Abstract
In order to gain a better understanding of the underlying biology of squamous cell carcinoma (SCC), we tested the hypothesis that SCC originating from different organs may possess common molecular alterations. SCC samples (N = 361) were examined using clinical-grade targeted next-generation sequencing (NGS). The most frequent SCC tumor types were head and neck, lung, cutaneous, gastrointestinal and gynecologic cancers. The most common gene alterations were TP53 (64.5% of patients), PIK3CA (28.5%), CDKN2A (24.4%), SOX2 (17.7%), and CCND1 (15.8%). By comparing NGS results of our SCC cohort to a non-SCC cohort (N = 277), we found that CDKN2A, SOX2, NOTCH1, TP53, PIK3CA, CCND1, and FBXW7 were significantly more frequently altered, unlike KRAS, which was less frequently altered in SCC specimens (all P < 0.05; multivariable analysis). Therefore, we identified “squamousness” gene signatures (TP53, PIK3CA, CCND1, CDKN2A, SOX2, NOTCH 1, and FBXW7 aberrations, and absence of KRAS alterations) that were significantly more frequent in SCC versus non-SCC histologies. A multivariable co-alteration analysis established 2 SCC subgroups: (i) patients in whom TP53 and cyclin pathway (CDKN2A and CCND1) alterations strongly correlated but in whom PIK3CA aberrations were less frequent; and (ii) patients with PIK3CA alterations in whom TP53 mutations were less frequent (all P ≤ 0 .001, multivariable analysis). In conclusion, we identified a set of 8 genes altered with significantly different frequencies when SCC and non-SCC were compared, suggesting the existence of patterns for “squamousness.” Targeting the PI3K-AKT-mTOR and/or cyclin pathway components in SCC may be warranted.
Keywords: cancer, gene signature, next-generation sequencing, squamous, SCC
Introduction
Squamous cell carcinomas (SCCs) occur in tissues normally covered with squamous epithelium, and may involve many different organs, although 4 major sites include head and neck, cutaneous, esophageal, and lung SCCs.1 SCCs, other than those in the skin, commonly metastasize,2,3 and the survival rates have not improved in decades. A better understanding of the underlying biology of SCCs is just beginning to be elucidated,4 but would be enhanced by the characterization of their common molecular alterations.
Defining the genetic landscape of SCCs would also facilitate the use of targeted therapies, which have been generally lacking for SCCs. For example, while targeted therapy employing EGFR inhibitors is commonly used to treat lung adenocarcinomas harboring EGFR alterations, no effective agents have been specifically developed for squamous cell carcinomas of the lung. Indeed, EGFR mutations are very rare in their squamous cell counterpart.5 Interestingly, it has been suggested that head and neck SCCs may share patterns of molecular alterations with SCCs of the lung.6-9 Further, esophageal and head and neck SCCs may also share some pathogenic mechanisms.10 Examining genetic aberrations of SCCs may shed light on additional common pathways underlying SCC tumorigenesis.
We hypothesized that squamous cell carcinomas may possess common biologic/molecular alterations, regardless of organ of origin. Herein, 361 tumor samples of patients with diverse squamous cell carcinomas including, but not limited to, lung, head and neck, cutaneous, gastrointestinal, and gynecologic/genitourinary SCCs, were analyzed using targeted next-generation sequencing (NGS) in order to determine if a pattern of “squamousness” alterations could be discerned.
Results
Genetic alterations identified in SCCs
Our squamous cell carcinomas (SCCs) cohort included 361 samples, the majority of them being SCCs of the head and neck (N = 107, 29.6%) and lung (N = 92, 25.5%), Table 1. The most frequent genetic alterations found were TP53 (64.5%), PIK3CA (28.5%), CDKN2A (24.4%), SOX2 (17.7%), and CCND1 (15.8%) (Fig. 1A). Most of the alterations were mutations (61.2%) or amplifications (30.5%) (Fig. 1B). A total of 1,382 alterations were identified in 132 different genes (775 distinct alterations), Table S1. Patients had a median number of 4 (range 1–11) abnormalities identified, Figure 1C.
Table 1.
Histologies breakdown of 361 squamous cell carcinomas
| Squamous Cell Carcinomas (N = 361 ) | |
|---|---|
| Histology | Number (%) |
| Head and neck | 107 (29.6%) |
| Lung | 92 (25.5%) |
| Cutaneous | 36 (10.0%) |
| Gynecologic/Genitourinarya | 40 (11.1%) |
| Gastrointestinal (GI)b | 51 (14.1%) |
| Unknown origin | 35 (9.7%) |
| TOTAL | 361 (100%) |
Included cervical, n = 22; vagina, n = 5; vulva, n = 3; penile, n = 6; bladder, n = 2; and urethra, n = 2.
Included esophageal, n = 21; anal, n = 21; rectal, n = 7; and gallbladder, n = 2.
Figure 1.
