A3A (65% identity). However, unlike A3A that has highly efficient mC deamination, A3GCD2 is reported to be deficient in mC deamination [35,36]. Even though the high-resolution structures of A3A and A3GCD2, as well as five other APOBEC domain structures, are available [44–49], the structural elements responsible for the efficient mC deamination activity observed for A3A or the lack of mC deamination for A3G have not been identified. Among all APOBECs, A3BCD2 shares the highest sequence homology with A3A, with 89% sequence identity between them. Moreover, A3B is the only APOBEC that shows constitutive nucleus localization [50,51], and its activity in editing nuclear DNA is implicated in various cancers [24–31,52]. However, whether the CD2 of A3B that shares high sequence homology with A3A can also deaminate mC is unclear.
To address these questions, we characterized the in vitro deamination activity of A3B using proteins purified from several A3B deletion/mutation constructs. Our results clarified that only A3BCD2 is the catalytically active domain. We also showed that A3B and A3BCD2 had detectible weak deamination activity on mC at a level approximately three orders of magnitude less than that of A3A. In order to reveal the factors(s) important for determining mC deamination activity, we performed structure-guided mutagenesis studies; these studies enabled us to successfully engineer A3BCD2 constructs with mutations resulting in increased mC deamination activity by about two orders of magnitude. This work has allowed us to identify the important elements that affect the activity and specificity of mC deamination for A3BCD2, and possibly for other APOBECs.
EXPERIMENTAL
Cloning, expression and protein purification
The full-length A3B, A3BCD1, A3BCD2 and their corresponding mutants were constructed in pMAL-c5X vector (New England Biolabs) and expressed as an N-terminal maltose-binding protein (MBP) fusion in Escherichia coli cells. A3BCD2 and its various mutant constructs were also cloned into pET-28a (+) vector (Novagen) with a C-terminal His-tag. All clones were sequenced to confirm the correct sequences before proceeding with protein expression, purification and activity assays.
Protein expression for MBP–A3B constructs was induced with 0.3 mM IPTG at 16°C for 18 h in a shaker incubator. Cell pellets were resuspended with lysis buffer A (20 mM Tris/HCl, pH 8.0, 250 mM NaCl and 2 mM DTT) and lysed by French press. The crude cell lysate was then centrifuged at 12000 rpm for 1 h. The MBP-fusion proteins in the supernatant were purified by passing through a column with Amylose resin (New England Biolabs), followed with extensive wash using 10 column volumes of wash buffer (20 mM Tris/HCl, pH 8.0, 1 M NaCl and 2 mM DTT). The A3B proteins eluted with elution buffer (20 mM Tris/HCl, pH 8.0, 250 mM NaCl, 2 mM DTT and 20 mM maltose) were concentrated and further purified using Superose 6 gel filtration chromatography (GE Healthcare). The fractions containing chromatographically homogeneous A3B proteins were pooled, concentrated, divided into aliquotes in multiple small tubes (20 μl/tube), flash-frozen in liquid nitrogen and stored at −80°C for activity assay.
For the His-tagged A3BCD2 mutant constructs and the wild -type (WT) A3A, the fusion proteins were initially purified by nickel resin column (Qiagen) from the supernatant fraction, followed by extensive washing with 10 column volumes of buffer B (20 mM Tris/HCl, pH 8.0, and 300 mM NaCl) plus 50 mM imidazole. The fusion proteins were then eluted from the nickel column with buffer B plus 500 mM imidazole. Elutions were combined and switched to protein buffer (20 mM Tris/HCl, pH 8.0, 250 mM NaCl, 1 mM DTT and 1 mM EDTA) by buffer exchange. Concentrated protein was divided into small aliquots, flash-frozen in liquid nitrogen and stored at −80°C for activity assay. All purified proteins used in the present study were quantified by UV absorption and the final concentrations were calibrated by SDS/PAGE as shown in Supplementary Figure S1.
Deamination assay
A3B and A3A proteins were reacted with 600 nM 5′ 6-carboxyfluorescein (FAM)-labelled ssDNA substrates (synthesized by Integrated DNA Technologies, Supplementary Table S1) in deamination buffer (25 mM HEPES, pH 6.5, 100 mM NaCl, 0.1% Triton X-100, 1 mM DTT and 0.1 μg/ml RNase). Reactions were incubated at 37°C for the designated times and terminated by heating to 90°C for 5 min. Deamination products were detected using protocols described previously [53]. Briefly, reactions with normal C deamination were treated with 2 units of uracil–DNA glycosylase (UDG) (New England Biolabs) for 1 h at 37°C; reactions with mC deamination were treated with 2 units of thymine–DNA glycosylase (TDG) (Trevigen) in the presence of 3 × complementary ssDNA for 12 h at 42°C. Samples were incubated at 90°C for 10 min in the presence of 0.1 M NaOH. Deamination products were separated on 20% denaturing PAGE gel, visualized with Molecular Imager FX (Bio-Rad Laboratories) and quantified with Quantity One® 1-D analysis software (Bio-Rad Laboratories). For calculation of the initial velocity, fixed concentrations of A3BCD2 and mutant proteins were incubated with ssDNA substrates for a series of incubations (with intervals of 1 min starting from 0 min) at 37°C. The deamination products were quantified and the data were fitted with linear regression using GraphPad Prism 6 software. The slope divided by protein concentration represents the initial velocity at concentration of 1 μM protein and was determined from three independent experiments.
Steady-state rotational anisotropy DNA-binding assay
5′ FAM-labelled 30 nt ssDNA containing a TCA motif was used as a substrate for A3B-binding assay monitored by change in steady-state fluorescence depolarization (rotational anisotropy). Increasing concentrations of A3B constructs and their inactive mutant proteins were incubated with 50 nM ssDNA in a 65 μl reaction volume containing 10 mM HEPES, pH 6.5, and 100 mM NaCl for 1 min at room temperature. The deaminase-inactive mutants were used in the DNA substrate-binding assay for some faster reactive A3B constructs to ensure the binding affinity are for substrate DNA binding instead of product DNA binding or a mixture of substrate and product DNA binding. The rotational anisotropy was measured using a QuantaMaster QM-1 fluorometer (Photon Technology International) with a single emission channel. Samples were excited with vertically polarized light at 495 nm, and both vertical and horizontal emissions were monitored at 520 nm (8-nm bandwidth). The apparent dissociation constant Kd was calculated by fitting the data to a one-site-specific binding curve using GraphPad Prism 6 software and was determined from three independent experiments.
Structural modelling
The homology modelling programme SWISS-MODEL was used to generate the A3BCD2 model using the structures of A3A (PDB: 2M65) and A3GCD2 (PDB: 3IQS and 3IR2) as templates. The C and mC docking on to A3A and the modelled A3BCD2 structures was performed using the programme Glide [54]. We also performed the structural superimposition of A3A and A3BCD2 with the complex structure of mouse free cytosine deaminase bound to cytidine (PDB: 2FR6), and used the resulting orientation/position of the C in the superimposition to guide the selection of the docked C/mC poses resulted from Glide.
