Abstract
Two pseudogenes for HMGA1, whose overexpression has a critical role in cancer progression, have been identified. They act as decoy for miRNAs that are able to target the HMGA1 gene then enhancing cell proliferation and migration. Moreover, these pseudogenes contain sequences that are potential target sites for cancer-related miRNAs. Interestingly, HMGA1 pseudogenes are highly expressed in human anaplastic thyroid carcinomas, that is one of the most aggressive tumor in mankind, but almost undetectable in well differentiated thyroid carcinomas.
The human genome has been sequenced, and no more than 2% codes for proteins building our bodies.1 Therefore, a key question is to identify which is the meaning of the other 98% of the genome. Many evidences show that the mammalian genome is able to generate numerous previously undiscovered transcripts called “non coding RNA” (ncRNA).1 However, the role of these ncRNAs remains largely unknown. The ncRNAs include different classes: the “short interfering RNAs” (siRNAs), the “Natural Antisense Transcripts” (NATs), the “microRNAs” (miRNAs), long non coding RNAs (lncRNAs), and pseudogenes.2
siRNAs are double-stranded small interfering RNAs of ∼21 base pairs in length that serve as effector molecules of sequence-specific gene silencing. They are highly conserved across species.3
NATs are RNAs that are at least in part complementary to other endogenous RNAs. They might be transcribed in cis from opposing DNA strands at the same genomic locus (cis-NATs) or in trans at separate loci (trans-NATs).4 NATs biological role is based on gene knockdown induced by base-pairing of sense and anti-sense strands. To date, in human have been identified cis-encoded exon-overlapping sense-antisense (SA) in a number of 7356 NATs.4
Among trans-NATs the most studied are miRNAs. They are small RNAs (18-21 nt) that can inhibit mRNA expression by binding to 3′ Untranslated Region (3′UTR) with perfect or imperfect match, suppressing mRNA translation or affecting RNA stability.5 Krek et al. reported that vertebrate microRNAs target, usually, almost 200 transcripts each one.6,7
Long non coding RNAs (lncRNAs) are non-protein coding transcripts longer than 200 nucleotides.8 This rather arbitrary limit distinguishes lncRNAs from small regulatory RNAs such as miRNAs, siRNAs, Piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), and other short RNAs.8
Pseudogenes are dysfunctional relatives of genes that have lost their protein-coding skill or are otherwise no longer expressed in the cell.9-11 They are characterized by a mixture of homology to a known gene and nonfunctionality: every pseudogene has a DNA sequence that is similar to some functional genes, but they are unable to produce functional final protein products.9-11
There are 3 major families of pseudogenes: processed (or retrotransposed), non-processed (or duplicated), and disabled pseudogenes. Processed pseudogenes originate from a segment of mRNA transcript of a gene that is spontaneously reverse transcribed back into DNA and inserted into chromosomal DNA. Once these pseudogenes are inserted back into the genome, they generally include a poly-A tail, and their introns are frequently spliced out.12 Non-processed pseudogenes arise as a consequence of a gene duplication event and then acquire mutations making them nonfunctional. Duplicated pseudogenes usually include all the same characteristics of genes from they originate, as well as an intact exon-intron structure and promoter sequences.12 Disabled genes (or unitary pseudogenes) present different mutations that stop a gene from being productively transcribed or translated.13
Recently, Esposito et al. have identified 2 processed pseudogenes, HMGA1P6 and HMGA1P7, belonging to the HMGA1 gene family. They are placed at 13q12.12 and 6q23.2, respectively, and have been reported to have a critical role in the process of carcinogenesis.14 The High Mobility Group A1 (HMGA1) gene codes for the HMGA1 proteins, HMGA1a and HMGA1b.15 These proteins are non-histone chromatinic proteins that bind to DNA and organize chromatin architecture, interacting with several transcription factors, and then regulate the expression of several genes, positively or negatively.15 Their role in carcinogenesis is widely accepted. Indeed, these proteins are expressed at high levels for the duration of embryogenesis and in malignancies where their expression levels point out a poor prognosis of the cancer patients, whereas their expression is brought down in adult normal tissues.16-18 Moreover, the knock-down of HMGA1 expression prevents thyroid cells transformation and leads cancer cells of diverse tissue origin to apoptosis.19-21 On the contrary, their overexpression in vitro induces mouse and rat fibroblast transformation,22 and transgenic mice overexpressing hmga1 develop several neoplasias including pituitary adenomas, Natural Killer (NK)/T-cell lymphomas,24 lipomas, cervix and body adenocarcinomas.15
HMGA1 pseudogenes, HMGA1P6 and HMGA1P7, show just few mismatches all over the coding sequence and the 5’ and 3’ UTRs of HMGA1. They have conserved seed matches for miRNAs that have been previously confirmed to target the HMGA1 gene. Subsequently, the authors show that these pseudogenes equally operate as decoys for HMGA1-targeting miRNAs (Fig. 1). Indeed, HMGA1P6 and HMGA1P7 overexpression increases HMGA1 protein levels whereas their silencing results in decreased HMGA1 mRNA and protein levels.14 Consistently with the HMGA1P6 and HMGA1P7 decoy function, cells overexpressing them show an enhanced migration, invasiveness, and a faster proliferation ability.14 Opposite results are obtained when these pseudogenes are silenced with also an increase in apoptotic cells following a reduced HMGA1 protein levels, as already observed when HMGA1 is knocked down in thyroid cells.21
Figure 1.