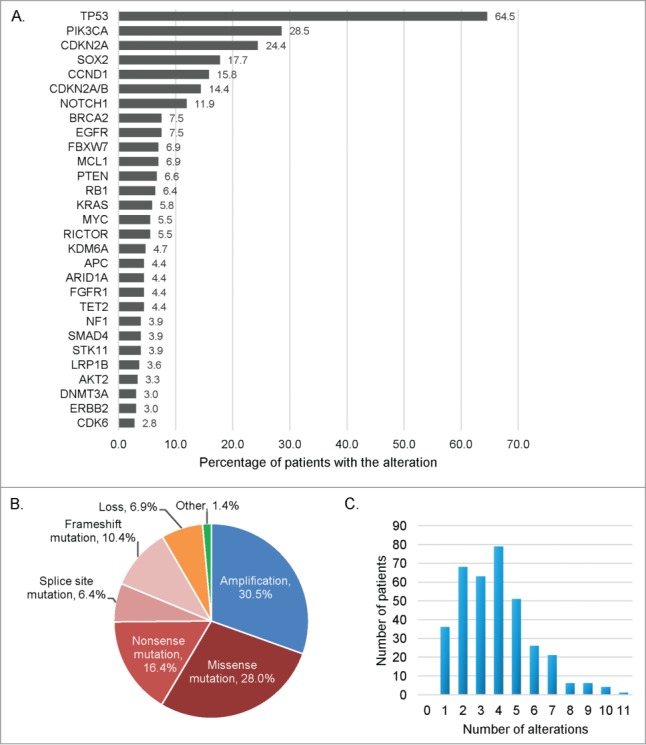
Frequency and type of molecular alterations identified in 361 patients with diverse squamous cell carcinomas. (A) Bar graph representing the alteration frequency. Only the more frequent alterations are represented (at least 10 patients with the alteration), and some patients had multiple alterations in the same gene. (B) Pie chart displaying the different type of alterations. Other refers to truncation (n = 11 ), fusion (n = 3 ), rearrangement (n = 3 ), deletion (n = 2 ), and duplication (n = 1 ). (C) Bar graph representing the number of patients by number of molecular alterations. Patients had a median of 4 alterations, (range 1–11).
Specificity of the alterations portfolio identified in SCC specimens
We next investigated if we could identify a particular set of genes whose alteration frequencies were different compared to non-SCC samples. For this purpose, we compared the alterations found in 361 SCC samples (SCCs cohort) to a separate cohort including 277 patients' samples with diverse malignancies, but none with SCCs (non-SCCs cohort), that had undergone similar next-generation sequencing testing. Organ of origin breakdown was as follows for the non-squamous tumors: head and neck, n = 32; lung, n = 30; cutaneous, n = 38; gynecologic/genitourinary (included n = 29 gynecologic cases), n = 37; gastrointestinal, n = 117; other, n = 23. We first performed a univariable analysis that highlighted 8 genes whose alteration frequency was statistically different between SCC vs. non-SCC specimens. Because there were imbalances in the number of patients for some tumor types (i.e. organs of origin) between the SCC and non-SCC cohorts (gastrointestinal, head and neck, and lung, all P < 0.05) we included these tumor types and the 8 genes identified in the univariable analysis as variables for a multivariable model. The results of the multivariable confirmed that 8 genes had statistically different alteration frequencies. More specifically, 7 genes were found to be significantly more altered in SCC specimens compared to non-SCCs, as follows: CDKN2A (24% versus 8%, P = 0 .003), SOX2 (18% vs. 1%, P = 0 .001), NOTCH1 (12% versus 1%, P = 0 .0002), TP53 (65% vs. 45%, P = 0 .004), PIK3CA (29% versus 9%, P < 0.0001), CCND1 (16% vs. 4%, P = 0 .001), and FBXW7 (7% versus 3%, P = 0 .048), Table 2 and Figure 2. Of note, the only gene that was statistically less frequently altered in SCC specimens was KRAS (6% in SCC vs. 15% in non-SCC cases, P = 0 .001).
Table 2.
Alteration frequency differences in squamous versus non-squamous cell carcinomas (multivariable analysis)
| Squamous cases (N = 361) | Non-Squamous cases (N = 277) | Wald statistic | Odds Ratio | CI 95% | P-Value | |
|---|---|---|---|---|---|---|
| Alterations | ||||||
| TP53 | 233 (65%) | 125 (45%) | 8.4 | 1.82 | 1.22–2.74 | 0.004 |
| PIK3CA | 103 (29%) | 26 (9%) | 20.7 | 3.86 | 2.16–6.91 | <0.0001 |
| CDKN2A | 88 (24%) | 23 (8%) | 8.8 | 2.44 | 1.35–4.39 | 0.003 |
| SOX2 | 64 (18%) | 3 (1%) | 10.8 | 7.94 | 2.31–27.3 | 0.001 |
| CCND1 | 57 (16%) | 12 (4%) | 10.7 | 3.57 | 1.67–7.65 | 0.001 |
| NOTCH1 | 43 (12%) | 4 (1%) | 13.9 | 8.38 | 2.74–25.6 | 0.0002 |
| FBXW7 | 25 (7%) | 7 (3%) | 3.9 | 2.70 | 1.01–7.20 | 0.048 |
| KRAS | 21 (6%) | 66 (24%) | 11.8 | 0.33 | 0.18–0.62 | 0.001 |
For all the included variables, the P-values in the univariable analysis were <0.001. To account for imbalances of number of cases in certain tumor types, lung, gastrointestinal, and head and neck histologies were also computed in the multivariable. All P-values remained significant in the multivariable analysis. Further, a bootstrapping analysis (5,000 bootstrap samples) confirmed the results, with all P-values ≤0 .005, except FBXW7, with P = 0 .029). Genes altered in more than 20 SCC cases were included.