RESULTS
Cytosine deamination by A3B
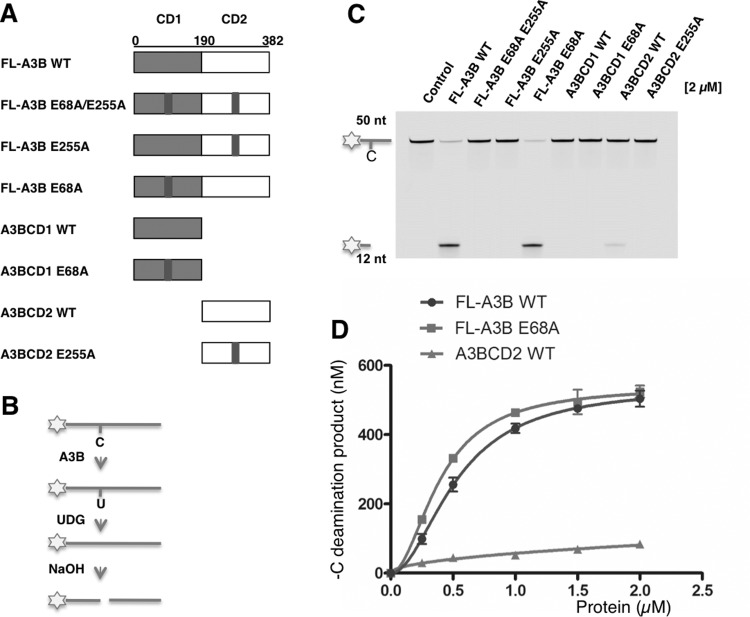
There are conflicting reports regarding whether both A3BCD1 and A3BCD2 are catalytically active [22,52,55]. To resolve this issue, we generated A3B mutants with inactivated mutations in the CD1 and/or the CD2 catalytic centre in the full-length A3B (FL-A3B) construct or in the separated CD1 and CD2 clones (Figure 1A). Specifically, either the putative catalytic residue Glu68 of CD1 was mutated to generated E68A mutant or the catalytic residue Glu255 of CD2 was mutated to generated E255A mutant or both residues mutated to generate a combined E68A/E255A mutant.
Figure 1. Deamination on normal cytidine (C) by A3B and its mutants.
(A) Schematic of various A3B constructs, expressed as an MBP fusion. The putative catalytic residue mutations E68A and E255A are marked by a black bar. (B) Flowchart of deamination assay on 5′ FAM (asterisk)-labelled ssDNA containing a single target C. The C is deaminated to uridine (U) by A3B, UDG releases uracil from ssDNA, and NaOH treatment converts the ssDNA to two shorter ssDNA fragments. Only the ssDNA linked to FAM is detected by fluorescent detector. (C) Gel image showing the C deamination activity of the eight A3B constructs. Protein (2 μM) was incubated with 600 nM 50 nt ssDNA substrate containing C for 2 h at 37°C. The reaction mixture was separated on a 20% TBE/urea polyacrylamide gel. The control reaction contained no A3B but with UDG and NaOH treatment. (D) Dose response of FL-A3B WT, FL-A3B E68A and A3BCD2 WT. Protein at concentration 0.25 μM, 0.5 μM, 1 μM, 1.5 μM and 2 μM was incubated with 600 nM 50 nt substrate for 2 h at 37°C. Error bars represent S.D. from the mean for three independent experiments.
The C deaminase activities were examined by a gel-based fluorescence assay using an ssDNA substrate containing a target C (Figure 1B). The sequence motif specificity of the ssDNA substrates for FL-A3B was examined using the ssDNA substrates 5-NCA and 5-TCN, and the results show that 5′-TCA/G is the preferred DNA sequence motif (Supplementary Figures S2A and S2B). Then the C deaminase activity of the purified proteins of these A3B constructs was compared using an ssDNA substrate containing the hotspot 5′-TCA (Figure 1C). The results show that FL-A3B at a concentration of 2 μM was able to deaminate approximately 90% of 600 nM substrate in 120 min. The FL-A3B mutant E68A/E255A showed no detectable deamination activity. However, FL-A3B E68A (with CD1 mutation) showed similar activity to that of FL-A3B, but FL-A3B E255A (with CD2 mutation) did not show any activity. These in vitro results clearly indicate that CD2 of A3B is catalytically active and CD1 inactive.
The individual WT CD2 (A3BCD2 WT) alone was also active in C deamination (Figure 1C), even though with approximately 6-fold lower activity than that of FL-A3B at a protein concentration of 2 μM (Figure 1D). These data suggest that even though CD1 has no C deamination activity, it enhances the activity of CD2 in the context of FL-A3B. In further dose-response studies, FL-A3B WT and FL-A3B E68A mutant (CD1 mutation) showed similar activity within the tested protein concentration range, whereas A3BCD2 WT alone showed much reduced activity in the tested concentration range (Figure 1D). These results clearly indicate that the FL-A3B E68A mutant in CD1 behaved similarly to WT CD1 in its role for enhancing the CD2 deamination activity in the context of the FL-A3B protein.
Given the fact that the overall charge of +4.5 for CD1 and −3.9 for CD2 at pH 6.5 (http://protcalc.sourceforge.net/), it is plausible that CD1 in FL-A3B may help bind ssDNA substrate and orient it for efficient deamination of the target C by CD2. Using a steady-state rotational anisotropy-binding assay we measured the apparent dissociation constant Kd of CD1, CD2 or FL-A3B with the ssDNA substrate containing the hotspot 5′-TCA. The results show that, whereas CD1 or CD2 alone has a relatively weak affinity for ssDNA (Kd=1 and 3 μM, respectively), FL-A3B has Kd=67.9 nM showing a much enhanced binding to ssDNA (Supplementary Figures S2C–S2E). These results suggest that the combination of CD1 and CD2 leads to a synergistically enhanced binding to ssDNA, which may directly contribute to the enhanced deamination activity. This synergistic effect could be the result of the 3D arrangement of CD1 and CD2 in the full-length structure which allows the involvement of different parts of two domains coming together to enhance the affinity and activity. The sequence specificity, on the other hand, is independent of CD1 since both the E68A mutant and the CD1 deletion mutant display similar sequence specificity that of FL-A3B (Supplementary Figures S3A–S3D).
Methylcytosine deamination by A3B
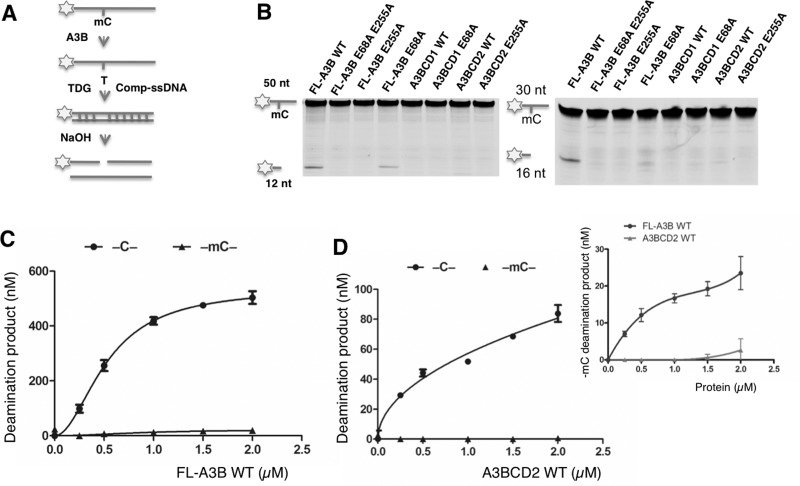
We examined mC deamination by A3B using an ssDNA containing a 5′-TmCA motif (Figure 2A). Although the inactive mutant FL-A3B E68A/E255A did not show any mC deamination activity, FL-A3B and FL-A3B E68A showed clearly detectable deaminase activity on mC with both 50 nt and 30 nt substrates (Figure 2B). This mC deamination activity by FL-A3B was much lower than the C deamination over a wide range of protein concentrations tested (Figure 2C). The A3BCD2 WT construct also showed detectable but lower mC deamination than that of FL-A3B (Figure 2D, inset in Figure 2D). When comparing the specific deamination activity in the linear range (Table 1), A3BCD2 had approximately 50-fold lower mC deamination than C deamination. Despite much lower mC deaminase activity, the sequence specificity of the mC substrates of FL-A3B appears similar to that of the C substrates, suggesting that the reduced deaminase activity with mC is not due to the change in the sequence motif specificity (Supplementary Figures S3E and S3F).