Figure 2.
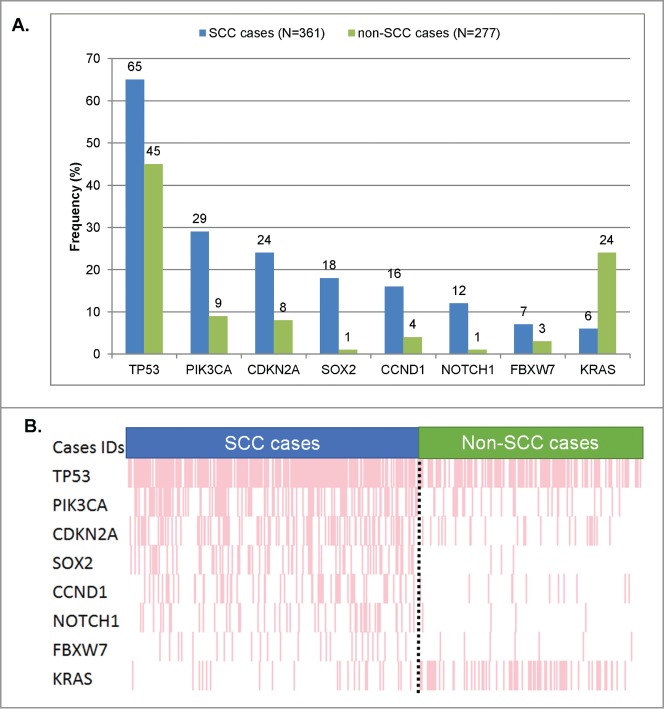
Significant differences in frequency of molecular alterations between squamous vs. non-squamous cases. Only genes that were found to be aberrant at statistically different rates (Table 2) in squamous versus non-squamous cancers were included. (A) Bar graph comparing the alteration frequencies between squamous vs. non-squamous cases. (B) Raw data on each patient in a “reverse array” fashion. Each pink bar corresponds to an alteration in the designated gene in one patient. Squamous cases have an over-representation of alterations in all the genes included, except KRAS (whose alterations are under-represented in SCCs). All the P-values were ≤0 .001, except for FBXW7 (P = 0 .048). All detailed P-values can be found in Table 2.
We then used the 8 genes found to be significantly differentially altered in squamous cases versus non-squamous cases (more frequent in SCC: TP53, PIK3CA, CCND1, CDKN2A, SOX2, NOTCH 1, FBXW7; less frequent in SCC: KRAS) as a “signature” set of altered genes (“squamousness” signature). To confirm the specificity of this signature in SCCs, we scored all tumor samples by allocating one point for each gene of the signature altered and for each patient. As KRAS alterations were found less frequently in squamous cases, it was the absence of KRAS that was given 1 point (as opposed to all the other genes of the signature for which the presence of the alteration gave 1 point). Since the signature comprised 8 genes, each patient could cumulate a maximum of 8 points. Overall, SCC cases had a median of 3 points vs. only 1 for non-SCC specimens (P < 0.0001). The overall population (SCC and non-SCC cases) had a median of 2 points. We found that the percentage of number of specimens with ≥ 2 points was significantly higher in squamous cases than in non-squamous cases (82% versus 43%, P < 0.0001). These results were also statistically significant within all primary site of origins (all P ≤ 0 .006), Figure 3. Further, when comparing squamous tumors of a particular site of origin to all other squamous tumors, no significant differences were discerned (Fig. 3 and Table S2). These results demonstrate that even though the tumor specimens share the same tissue location, their genetic characteristics were statistically different depending on their status as a squamous tumor or not.
Figure 3.
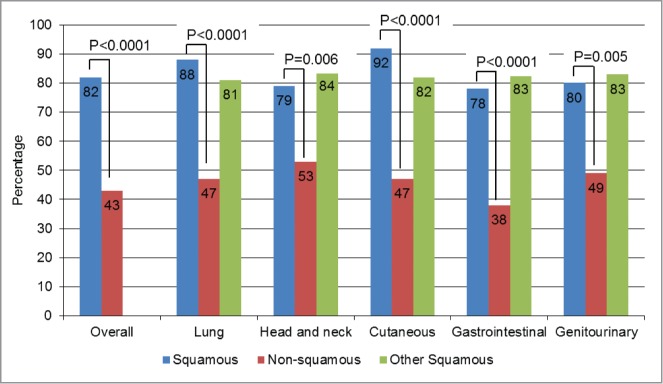
Squamousness signature frequency: comparison by histology. For this analysis, one point (cumulative) was given for each of the genes where abnormalities were more prevalent in squamous tumors–TP53, PIK3CA, CCND1, CDKN2A, SOX2, NOTCH1, and FBXW7– each time they were present. Because KRAS aberrations were significantly less prevalent in squamous tumors, 1 point was also assigned for the absence of KRAS. The numbers were then added up for each case. Overall, squamous cases had a median of 3 points versus only 1 point for non-squamous cases, P < 0.0001. In this bar graph, we represented the percentages of patients with ≥2 points (2 points was the median for the overall population (squamous and non-squamous cases)). All the P-values comparing squamous (blue bars) vs. non-squamous cases (red bars) were ≤0 .006. When comparing squamous tumors of a particular histology with all other squamous tumors (blue versus green bars), no significant differences were seen. For details on numbers and P-values refer to Table S2.