Figure 2. Deamination on methylcytidine (mC) by A3B and its mutants.
(A) Flowchart of deamination assay on 5′ FAM (asterisk)-labelled ssDNA containing a single target mC. The mC was deaminated to thymine (T) by A3B. A 3-fold excess of complement strand (Comp-ssDNA) was added to form a T:G mismatch. TDG releases T from a T:G mismatch and NaOH treatment converts the product to two shorter ssDNA fragments. (B) Gel image showing the mC deamination activity of eight A3B constructs on 50 nt (left) and 30 nt (right) ssDNA substrates. Protein (2 μM) was incubated with 600 nM ssDNA substrates containing mC for 2 h at 37°C. (C) Comparison of the dose-response deamination activity on C and mC by FL-A3B WT. (D) Comparison of the dose-response deamination activity on C and mC by A3BCD2 WT. The inset is an amplified chart that compares the mC deamination activity by FL-A3B WT and A3BCD2 WT. For (C) and (D), protein at various concentrations was incubated with 600 nM 50 nt ssDNA substrate for 2 h at 37°C. Error bars represent S.D. from the mean for three independent experiments.
Table 1. Comparison of the deaminase activity and mC selectivity factor of various A3BCD2 mutants.
| A3BCD2 | A3A | Mt0 | M3 | M4 | M3M4 | Y313F | |
|---|---|---|---|---|---|---|---|
| C deamination activity* (nM product/μM enzyme) | 506.4±30.1 | 401900±20230 | 3733±230.5 | 498.7±65.7 | 3605±449.0 | 6943±124.8 | 55.32±7.7 |
| mC deamination activity* (nM product/μM enzyme) | 10.22±0.4 | 51030±1480 | 566.8±72.5 | 41.81±2.2 | 353.4±27.8 | 1309±70.8 | 7.572±0.5 |
| mC selectivity factor† [(mC/C activity) × 100] | 2.01 | 12.70 | 15.18 | 8.38 | 9.80 | 18.85 | 13.69 |
*Deaminase activity (nM product/μM enzyme) for the C and mC deamination was calculated from the initial linear range of dose-response assays for each protein construct (Supplementary Figures S3B, S3C, S6B, S7C, S7D and S8A). S.D. was estimated from data collected in three independent experiments.
†The mC selectivity factor was calculated as [mC/C activity × 100], where the deaminase activity for C and mC was used in this calculation.
Engineering A3BCD2 to achieve increased mC deamination activity
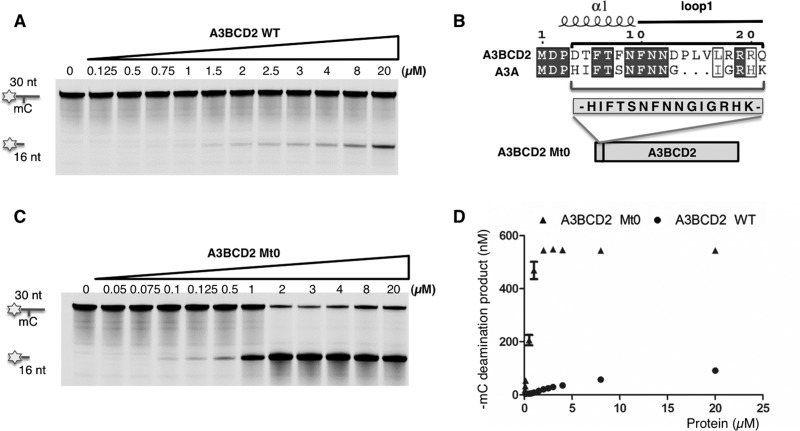
We also have a highly purified A3BCD2 WT protein with a His-tag fused to its C-terminus, and the dose-response deamination assay showed that this construct had roughly similar activity for mC (Figure 3A) and normal C deamination compared with the previous MBP-fused construct (Supplementary Figures S4A and S4B). Additional controls in Supplementary Figure S4C confirmed the deamination activity on C and mC substrates. These deamination activity levels for C and mC are similar to that reported for AID [40]. However, when comparing the deamination activity between A3BCD2 and A3A (expressed as nM product/μM enzyme characterized in the linear protein concentration range), A3A has 525-fold higher activity for C deamination, but a remarkable 3317-fold higher activity for mC deamination (Table 1 and Supplementary Figure S4D). Because of the large difference in specific activities between A3BCD2 and A3A, to get a better sense of the relative activity for mC deamination, we also compared the ratio of the mC/C activity × 100, which is defined as the selectivity factor for mC deamination. In other words, this selectivity factor for mC deamination can be thought of as the number of mC deaminations per every 100 C deaminations under the same conditions. When using this standard to compare the two proteins, the selectivity factor for mC deamination is 2.01 for WT A3BCD2, which is approximately 6-fold lower than the 12.70 for A3A (Table 1, Supplementary Figure S4E). The selectivity factor for mC deamination of A3A obtained in the present study is similar to the value calculated from the published data for A3A [35].
Figure 3. An engineered A3BCD2 mutant with much higher mC deamination activity.
(A) Gel image showing the mC deamination activity by A3BCD2 WT construct. A3BCD2 at various concentrations was incubated with 600 nM 30 nt ssDNA substrate containing mC at 37°C for 2 h. (B) Design of A3BCD2 Mt0 construct. Sequence alignment of A3BCD2 and A3A shows the difference around the loop-1 region (see Supplementary Figure S4 for a full alignment). The 15-amino-acid sequence in the loop-1 region of A3A was inserted into the corresponding region in A3BCD2 to make A3BCD2 Mt0. (C) Gel image showing the mC deamination activity by A3BCD2 Mt0. A3BCD2 Mt0 at various concentrations was incubated with 600 nM 30 nt ssDNA substrate at 37°C for 2 h. (D) Quantification of the mC deamination by A3BCD2 WT and Mt0, showing significantly increased activity on mC by Mt0 mutant.
Currently, A3A and a zebra AID in the APOBEC family are the only two members that have been reported to have relatively high activity on mC with over 10% of their activity on regular C [35,56]. Two previous studies on mutagenesis and domain-swapping of A3A and fish AID tried to identify the domains contributing to efficient deamination activities on mC; however, it is still not resolved [35,56]. Here, given the high similarity of A3BCD2 to A3A but its small mC deamination activity, we tried to understand why there is such a big difference in mC deamination activity by analysing the available structural data. The available structures of A3A and four other APOBECs [44–48] show no obvious features within the C- or mC-binding pocket that could explain the observed difference in mC deamination activity of A3A, A3BCD2 and other APOBECs. However, the superimposition of the five structures reveals many differences in the peripheral loops around the active site (Figure 4A). Among these differences, two features caught our attention. One is that, although the loop-1 conformations of the four APOBECs (excluding A3A) are more or less similar to each other, the A3A loop-1 conformation is noticeably different from the other APOBECs (Figure 4A). The second feature is that, on loop-7, a highly conserved tyrosine residue (the equivalent of Tyr130 in A3A) has a similar conformation for the four non-A3A APOBECs, occupying a space near the active site as a half-opened ‘lid’ next to the active site pocket (Figure 4B), which may present partial steric hindrance for the methyl group on mC at the active site pocket. However, the side chain of this Tyr130 on A3A loop-7 is oriented away from the active site pocket (Figure 4C), which could potentially reduce the partial hindrance and allow the bulkier and more hydrophobic mC to get to the active site pocket. At the sequence level, A3A and A3B-CD2 share the highest identity (89%) and homology (92%) among all APOBECs, and the differences between the two proteins are mostly concentrated around the loop-1, β2 and α5 regions (Supplementary Figure S5), among which the loop-1 is the only region located around the active site.
Figure 4. Superimposition of the known APOBEC structures around the Zn active site.

A3A (green, PDB: 2M65), A3C (red, PDB: 3VOW), A3FCD2 (purple, PDB: 4J4J and 4IOU), A3GCD2 (yellow, PDB: 3IQS and 3IR2), A3BCD2 (pink, modelled structure). (A) A view of the superimposition of loop-1, loop-3 and loop-7 around the Zn active site. The conserved Tyr130 in A3A can adopt a conformation different from its equivalent tyrosine residues in other APOBECs (in sticks), which is probably permitted by the different loop-1 conformation in A3A (green). The loop-1 conformations in other APOBECs should prevent their tyrosine residues to assume the conformation of Tyr130 in A3A. (B) A closer view of the active site for the non-A3A APOBECs, showing the conserved tyrosine residue as a partial ‘lid’ on the edge of the mC at the active site pocket. (C) A closer view of the active site for A3A and the modelled A3BCD2, showing the different conformation for loop-1, and for the conserved tyrosine residue (Tyr130 for A3A, Tyr313 for A3BCD2) next to the mC at the active site. Tyr313 of A3BCD2 is closer to the mC, causing some clashes with the methyl group.
Guided by this structural and sequence analysis, we engineered novel A3BCD2 constructs in an attempt to identify the residues that can critically regulate the low or high activity and specificity for mC deamination in A3BCD2 or in A3A. The first construct we made was to graft a 15-amino-acid segment around the loop-1 region of A3A (H16IFTSNFNNGIGRHK30) to replace the corresponding region in A3BCD2, generating mutant A3BCD2 Mt0 (Figure 3B). Interestingly, the A3BCD2 Mt0 construct gained 7-fold increase in activity for C deamination, but a much more dramatic 56-fold increase for mC deamination, when compared with A3BCD2 WT (Table 1, Figures 3A, 3C and 3D). The significantly increased C deamination activity of A3BCD2 Mt0 is also shown in Supplementary Figures S6A and S6B. From the dose–response curves of both mC and C deamination for A3BCD2 Mt0 in Supplementary Figure S6A and for A3BCD2 WT in Supplementary Figure S4B, it is clear that A3BCD2 Mt0 showed a much higher activity on mC deamination relative to C deamination than A3BCD2 WT. When the selectivity factor for mC deamination is compared, A3BCD2 Mt0 has a selectivity factor of 15.18 (Table 1), 7-fold higher than that of WT A3BCD2, which is even slightly higher than the 12.7 for WT A3A. These results indicate that the short 15-residue loop-1 region of A3A and A3BCD2 can greatly influence the deamination activity and the selectivity factor for mC deamination.
Specific residues on the loop-1 region important for mC deamination
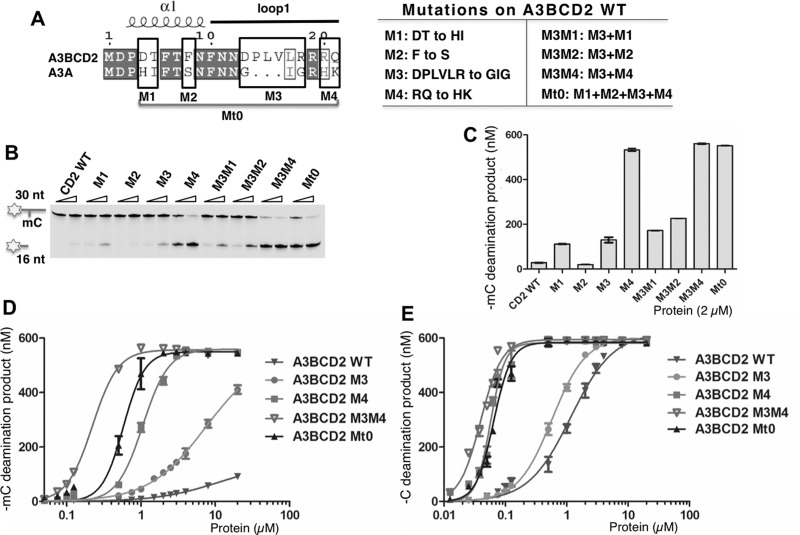
Seven out of the 15 residues from the grafted loop-1 region in the A3BCD2 Mt0 mutant are actually conserved between A3BCD2 and A3A (Figure 5A). The remaining non-conserved eight residues may therefore contribute to the increased activity and specificity for mC deamination observed in the A3BCD2 Mt0 construct. In order to evaluate the contributions of these non-conserved residues to the increased activity and specificity for mC, we divided the eight non-conserved residues into four groups and generated mutants (M1–M4) on the A3BCD2 WT construct, mutating the residues in each group to the corresponding A3A residues (Figure 5A). The results of the mC deamination assay showed that, with the exception of M2 mutant (F8S), mutants M1 (–DT–to–HI–), M3 (–DPLVLR–to–GIG–) and M4 (–RQ–to–HK–) all displayed greatly increased activity on mC (Figures 5B and 5C), with M4 showing the highest mC deamination activity. These results indicate that the mutated residues in M1, M3 and M4 can influence mC deamination activity.
Figure 5. Dissecting loop-1 residues important for increasing mC deamination.
Error bars represent S.D. from the mean for three independent experiments. (A) Design of the mutants on A3BCD2 WT. Sequence alignment of A3BCD2 and A3A showed four groups (M1–M4) around loop-1 that are not conserved (left). A3BCD2 was mutated to contain the corresponding residues of A3A individually to generated mutant M1–M4. A3BCD2 M3 was combined with others to generate three additional combined mutants. (B) Gel image showing the different mC deamination activities by the mutants. Each protein (0.5 μM and 2 μM) was incubated with 600 nM 30 nt ssDNA substrates at 37°C for 2 h. (C) Quantification of the activity on mC. The activity of the mutants at the concentration of 2 μM was quantified. (D) Dose-response of mC deamination activity by A3BCD2 WT, M3, M4, M3M4 and Mt0 constructs. (E) Dose-response of C deamination activity by A3BCD2 WT, M3, M4, M3M4 and Mt0 constructs.
We next examined the mC deamination activity of the combined mutants M3M1, M3M2 and M3M4, in order to evaluate the potential additive effects of the mutated residues (Figure 5A right panel). Each of these combined mutants showed slightly higher mC deamination activity than the corresponding individual mutants (Figures 5B and 5C), with M3M4 showing the highest activity (Figure 5C), even slightly higher than Mt0 that has the combined M1–M4 mutations. Despite that M2 alone did not show increased mC deamination activity, the combined M3M2 mutant had higher activity than M3 alone (Figure 5C). A similar trend of activity changes for the various mutants is also observed when the initial velocity for the deamination assay was performed (Supplementary Table S2).
In order to evaluate the relative increase in mC deamination compared with C deamination activity (or the selectivity factor for mC), we carried out a dose-response assay on both mC and C deamination activities for the four mutants (M3, M4, M3M4, Mt0) that showed significantly increased mC deamination activity. The results clearly indicated that a significant increase for mC deamination is also linked to the significant increase for C deamination as well (Figures 5D and 5E, Supplementary Figures S7A and S7B). However, the fold increase for mC deamination is much higher than that for C deamination, indicating a biased increase towards mC deamination. This phenomenon becomes more obvious when the selectivity factor for mC deamination are calculated for the four mutants, which shows a 4–9-fold higher mC selectivity factor than WT A3BCD2 (Table 1, Supplementary Figures S7C and S7D). Among them, the two combined mutants Mt0 and M3M4 have the highest specificity factor for mC deamination, with an increase of 7- and 9-fold higher than that of WT A3BCD2 (Table 1). These results indicate that the mutated residues of M3M4 from the loop-1 region of A3A (i.e.–G25I26G27–in M3 and–H29K30–in M4, Figure 5A) collectively play a critical role in the activity and specificity for mC deamination.