Co-alteration analysis
We then conducted a multivariable co-alteration analysis for TP53, which was by far the most altered gene in SCCs (64.5% among the SCC cases, Fig. 1A). This analysis established that TP53 alterations strongly correlated with aberrations in the cyclin pathway components CDKN2A and CCND1 (P < 0.0001), Figure 4A. TP53 and CDKN2A/CCND1 co-alteration was more frequently observed in squamous cases than in non-squamous cases (31% vs. 9%, P<0.0001), though it was also observed in non-squamous cases (P = 0 .003, Fig. 4B). More specifically, the co-alteration frequencies of TP53 and CDKN2A/CCND1 were as follows: 37.4% for head and neck SCC (vs. 15.6% in non-squamous head and neck cancers), 29.3% for SCC of the lung (vs. 6.7 in non- squamous lung cancers), 23.5% for gastro-intestinal SCC (vs. 9.4% for non-squamous gastro-intestinal cancers), and 7.5% for gynecologic/genitourinary SCC (vs. 0% of 37 cases in non-squamous gynecologic/genitourinary cancers.
Figure 4.
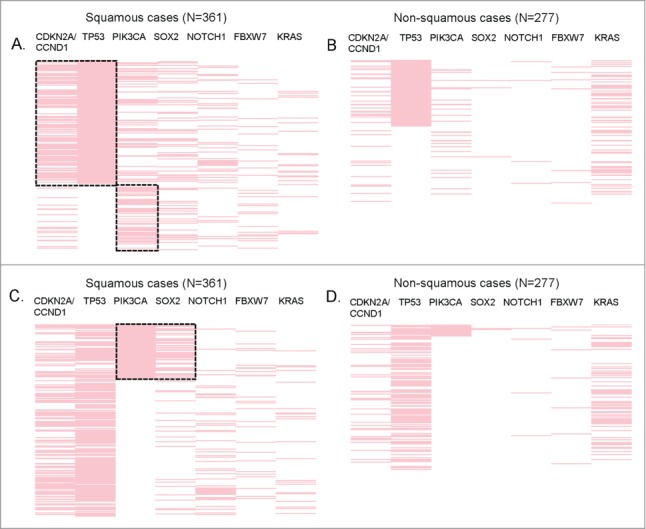
Significant co-alterations in patients with SCCs. Reverse array plot displaying raw data. Each pink bar corresponds to an alteration in the designated gene in one patient (total N = 361 SCC and N = 277 non-SCC specimens). (A) (squamous cases) and (B) (non-squamous cases) have been sorted according to the presence of TP53 mutations. Panel A shows that within squamous tumors, TP53 is co-altered with CDKN2A/CCND1, P < 0.0001 (P-values were also ≤0 .001 if CDKN2A and CCND1 were considered separately). There also was a negative association between TP53 and PIK3CA alteration, P = 0.001. (B) shows that within non-squamous tumors, TP53 alterations were also associated with CDKN2A/CCND1, P = 0 .003 (P-value = 0 .008 for CCND1 and P = 0 .129 for CDKN2A when considered separately). However, the co-alteration frequency of TP53 and CDKN2A/CCND1 was significantly higher in squamous tumors (31% vs. 9%, P < 0.0001). No association (positive or negative) was found between TP53 and PIK3CA in non-squamous cases. (C) (squamous cases) and (D) (non-squamous cases) display the same data as panel A and B, but sorted by the presence of PIK3CA alteration. Within squamous tumors, PIK3CA is co-altered with SOX2 (P < 0.0001). (D) PIK3CA alterations are not significantly associated with SOX2 in non-squamous tumors.
In contrast, cases with TP53 alterations (versus wild-type TP53) had less PIK3CA alterations in squamous cases (P = 0 .001); this difference was not observed in non-squamous specimens, Figure 4A. In addition, we detected that SOX2 alterations were statistically associated with PIK3CA alterations (P < 0.0001) in squamous cases (Fig. 4C), but not in non-squamous cases (Fig. 4D). PIK3CA and SOX2 co-alterations were the most frequently observed in lung SCC (24% vs. 3.3 in lung non-SCC), followed by 11.8% in gastro-intestinal SCC (vs. 0% of 117 non-squamous gastro-intestinal cases), 10% in gynecologic/genitourinary SCC (vs. 2.7% in non-squamous gynecologic/genitourinary cancers), 9.3% in head and neck SCC (vs. 3.1% in non-squamous head and neck cancers). Of note, none of the cutaneous cases (squamous or not) harbored PIK3CA/SOX2 co-alterations.