We also examined the preferred trinucleotide sequence motif for deamination and the substrate ssDNA-binding affinity of A3BCD2, and the three mutants M3, M4 and Mt0. Our results show that A3BCD2 and the three mutants have unchanged sequence motif specificity for deamination (Supplementary Figures S8 and S9) and had similar binding affinity to substrate ssDNA (Supplementary Figure S10), indicating that the increased deamination activity and selectivity factor for mC deamination of these engineered mutants is not a result of altered DNA sequence motif specificity or the altered overall binding affinity for the substrate ssDNA.
Flexibility of loop-1 important for mC specificity
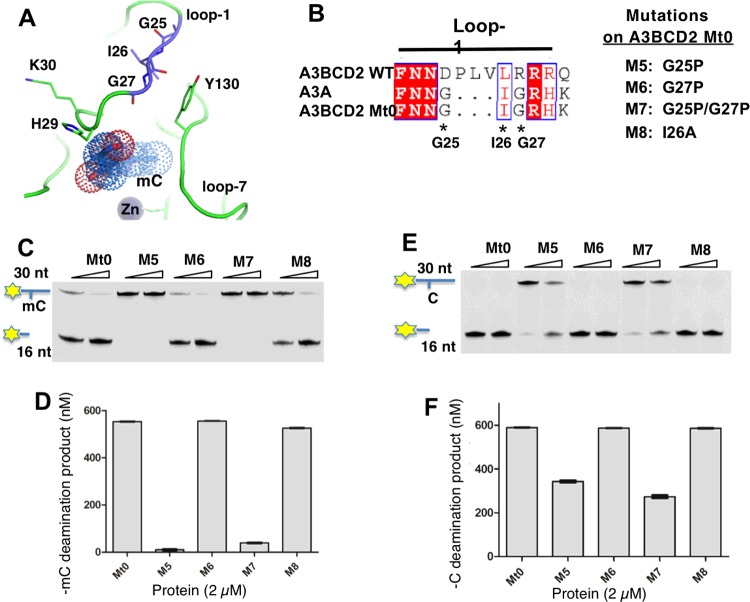
The available A3A NMR structure shows that the conformation of loop-1 differs from that of other known APOBECs and is very flexible in adopting several conformations [47]. The two glycine residues–GIG–on loop-1 of A3A and of the A3BCD2 M3M4 mutant may be important for the flexible conformation of the loop (Figures 6A and 6B), which may contribute to the higher mC activity and specificity. To test this hypothesis, we used A3BCD2 Mt0 that contains the–GIG–sequence in the loop-1 region to perform further mutational studies, in which the two highly flexible glycine residues (Gly25, Gly27) in the–G25I26G27–of loop-1 were mutated to proline, and also the isoleucine residue (Ile26) was mutated to alanine (Figures 6A and 6B, mutants M5–M8), and then examined their effects on mC and C deamination activity. The activity assay results showed that, at a protein concentration of 2 μM, mutants M5 and M7 containing the G25P mutation had essentially lost mC deamination activity, whereas mutant M6 and M8 displayed similar mC deamination as A3BCD2 Mt0 (Figures 6C and 6D). Interestingly, the C deamination of M5 and M7 is only partially affected by this G25P mutation, retaining approximately 50% of the activity level of the A3BCD2 Mt0 construct (Figures 6E and 6F). These results indicate that Gly25 in the–G25I26G27–motif is indeed critical for the activity and specificity for mC deamination, possibly because this Gly25 could allow the loop-1 to adopt the conformation necessary for positioning mC for a more optimal deamination reaction at the active site pocket.
Figure 6. The flexibility of loop-1 conformation in A3BCD2 Mt0 affects mC deamination activity.
Error bars represent S.D. from the mean for three independent experiments. (A) Structure of A3A loop-1 and Tyr130 on loop-7 around the Zn active site. Residues–G25I26G27–and–H29K30–on loop-1 are drawn as sticks. The mC modelled into the active site is shown as dots. In the NMR structure of A3A (PDB: 2M65), Tyr130 is close to–GIG–. (B) Design of four mutants on loop-1 of A3BCD2 Mt0. (C) Gel image showing the mC deamination activity of the mutants. Each protein (0.5 μM and 2 μM) was incubated with 600 nM 30 nt ssDNA substrates at 37°C for 2 h. (D) Quantification of the mC deamination activity at a protein concentration of 2 μM. (E) Gel image showing the C deamination activity of the mutants. Each protein (0.5 μM and 2 μM) was incubated with 600 nM 30 nt ssDNA substrates at 37°C for 2 h. (F) Quantification of the C deamination activity at a protein concentration of 2 μM.
The conserved loop-7 tyrosine residue for mC specificity
A highly conserved loop-7 tyrosine residue among all APOBECs corresponds to Tyr313 in A3BCD2 and Tyr130 in A3A. As mentioned above, the structural alignment reveals that the loop-7 Tyr130 in A3A adopts a different conformation from all of the equivalent tyrosine residues in other APOBECs (Figures 4A–4C), possibly resulting in a more accessible active site for accommodating the bulkier mC residue in A3A than in other APOBECs (such as A3B, A3G), and as such accounting for the high activity and selectivity for mC deamination. In other words, the less accessible active site due to the conformations of the conserved Tyr313 in A3BCD2 (Figure 4C) may contribute to the observed low activity and specificity for mC deamination.
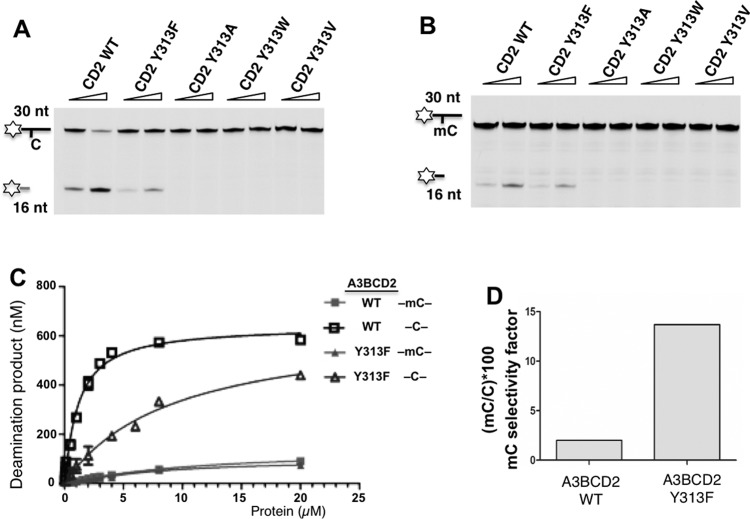
To test this hypothesis, we mutated Tyr313 in the A3BCD2 WT template to different hydrophobic residues, including a larger tryptophan residue (Y313W) or a slightly smaller phenylalanine residue (Y313F) or the much smaller valine and alanine residues (Y313V, Y313A) and then evaluated their deamination activity and specificity for mC. The results indicated that A3BCD2 constructs with mutations Y313W, Y313V and Y313A had no detectible deamination activity for C or mC substrates (Figures 7A and 7B), suggesting that too large a side chain like tryptophan may completely block the access to the active site in A3BCD2, and too small a side chain may not be sufficient to stably hold the C or mC in the active site for deamination reaction. However, Y313F retained essentially the same level of mC deamination, but caused a significant decrease in C deamination activity (Figure 7C). If the selectivity factor for mC deamination is calculated, the Y313F mutant is 13.69, which is similar to that of A3A and had an increase of over 6-fold compared with the 2.01 of WT A3BCD2 (Figure 7D, Table 1 and Supplementary Figure S11). We postulate that the loss of a hydroxy group in the Y313F mutant may result in a slightly more open ‘gate’ to the active site of A3BCD2 (Figure 4C), possibly allowing a better access for the bulkier and more hydrophobic mC to the active site yet with only a small penalty of C stability and deamination, which might be the reason for the observed 6-fold increase in selectivity factor for mC deamination of the Y313F mutant.
Figure 7. A3BCD2 Tyr313 mutation enhances the substrate specificity on mC.
(A) Gel image showing the C deamination activity of A3BCD2 mutants. Each protein (0.5 μM and 2 μM) was incubated with 600 nM 30 nt ssDNA substrates at 37°C for 2 h. (B) Gel image showing the mC deamination activity of A3BCD2 mutants. Each protein (0.5 μM and 2 μM) was incubated with 600 nM 30 nt ssDNA substrates at 37°C for 2 h. (C) Dose-response of A3BCD2 WT and Y313F for C and mC deamination. (D) Comparison of the mC selectivity factor for A3BCD2 WT and Y313F. The data were calculated from Table 1. Error bars represent S.D. from the mean for three independent experiments.
DISCUSSION
Previous literature on A3B reported conflicting results regarding whether both CD1 and CD2 are enzymatically active [22,52,55]. In addition, there is precedent for an active CD1 in the mouse APOBEC3 protein [57]. In the present study, we performed in vitro biochemical analysis using purified proteins of various mutants of A3B and showed that CD2 is the enzymatically active domain and CD1 is inactive. However, the presence of CD1 can greatly enhance the deamination activity of CD2 in the context of the FL-A3B protein, which is probably due in part to the 3D arrangement of CD1 and CD2 in the native full-length structure which allow the synergistic binding of the two domains to ssDNA substrate to achieve higher deamination activity. We also show that FL-A3B and A3BCD2 can deaminate mC, even though this mC deamination activity is very weak which is approximately 50-fold lower than C deamination.
A3B's overall activities of C and mC deamination reported here appear to be quite similar to those reported for human AID, both with much lower mC deamination relative to C deamination. A3A, on the other hand, is the only human APOBEC protein that has been reported to have clearly much higher deamination activity for both C and mC based on in vitro assay [35]. Another extreme is A3G that is reported to have no detectible mC deamination [35]. What is the structural basis for these closely related APOBEC deaminases to discriminate the subtle differences between C and mC for deamination? To date, no amino acid sequences or/and structural elements of an APOBEC deaminase have been shown to be responsible for the observed discrimination for mC. In the present study we found that the loop-1 of A3BCD2 plays a major role in the activity and selectivity of mC deamination.
In particular, by substituting a few residues on the loop-1 of A3BCD2 to that of A3A (i.e.–G25I26G27–and–H29K30–) the resulting A3BCD2 mutants gained over two orders of magnitude higher mC deamination activity than WT A3BCD2 (Table 1). Moreover, our point mutagenesis studies indicated that a flexible Gly25 on loop-1, likely to be important to allow loop-1 to adopt certain conformations, plays an important role in mC deamination. Previously extensive studies with loop switches and mutations are limited to investigating sequence motif specificity surrounding the target C. These studies show that loops 1 and 7, especially loop-7 (previously named the ‘specificity’ loop), affect the sequence motif specificity [58–66]. For example, the sequence motif for A3G is–CC–, a graft of its loop-7 to that of A3A can change its sequence motif specificity of–TC–[65]. Depending on the specific pair of the loop-7 donor and receiver, the outcome can range between loss of activity, less stringent specificity or near complete conversion to the donor sequence motif specificity. Loop-1 graft mutants, on the other hand, can result in less stringent sequence specificity [62,66] or enhanced/reduced activity [62,65]. The–HK–equivalent residues have not been reported in the loop-grafting studies. However, our results showed that they have no effect on the sequence motif specificity, rather they have a significant impact on increasing the mC deamination activity and selectivity. Our study adds another layer of complexity as to how individual loops assist the recognition and reactivity of the target mC compared with C.
Even though the exact mechanism remains to be elucidated, two possibilities could explain the important role of loop-1 for mC deamination of A3BCD2. The first possibility is that the loop-1 sequence/conformation may influence the conformation of the highly conserved Tyr313 on loop-7 of A3BCD2, which may determine how well the target mC residue can get to and bind at the active site. Previous reports suggest that Tyr114 of AID and Tyr315 of A3G, which are equivalent to Tyr313 of A3B, may also participate in positioning the target C [48,67]. Therefore, Tyr313 of A3B may bear dual functions: stabilize target C and discriminate mC at the active pocket. For the loop-1 of A3A, it may allow its corresponding Tyr130 to adopt a conformation that is more accessible to the active pocket for a bulkier mC than the equivalent tyrosine residue of other APOBECs (Figure 4A). The second possibility is that loop-1 needs to have a certain flexibility that allows it to change conformations in order to bind ssDNA in certain way to present the target mC in a better position for deamination reaction. The two substituted loop-1 residues–H29K30–that impact significantly on mC deamination are actually located outside the active site pocket. Such positioning of the–H29K30–residues suggests that they are unlikely to interact directly with the target mC or C bound inside the active site pocket. They may interact with the base at the 3′- or 5′-side of the target C/mC to allow better presentation the target mC/C to the active Zn for efficient deamination [64,68]. The evidence that Lys30 is involved in DNA binding is consistent with this hypothesis [64]. It is also likely that both possibilities described above work together to regulate mC/C deamination.
In summary, our study on A3B indicates that only CD2 is catalytically active in vitro and that A3B has a very weak but clearly detectable activity on mC deamination. This mC deamination activity of A3B is much lower (roughly by three orders of magnitude) than that of A3A, the only APOBEC reported to have a robust mC deamination to date. Through structural and systematic mutational analysis, we have successfully engineered a mutant version of A3BCD2 that has gained over two orders of magnitude higher activity for mC deamination and has achieved an mC selectivity factor comparable to that of A3A. Important elements around the active site that contribute to the activity and specificity for mC deamination have been identified for the first time. It is clear from the present study that multiple determinants, rather than a single factor, contribute to the mC deamination activity and specificity by APOBEC deaminases. A thorough understanding of the detailed mechanism of the mC substrate selectivity as well as the nucleotide sequence motif specificity will require critical information from high-resolution complex structure(s) containing the enzyme bound to a series of ssDNA substrates.
Acknowledgments
We thank the core laboratory in the Center of Excellence in NanoBiophysics at University of Southern California for support in the related biophysical characterization of the purified protein and Aaron Wolf for a critical reading of the manuscript before submission.
Abbreviations
- A3A
APOBEC3A
- A3B
APOBEC3B
- A3G
APOBEC3G
- AID
activation-induced cytidine deaminase
- APOBEC
apolipoprotein B mRNA- editing enzyme catalytic polypeptide-like
- cccDNA
covalently closed circular DNA
- CD
cytosine deaminase domain
- FAM
6-carboxyfluorescein
- HBV
hepatitis B virus
- MBP
maltose-binding protein
- mC
methylcytosine
- TDG
thymine–DNA glycosylase
- UDG
uracil–DNA glycosylase
- WT
wild-type
AUTHOR CONTRIBUTION
Yang Fu, Hanjing Yang and Xiaojiang Chen designed the experiments; Yang Fu, Fumiaki Ito, Gewen Zhang, Braulio Fernandez and Hanjing Yang performed the experiments; Yang Fu, Hanjing Yang and Xiaojiang Chen wrote the manuscript.
FUNDING
This work was supported by the Nakajima Foundation (to F.I.); the Summer Undergraduate Research Fund (SURF) and URAP from University of Southern California (to B.F.); and the National Institutes of Health [grant number R01GM087986 (to X.S.C.)].