Discussion
Our study examined different types of SCC specimens by targeted next-generation sequencing and demonstrated that the most commonly altered genes across squamous tumors were TP53 (64.5%), PIK3CA (28.5%), CDKN2A (24.4%), SOX2 (17.7%), and CCND1 (15.8%), Figure 1. Furthermore, when comparing the alterations portfolio of SCC specimens with alterations found in non-SCC cases, we established that 8 genes had alteration frequencies that were statistically different when SCCs and non-SCC cases were compared in a multivariable analysis. While TP53, PIK3CA, CDKN2A, CCND1, SOX2, NOTCH1 and FBXW7 were found to be significantly more frequently altered in SCCs vs. non-SCCs, KRAS appeared to be less altered. In a review recently published11 and focusing on head and neck SCCs, the most commonly altered genes were TP53, NOTCH1, HRAS, PIK3CA, as well as CDKN2A; additional studies focusing on other pathologic diagnoses, such as esophageal,10 or lung cancer,12,13 reported alterations in genes overlapping with our findings. Of note, 5.6% of our head and neck specimens carried HRAS alterations (6/107 cases), while only one single specimen harbored a KRAS alteration (0.9%), consistent with what has been previously described in head and neck SCCs.7,8
More than half of the specimens carried TP53 alterations (64.5%), making this tumor suppressor gene the most commonly mutated gene in SCCs. TP53 is the gatekeeper of the genome and has crucial function such as DNA repair, apoptosis, and cell cycle arrest. TP53 alterations have been identified as early events in the disease and have been associated with disease progression and a poorer outcome.11 Therapies targeting TP53 loss of function are currently being explored in clinical trials, and several studies suggest that patients harboring TP53 alterations might respond better to angiogenesis inhibitors.14,15 Of note, a TP53 adenoviral-based treatment (rAd-p53; Gendicine) for patients with HNSCC has recently been approved for use in China.16
Cyclin-dependent kinase inhibitor 2A (CDKN2A), located at chromosome 9p21, is another well-known tumor suppressor gene involved in the regulation of cell cycle progression and was altered in 24.5% of our SCC cases. Cyclin D1 (CCND1), another component of this pathway, was also found highly altered in SCCs (15.8%). CCND1 interacts with CDK4/6 and forms complexes that play a central role in the G1/S transition of the cell cycle, acting by phosphorylating Rb.17-19 Several agents targeting the cyclin pathway are undergoing clinical phases. Studies of one such agent, palbociclib, have produced encouraging data in women with breast cancer20 and has been recently approved by the FDA (Feb 2015). Given the high incidence of cyclin pathway alterations in SCCs, testing cyclin-pathway inhibitors in clinical trials may be warranted in patients with SCCs.
Another highly activated pathway in SCCs appeared to be the PI3K-Akt-mTOR axis, as reflected by the high incidence of PIK3CA alterations. This pathway also leads to an increased translation of CCND1 and plays a crucial role in the cell cycle,21-23 as well as migration and invasion.24 Further, alteration of this pathway components, such as PIK3CA, may confer resistance to agents targeting upstream constituents, such as EGFR inhibitors, as many receptor tyrosine kinases (RTKs) activate PI3K.24 Of note, a case series investigating the use of an mTOR inhibitor for patients with head and neck SCCs and PTEN loss or PIK3CA mutations showed significant tumor regression in 3 of 5 patients.25
SOX2 is a transcription factor encoded at 3q26.33, which can act either as a suppressor or activator of gene transcription. SOX2 has been implicated in cellular reprogramming and normal embryogenesis mechanisms,26 and may represent an important player in tumor initiation and progression of SCCs.27,28 We have found that 18% of our specimens harbored SOX2 amplifications (compared to only 1% in non-squamous cases), making this gene a hallmark candidate for SCCs. Further, we found that SOX2 and PIK3CA were highly co-altered (P < 0.0001 in multivariable analysis), which may be explained by their chromosomal co-localization at 3q26.33.29
NOTCH1 is believed to play important roles in different biological processes disturbed in cancer, and NOTCH1 alterations were identified in 12% of our SCC samples (versus 1% in non SCCs). Murine models have highlighted the importance of NOTCH1 in squamous epithelial differentiation, and in cutaneous SCC, recent sequencing efforts also suggest loss of function mutational pattern of NOTCH1.30-32 Indeed, in our subset of cutaneous SCCs, NOTCH1 alterations were found in 33% of cases, vs. 10% in the other types of SCCs, (P = 0 .016 in a multivariable analysis, data not shown). Interestingly, in addition to NOTCH1 alterations, FBXW7 alterations were identified in 7% of our SCC specimens, versus 3% in non-SCC cases (P = 0 .048 in multivariable). FBXW7 forms part of the ubiquitin ligase complex that can mediate NOTCH1 degradation; consequently, FBXW7 mutations could also be affecting the NOTCH1 pathway,11 although FBXW7 is also known to target other oncogenic pathways by degrading cyclin E and c-myc.33
In the recent years, human papilloma virus (HPV) has emerged as a primary etiology of oropharyngeal cancers,13 as well as cutaneous and lung SCCs.1 HPV positive–associated cancers exhibit different characteristics (e.g. risk factors, epidemiology) but also a distinct clinical course. Not surprisingly, the alterations identified in these subgroups are also different. Several studies described that most HPV-negative samples carried TP53 as well as cyclin pathway alterations,34-36 as opposed to HPV-positive samples which appeared to harbor more PIK3CA alterations.35,37 Of potential interest, another study showed that squamous-like tumors without TP53 mutations had a higher density of PIK3CA mutations.38
There are several limitations to our study. There may have been selection bias of the specimens, as patients/physicians may have elected to submit tissue when there were fewer standard therapeutic options left. Also, the samples came from diverse institutions and practices. However, the fact that all the samples had undergone the same next-generation sequencing test by the same laboratory improved the homogeneity of our analysis. The size of the test panel changed from 182 to 236 genes over the course of the study sample. However, the markers identified as part of the squamousness signature were all present in both the original and expanded panels. The pool of samples also originated from different tissue types, and this heterogeneity may have confounded some of the analysis, though it also was crucial to our ability to compare squamous molecular portfolios in different malignancies. The HPV status of our patients was not known. In future studies, the existence of different squamousness signatures depending on HPV status should be investigated. Finally, a confirmatory cohort of patients should be tested to confirm our results.
In summary, we have analyzed 361 SCC specimens and highlighted a set of 8 genes whose alteration frequency was statistically different when compared to non-SCC cases (N = 277 ). Indeed, we observed an increased frequency of TP53, PIK3CA, CDKN2A, CCND1, SOX2, NOTCH1, and FBXW7 alterations, while a decreased frequency of KRAS alterations was noted. These alterations were used to generate a “squamousness signature” that was significantly more prevalent in SCCs across malignancies originating in different organs compared to patients with non-squamous tumors of the same organ of origin (Fig. 3). We also observed co-alteration patterns and report that TP53 mutations associated positively with CDKN2A and CCND1 alterations and negatively with PIK3CA in SCC specimens. Of interest, KRAS, a frequently aberrant gene in non-squamous tumors that leads to resistance to PI3K pathway inhibitors,39-41 was significantly less often aberrant in SCCs. A multivariable co-alteration analysis identified 2 SCC subtypes: (i) patients in whom TP53 and cyclin pathway (CDKN2A and CCND1) aberrations were associated but in whom PIK3CA anomalies were less common; and (ii) individuals with PIK3CA abnormalities, in whom TP53 mutations were less frequent (all P ≤ 0 .001, multivariable analysis). Together, this data suggests a direction for a more precise approach for treatment of SCCs. Targeting the PI3K-AKT-mTOR and/or cyclin pathway components may represent valuable investigational avenues for clinical trials in subgroups of patients with SCCs. Future studies should concentrate on large datasets that include full phenotypic annotation. Importantly, NGS provides a potentially important diagnostic test that permits recognition of the abnormalities present in each patient, hence enabling the therapeutic decision-making process.
Patients and Methods
Patients
This study included 361 SCC samples that were tested using NGS over a 2-year period of time (from December 2011 until November 2013). All consecutive patients for whom we had data were included in the analysis. Patients were tested on physician request. This cohort was compared to 277 other patient samples (separate population) comprising individuals that had diverse cancers (other than SCCs) who were seen at the UC San Diego Moores Cancer Center. This non-SCCs cohort included the same tumor sites of origin as the SCC cohort. This study was performed in accordance with UCSD Institutional Review Board guidelines.
Next-generation sequencing
All patients (both SCC and non-SCC cohorts) had formalin-fixed, paraffin-embedded tumor samples that were submitted to Foundation Medicine (FoundationOne™, Cambridge, Massachusetts, http://www.foundationone.com, which is a clinical grade CLIA-approved next-generation sequencing test). Samples were reviewed by a pathologist to ensure the correct diagnosis. Next-generation sequencing data were collected and interpreted by N-of-One®, Inc.. (Lexington, MA, www.n-of-one.com). For this study, patients' samples were tested with a 182 or 236 gene panel; the latter sequences the entire coding sequence of 236 cancer-related genes and 47 introns from 19 genes often rearranged in cancer. The entire coding sequence of the 182 or 236 cancer-related genes was generated with an average sequencing depth of coverage greater than 250x (full list available at http://foundationone.com/genelist1.php). This method of sequencing allows for detection of copy number alterations, gene rearrangements, and somatic mutations with >99% specificity and >99% sensitivity for base substitutions at ≥5 mutant allele frequency and >95% sensitivity for copy number alterations. Foundation Medicine uses a threshold of ≥8 copies for gene amplification.
Statistical analysis
When appropriate, Fisher's exact tests and binary logistic regressions (univariable and multivariable) were performed. A bootstrapping analysis (5,000 bootstrap samples) was performed for internal validation of the multiple logistic regression model results used to select the genes for the “squamousness” signature. Whenever possible, 95% confidence intervals (95% CI) were reported. All statistical analysis were performed by author MS with SPSS version 22.0.
Disclosure of Potential Conflicts of Interest
Dr. Kurzrock has consultant fees from Sequenom and has an ownership interest in RScueRx Inc. Sheryl K. Elkin, Brett N. Tomson, and Jennifer Levin Carter are employees of N-of-One, Inc. Jennifer Levin Carter had a leadership position and holds ownership interest at N-Of-One, Inc. Sheryl K. Elkin holds ownership interest at N-Of-One, Inc.