References
- 1.Macduff D.A., Harris R.S. Directed DNA deamination by AID/APOBEC3 in immunity. Curr. Biol. 2006;16:R186–R189. doi: 10.1016/j.cub.2006.02.035. [DOI] [PubMed] [Google Scholar]
- 2.Conticello S.G. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/S0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 4.Di Noia J.M., Neuberger M.S. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 5.Stenglein M.D., Burns M.B., Li M., Lengyel J., Harris R.S. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat. Struct. Mol. Biol. 2010;17:222–229. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiu Y.L., Greene W.C. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 7.Sheehy A.M., Gaddis N.C., Choi J.D., Malim M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 8.Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 9.Chen H., Lilley C.E., Yu Q., Lee D.V., Chou J., Narvaiza I., Landau N.R., Weitzman M.D. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 10.Love R.P., Xu H., Chelico L. Biochemical analysis of hypermutation by the deoxycytidine deaminase APOBEC3A. J. Biol. Chem. 2012;287:30812–30822. doi: 10.1074/jbc.M112.393181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillick K., Pollpeter D., Phalora P., Kim E.Y., Wolinsky S.M., Malim M.H. Suppression of HIV-1 infection by APOBEC3 proteins in primary human CD4(+) T cells is associated with inhibition of processive reverse transcription as well as excessive cytidine deamination. J. Virol. 2013;87:1508–1517. doi: 10.1128/JVI.02587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaipan C., Smith J.L., Hu W.S., Pathak V.K. APOBEC3G restricts HIV-1 to a greater extent than APOBEC3F and APOBEC3DE in human primary CD4+ T cells and macrophages. J. Virol. 2013;87:444–453. doi: 10.1128/JVI.00676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ooms M., Krikoni A., Kress A.K., Simon V., Munk C. APOBEC3A, APOBEC3B, and APOBEC3H haplotype 2 restrict human T-lymphotropic virus type 1. J. Virol. 2012;86:6097–6108. doi: 10.1128/JVI.06570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romani B., Engelbrecht S., Glashoff R.H. Antiviral roles of APOBEC proteins against HIV-1 and suppression by Vif. Arch. Virol. 2009;154:1579–1588. doi: 10.1007/s00705-009-0481-y. [DOI] [PubMed] [Google Scholar]
- 15.Bonvin M., Achermann F., Greeve I., Stroka D., Keogh A., Inderbitzin D., Candinas D., Sommer P., Wain-Hobson S., Vartanian J.P., Greeve J. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology. 2006;43:1364–1374. doi: 10.1002/hep.21187. [DOI] [PubMed] [Google Scholar]
- 16.Bogerd H.P., Wiegand H.L., Hulme A.E., Garcia-Perez J.L., O'Shea K.S., Moran J.V., Cullen B.R. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doehle B.P., Schafer A., Cullen B.R. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology. 2005;339:281–288. doi: 10.1016/j.virol.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Doehle B.P., Schafer A., Wiegand H.L., Bogerd H.P., Cullen B.R. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J. Virol. 2005;79:8201–8207. doi: 10.1128/JVI.79.13.8201-8207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Q., Chen D., Konig R., Mariani R., Unutmaz D., Landau N.R. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 2004;279:53379–53386. doi: 10.1074/jbc.M408802200. [DOI] [PubMed] [Google Scholar]
- 20.Vieira V.C., Leonard B., White E.A., Starrett G.J., Temiz N.A., Lorenz L.D., Lee D., Soares M.A., Lambert P.F., Howley P.M., Harris R.S. Human papillomavirus E6 triggers upregulation of the antiviral and cancer genomic DNA deaminase APOBEC3B. MBio. 2014;5:e02234-14. doi: 10.1128/mBio.02234-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu R., Zhang X., Zhang W., Fang Y., Zheng S., Yu X.F. Association of human APOBEC3 cytidine deaminases with the generation of hepatitis virus B × antigen mutants and hepatocellular carcinoma. Hepatology. 2007;46:1810–1820. doi: 10.1002/hep.21893. [DOI] [PubMed] [Google Scholar]
- 22.Bonvin M., Greeve J. Effects of point mutations in the cytidine deaminase domains of APOBEC3B on replication and hypermutation of hepatitis B virus in vitro. J. Gen. Virol. 2007;88:3270–3274. doi: 10.1099/vir.0.83149-0. [DOI] [PubMed] [Google Scholar]
- 23.Lucifora J., Xia Y., Reisinger F., Zhang K., Stadler D., Cheng X., Sprinzl M.F., Koppensteiner H., Makowska Z., Volz T., et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221–1228. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burns M.B., Lackey L., Carpenter M.A., Rathore A., Land A.M., Leonard B., Refsland E.W., Kotandeniya D., Tretyakova N., Nikas J.B., et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burns M.B., Temiz N.A., Harris R.S. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 2013;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leonard B., Hart S.N., Burns M.B., Carpenter M.A., Temiz N.A., Rathore A., Vogel R.I., Nikas J.B., Law E.K., Brown W.L., et al. APOBEC3B upregulation and genomic mutation patterns in serous ovarian carcinoma. Cancer Res. 2013;73:7222–7231. doi: 10.1158/0008-5472.CAN-13-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gwak M., Choi Y.J., Yoo N.J., Lee S. Expression of DNA cytosine deaminase APOBEC3 proteins, a potential source for producing mutations, in gastric, colorectal and prostate cancers. Tumori. 2014;100:112e–117e. doi: 10.1700/1636.17922. [DOI] [PubMed] [Google Scholar]
- 28.Jin Z., Han Y.X., Han X.R. The role of APOBEC3B in chondrosarcoma. Oncol. Rep. 2014;32:1867–1872. doi: 10.3892/or.2014.3437. [DOI] [PubMed] [Google Scholar]
- 29.Sieuwerts A.M., Willis S., Burns M.B., Look M.P., Meijer-Van Gelder M.E., Schlicker A., Heideman M.R., Jacobs H., Wessels L., Leyland-Jones B., et al. Elevated APOBEC3B correlates with poor outcomes for estrogen-receptor-positive breast cancers. Horm. Cancer. 2014;5:405–413. doi: 10.1007/s12672-014-0196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasaki H., Suzuki A., Tatematsu T., Shitara M., Hikosaka Y., Okuda K., Moriyama S., Yano M., Fujii Y. gene overexpression in non-small-cell lung cancer. Biomed. Rep. 2014;2:392–395. doi: 10.3892/br.2014.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor B.J., Nik-Zainal S., Wu Y.L., Stebbings L.A., Raine K., Campbell P.J., Rada C., Stratton M.R., Neuberger M.S. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris R.S. Molecular mechanism and clinical impact of APOBEC3B-catalyzed mutagenesis in breast cancer. Breast Cancer Res. 2015;17:498. doi: 10.1186/s13058-014-0498-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burns M.B., Leonard B., Harris R.S. APOBEC3B: pathological consequences of an innate immune DNA mutator. Biomed. J. 2015;35:102–110. doi: 10.4103/2319-4170.148904. [DOI] [PubMed] [Google Scholar]