Funding
Funded in part by the Joan and Irwin Jacobs Fund.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Yan W, Wistuba II, Emmert-Buck MR, Erickson HS. Squamous cell carcinoma - similarities and differences among anatomical sites. Am J Cancer Res 2010; 1:275-300. [PMC free article] [PubMed] [Google Scholar]
- 2.Miller DL, Weinstock MA. Nonmelanoma skin cancer in the United States: incidence. J Am Acad Dermatol 1994; 30:774-8; PMID:8176018 [DOI] [PubMed] [Google Scholar]
- 3.Czarnecki D, Staples M, Mar A, Giles G, Meehan C. Metastases from squamous cell carcinoma of the skin in southern Australia. Dermatol Basel Switz 1994; 189:52-4. [DOI] [PubMed] [Google Scholar]
- 4.Agada FO, Patmore H, Alhamarneh O, Stafford ND, Greenman J. Genetic profile of head and neck squamous cell carcinoma: clinical implications. J Laryngol Otol 2009; 123:266-72; PMID:18694533 [DOI] [PubMed] [Google Scholar]
- 5.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong K-K. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 2014; 14:535-46; PMID:25056707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network . Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012; 489:519-25; PMID:22960745; http://dx.doi.org/ 10.1038/nature11404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al.. The mutational landscape of head and neck squamous cell carcinoma. Science 2011; 333:1157-60; PMID:21798893; http://dx.doi.org/ 10.1126/science.1208130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie T-X, Zhang J, Wang J, et al.. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 2011; 333:1154-7; PMID:21798897; http://dx.doi.org/ 10.1126/science.1206923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, Leiserson MDM, Niu B, McLellan MD, Uzunangelov V, et al.. Multiplatform Analysis of 12 Cancer Types Reveals Molecular Classification within and across Tissues of Origin. Cell 2014; 158:929-44; PMID:25109877; http://dx.doi.org/ 10.1016/j.cell.2014.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song Y, Li L, Ou Y, Gao Z, Li E, Li X, Zhang W, Wang J, Xu L, Zhou Y, et al.. Identification of genomic alterations in oesophageal squamous cell cancer. Nature 2014; 509:91-5; PMID:24670651; http://dx.doi.org/ 10.1038/nature13176 [DOI] [PubMed] [Google Scholar]
- 11.Loyo M, Li RJ, Bettegowda C, Pickering CR, Frederick MJ, Myers JN, Agrawal N. Lessons learned from next-generation sequencing in head and neck cancer. Head Neck 2013; 35:454-63; PMID:22907887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgensztern D, Devarakonda S, Govindan R. Genomic landscape of squamous cell carcinoma of the lung. Am Soc Clin Oncol Educ Book ASCO Am Soc Clin Oncol Meet 2013:348-53. [DOI] [PubMed] [Google Scholar]
- 13.Rooney M, Devarakonda S, Govindan R. Genomics of squamous cell lung cancer. The Oncologist 2013; 18:707-16; PMID:23728941; http://dx.doi.org/ 10.1634/theoncologist.2013-0063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwaederle M, Vladimir L, Validire P, Hansson J, Lacroix L, Soria J-C, Pawitan Y, Kurzrock R. VEGF-A Expression Correlates with TP53 Mutations in Non-Small Cell Lung Cancer: Implications for Anti-Angiogenesis Therapy. Cancer Res 2015; 75:1187–90; PMID:25672981 [DOI] [PubMed] [Google Scholar]
- 15.Said R, Hong DS, Warneke CL, Lee JJ, Wheler JJ, Janku F, Naing A, Falchook GS, Fu S, Piha-Paul S, et al.. P53 Mutations in Advanced Cancers: Clinical Characteristics, Outcomes, and Correlation between Progression-Free Survival and Bevacizumab-Containing Therapy. Oncotarget 2013; 4:705-14; PMID:23670029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nemunaitis J, Nemunaitis J. Head and neck cancer: response to p53-based therapeutics. Head Neck 2011; 33:131-4; PMID:20222046 [DOI] [PubMed] [Google Scholar]
- 17.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer 2011; 11:558-72; PMID:21734724 [DOI] [PubMed] [Google Scholar]
- 18.Caldon CE, Musgrove EA. Distinct and redundant functions of cyclin E1 and cyclin E2 in development and cancer. Cell Div 2010; 5:2; PMID:20180967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwaederlé M, Daniels GA, Piccioni DE, Fanta PT, Schwab RB, Shimabukuro KA, Parker BA, Kurzrock R. Cyclin alterations in diverse cancers: Outcome and co-amplification network. Oncotarget 2015; 6:3033–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.palblociclib press release [Internet] Available from: http://www.pfizer.com/news/press-release/press-release-detail/pfizer_announces_positive_top_line_results_from_paloma_1_evaluating_palbociclib_plus_letrozole_in_women_with_advanced_breast_cancer. [Google Scholar]
- 21.Liang J, Slingerland JM. Multiple roles of the PI3K/PKB (Akt) pathway in cell cycle progression. Cell Cycle Georget Tex 2003; 2:339-45. [PubMed] [Google Scholar]
- 22.Averous J, Fonseca BD, Proud CG. Regulation of cyclin D1 expression by mTORC1 signaling requires eukaryotic initiation factor 4E-binding protein 1. Oncogene 2007; 27:1106-13; PMID:17724476; http://dx.doi.org/ 10.1038/sj.onc.1210715 [DOI] [PubMed] [Google Scholar]
- 23.Steelman LS, Chappell WH, Abrams SL, Kempf RC, Long J, Laidler P, Mijatovic S, Maksimovic-Ivanic D, Stivala F, Mazzarino MC, et al.. Roles of the Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR pathways in controlling growth and sensitivity to therapy-implications for cancer and aging. Aging 2011; 3:192-222; PMID:21422497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers SJ, Box C, Harrington KJ, Nutting C, Rhys-Evans P, Eccles SA. The phosphoinositide 3-kinase signalling pathway as a therapeutic target in squamous cell carcinoma of the head and neck. Expert Opin Ther Targets 2005; 9:769-90; PMID:16083342 [DOI] [PubMed] [Google Scholar]
- 25.Holsinger FC, Piha-Paul SA, Janku F, Hong DS, Atkins JT, Tsimberidou AM, Kurzrock R. Biomarker-directed therapy of squamous carcinomas of the head and neck: targeting PI3K/PTEN/mTOR pathway. J Clin Oncol Off J Am Soc Clin Oncol 2013; 31:e137-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weina K, Utikal J. SOX2 and cancer: current research and its implications in the clinic. Clin Transl Med 2014; 3:19; PMID:25114775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maier S, Wilbertz T, Braun M, Scheble V, Reischl M, Mikut R, Menon R, Nikolov P, Petersen K, Beschorner C, et al.. SOX2 amplification is a common event in squamous cell carcinomas of different organ sites. Hum Pathol 2011; 42:1078-88; PMID:21334718 [DOI] [PubMed] [Google Scholar]
- 28.Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, Delatte B, Caauwe A, Lenglez S, Nkusi E, et al.. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 2014; 511:246-50; PMID:24909994; http://dx.doi.org/ 10.1038/nature13305 [DOI] [PubMed] [Google Scholar]
- 29.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, et al.. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 2009; 41:1238-42; PMID:19801978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, Hui C, Clevers H, Dotto GP, Radtke F. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet 2003; 33:416-21; PMID:12590261 [DOI] [PubMed] [Google Scholar]
- 31.Dotto GP. Notch tumor suppressor function. Oncogene 2008; 27:5115-23; PMID:18758480; http://dx.doi.org/ 10.1038/onc.2008.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen B-C, Lefort K, Mandinova A, Antonini D, Devgan V, Della Gatta G, Koster MI, Zhang Z, Wang J, Tommasi di Vignano A, et al.. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev 2006; 20:1028-42; PMID:16618808; http://dx.doi.org/ 10.1101/gad.1406006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujii Y, Yada M, Nishiyama M, Kamura T, Takahashi H, Tsunematsu R, Susaki E, Nakagawa T, Matsumoto A, Nakayama KI. Fbxw7 contributes to tumor suppression by targeting multiple proteins for ubiquitin-dependent degradation. Cancer Sci 2006; 97:729-36; PMID:16863506; http://dx.doi.org/ 10.1111/j.1349-7006.2006.00239.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lechner M, Frampton GM, Fenton T, Feber A, Palmer G, Jay A, Pillay N, Forster M, Cronin MT, Lipson D, et al.. Targeted next-generation sequencing of head and neck squamous cell carcinoma identifies novel genetic alterations in HPV+ and HPV- tumors. Genome Med 2013; 5:49; PMID:23718828; http://dx.doi.org/ 10.1186/gm453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suh Y, Amelio I, Guerrero Urbano T, Tavassoli M. Clinical update on cancer: molecular oncology of head and neck cancer. Cell Death Dis 2014; 5:e1018; PMID:24457962; http://dx.doi.org/ 10.1038/cddis.2013.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabatabaeifar S, Kruse TA, Thomassen M, Larsen MJ, Sørensen JA. Use of next generation sequencing in head and neck squamous cell carcinomas: A review. Oral Oncol 2014; 50:1035–40; PMID:25223596 [DOI] [PubMed] [Google Scholar]
- 37.Gross AM, Orosco RK, Shen JP, Egloff AM, Carter H, Hofree M, Choueiri M, Coffey CS, Lippman SM, Hayes DN, et al.. Multi-tiered genomic analysis of head and neck cancer ties TP53 mutation to 3p loss. Nat Genet 2014; 46:939-43; PMID:25086664; http://dx.doi.org/ 10.1038/ng.3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, Leiserson MDM, Niu B, McLellan MD, Uzunangelov V, et al.. Multiplatform Analysis of 12 Cancer Types Reveals Molecular Classification within and across Tissues of Origin. Cell 2014; 158:929–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janku F, Hong DS, Fu S, Piha-Paul SA, Naing A, Falchook GS, Tsimberidou AM, Stepanek VM, Moulder SL, Lee JJ, et al.. Assessing PIK3CA and PTEN in early-phase trials with PI3K/AKT/mTOR inhibitors. Cell Rep 2014; 6:377-87; PMID:24440717; http://dx.doi.org/ 10.1016/j.celrep.2013.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garrido-Laguna I, Hong DS, Janku F, Nguyen LM, Falchook GS, Fu S, Wheler JJ, Luthra R, Naing A, Wang X, et al.. KRASness and PIK3CAness in patients with advanced colorectal cancer: outcome after treatment with early-phase trials with targeted pathway inhibitors. PloS One 2012; 7:e38033; PMID:22675430; http://dx.doi.org/ 10.1371/journal.pone.0038033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janku F, Wheler JJ, Westin SN, Moulder SL, Naing A, Tsimberidou AM, Fu S, Falchook GS, Hong DS, Garrido-Laguna I, et al.. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. J Clin Oncol Off J Am Soc Clin Oncol 2012; 30:777-82; http://dx.doi.org/ 10.1200/JCO.2011.36.1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