- 34.Cescon D.W., Haibe-Kains B., Mak T.W. APOBEC3B expression in breast cancer reflects cellular proliferation, while a deletion polymorphism is associated with immune activation. Proc. Natl. Acad. Sci. U.S.A. 2015;112:2841–2846. doi: 10.1073/pnas.1424869112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carpenter M.A., Li M., Rathore A., Lackey L., Law E.K., Land A.M., Leonard B., Shandilya S.M., Bohn M.F., Schiffer C.A., et al. Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J. Biol. Chem. 2012;287:34801–34808. doi: 10.1074/jbc.M112.385161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wijesinghe P., Bhagwat A.S. Efficient deamination of 5-methylcytosines in DNA by human APOBEC3A, but not by AID or APOBEC3G. Nucleic Acids Res. 2012;40:9206–9217. doi: 10.1093/nar/gks685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morgan H.D., Dean W., Coker H.A., Reik W., Petersen-Mahrt S.K. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J. Biol. Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 38.Bransteitter R., Pham P., Scharff M.D., Goodman M.F. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larijani M., Frieder D., Sonbuchner T.M., Bransteitter R., Goodman M.F., Bouhassira E.E., Scharff M.D., Martin A. Methylation protects cytidines from AID-mediated deamination. Mol. Immunol. 2005;42:599–604. doi: 10.1016/j.molimm.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 40.Nabel C.S., Jia H., Ye Y., Shen L., Goldschmidt H.L., Stivers J.T., Zhang Y., Kohli R.M. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat. Chem. Biol. 2012;8:751–758. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Popp C., Dean W., Feng S., Cokus S.J., Andrews S., Pellegrini M., Jacobsen S.E., Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhutani N., Brady J.J., Damian M., Sacco A., Corbel S.Y., Blau H.M. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bohn M.F., Shandilya S.M., Albin J.S., Kouno T., Anderson B.D., McDougle R.M., Carpenter M.A., Rathore A., Evans L., Davis A.N., et al. Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain. Structure. 2013;21:1042–1050. doi: 10.1016/j.str.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siu K.K., Sultana A., Azimi F.C., Lee J.E. Structural determinants of HIV-1 Vif susceptibility and DNA binding in APOBEC3F. Nat. Commun. 2013;4:2593. doi: 10.1038/ncomms3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kitamura S., Ode H., Nakashima M., Imahashi M., Naganawa Y., Kurosawa T., Yokomaku Y., Yamane T., Watanabe N., Suzuki A., et al. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat. Struct. Mol. Biol. 2012;19:1005–1010. doi: 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- 47.Byeon I.J., Ahn J., Mitra M., Byeon C.H., Hercik K., Hritz J., Charlton L.M., Levin J.G., Gronenborn A.M. NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nat. Commun. 2013;4:1890. doi: 10.1038/ncomms2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holden L.G., Prochnow C., Chang Y.P., Bransteitter R., Chelico L., Sen U., Stevens R.C., Goodman M.F., Chen X.S. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prochnow C., Bransteitter R., Klein M.G., Goodman M.F., Chen X.S. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007;445:447–451. doi: 10.1038/nature05492. [DOI] [PubMed] [Google Scholar]
- 50.Pak V., Heidecker G., Pathak V.K., Derse D. The role of amino-terminal sequences in cellular localization and antiviral activity of APOBEC3B. J. Virol. 2011;85:8538–8547. doi: 10.1128/JVI.02645-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lackey L., Demorest Z.L., Land A.M., Hultquist J.F., Brown W.L., Harris R.S. APOBEC3B and AID have similar nuclear import mechanisms. J. Mol. Biol. 2012;419:301–314. doi: 10.1016/j.jmb.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shinohara M., Io K., Shindo K., Matsui M., Sakamoto T., Tada K., Kobayashi M., Kadowaki N., Takaori-Kondo A. APOBEC3B can impair genomic stability by inducing base substitutions in genomic DNA in human cells. Sci. Rep. 2012;2:806. doi: 10.1038/srep00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chelico L., Pham P., Calabrese P., Goodman M.F. APOBEC3G DNA deaminase acts processively 3′ → 5′ on single-stranded DNA. Nat. Struct. Mol. Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- 54.Yoo J., Medina-Franco J.L. Homology modeling, docking and structure-based pharmacophore of inhibitors of DNA methyltransferase. J. Comput. Aided Mol. Des. 2011;25:555–567. doi: 10.1007/s10822-011-9441-1. [DOI] [PubMed] [Google Scholar]
- 55.Bogerd H.P., Wiegand H.L., Doehle B.P., Cullen B.R. The intrinsic antiretroviral factor APOBEC3B contains two enzymatically active cytidine deaminase domains. Virology. 2007;364:486–493. doi: 10.1016/j.virol.2007.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abdouni H., King J.J., Suliman M., Quinlan M., Fifield H., Larijani M. Zebrafish AID is capable of deaminating methylated deoxycytidines. Nucleic Acids Res. 2013;41:5457–5468. doi: 10.1093/nar/gkt212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hakata Y., Landau N.R. Reversed functional organization of mouse and human APOBEC3 cytidine deaminase domains. J. Biol. Chem. 2006;281:36624–36631. doi: 10.1074/jbc.M604980200. [DOI] [PubMed] [Google Scholar]
- 58.Langlois M.A., Beale R.C., Conticello S.G., Neuberger M.S. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 2005;33:1913–1923. doi: 10.1093/nar/gki343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Narvaiza I., Linfesty D.C., Greener B.N., Hakata Y., Pintel D.J., Logue E., Landau N.R., Weitzman M.D. Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase. PLoS Pathog. 2009;5:e1000439. doi: 10.1371/journal.ppat.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kohli R.M., Abrams S.R., Gajula K.S., Maul R.W., Gearhart P.J., Stivers J.T. A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. J. Biol. Chem. 2009;284:22898–22904. doi: 10.1074/jbc.M109.025536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kohli R.M., Maul R.W., Guminski A.F., McClure R.L., Gajula K.S., Saribasak H., McMahon M.A., Siliciano R.F., Gearhart P.J., Stivers J.T. Local sequence targeting in the AID/APOBEC family differentially impacts retroviral restriction and antibody diversification. J. Biol. Chem. 2010;285:40956–40964. doi: 10.1074/jbc.M110.177402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carpenter M.A., Rajagurubandara E., Wijesinghe P., Bhagwat A.S. Determinants of sequence-specificity within human AID and APOBEC3G. DNA Repair (Amst.) 2010;9:579–587. doi: 10.1016/j.dnarep.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang M., Rada C., Neuberger M.S. Altering the spectrum of immunoglobulin V gene somatic hypermutation by modifying the active site of AID. J. Exp. Med. 2010;207:141–153. doi: 10.1084/jem.20092238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mitra M., Hercik K., Byeon I.J., Ahn J., Hill S., Hinchee-Rodriguez K., Singer D., Byeon C.H., Charlton L.M., Nam G., et al. Structural determinants of human APOBEC3A enzymatic and nucleic acid binding properties. Nucleic Acids Res. 2014;42:1095–1110. doi: 10.1093/nar/gkt945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rathore A., Carpenter M.A., Demir O., Ikeda T., Li M., Shaban N.M., Law E.K., Anokhin D., Brown W.L., Amaro R.E., Harris R.S. The local dinucleotide preference of APOBEC3G can be altered from 5′-CC to 5′-TC by a single amino acid substitution. J. Mol. Biol. 2013;425:4442–4454. doi: 10.1016/j.jmb.2013.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Logue E.C., Bloch N., Dhuey E., Zhang R., Cao P., Herate C., Chauveau L., Hubbard S.R., Landau N.R. A DNA sequence recognition loop on APOBEC3A controls substrate specificity. PLoS One. 2014;9:e97062. doi: 10.1371/journal.pone.0097062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gajula K.S., Huwe P.J., Mo C.Y., Crawford D.J., Stivers J.T., Radhakrishnan R., Kohli R.M. High-throughput mutagenesis reveals functional determinants for DNA targeting by activation-induced deaminase. Nucleic Acids Res. 2014;42:9964–9975. doi: 10.1093/nar/gku689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bulliard Y., Narvaiza I., Bertero A., Peddi S., Rohrig U.F., Ortiz M., Zoete V., Castro-Diaz N., Turelli P., Telenti A., et al. Structure–function analyses point to a polynucleotide-accommodating groove essential for APOBEC3A restriction activities. J. Virol. 2011;85:1765–1776. doi: 10.1128/JVI.01651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]