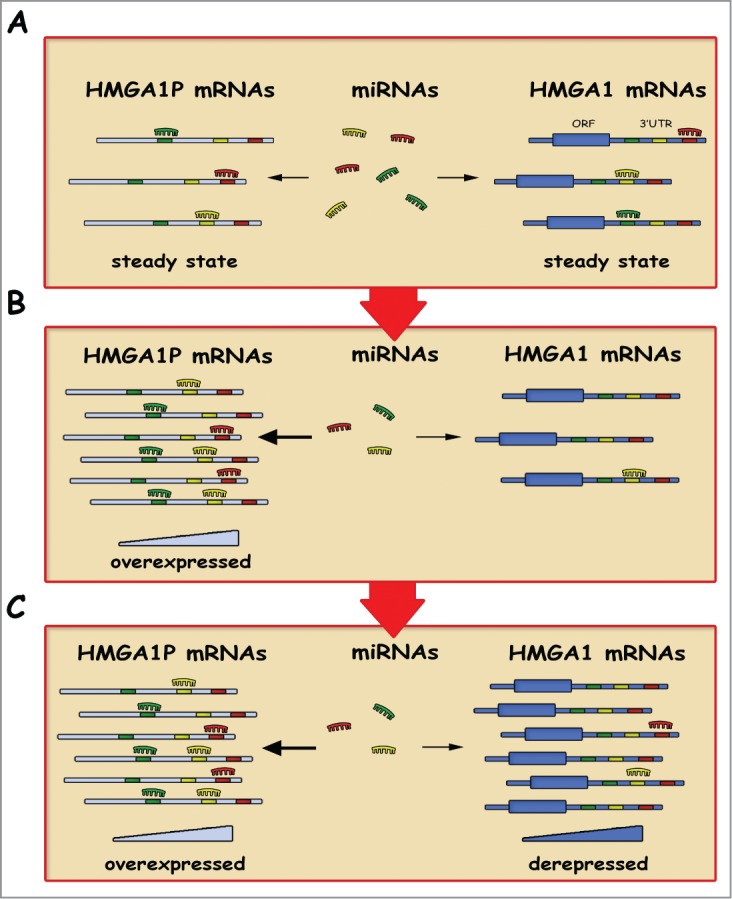
Decoy function of HMGA1Ps. Several RNA transcripts have seed sequences for the same miRNAs (A). As proposed by Esposito et al., overexpression of HMGA1Ps mRNAs increases cellular concentrations of particular seed sequences (B), resulting in the derepression of HMGA1 transcripts that contain the same seed sequences (C).
The generation of transgenic mice overexpressing HMGA1P6 or HMGA1P7 has confirmed their oncogenic activity. Indeed, mouse embryonic fibroblasts (MEFs) deriving from HMGA1P6 or HMGA1P714 overexpressing mice grow faster and senesce later than their wild-type counterparts.
However, we retain that the key point of this study is the finding of a role of these pseudogenes in human carcinogenesis. In fact, anaplastic thyroid carcinoma (ATC), that represents one of the most aggressive tumors in the mankind, evidenced a exceptionally high expression of the HMGA1 pseudogenes essentially correlating with the HMGA1 protein levels.14 Conversely, their expression is almost undetectable in papillary and follicular thyroid carcinomas, that are well differentiated and much less aggressive. Analogous results were obtained when HMGA1Ps expression was analyzed in human ovarian14 and larynx carcinomas, and pituitary adenomas (manuscript in preparation).
Remarkably, the presence in the HMGA1P6, HMGA1P7, and also HMGA1 UTR regions of sequences that are potential target sites for cancer-related miRNAs targeting genes such as High Mobility Group A2 (HMGA2), Enhancer of Zeste Homolog 2 (EZH2), Vascular Endothelial Growth Factor (VEGF), and Ephrin Type-A Receptor 3 (Epha3), that are effectively upregulated in HMGA1P6 and HMGA1P714 overexpressing cells and MEFs with respect to the control cells. This has important consequences. In fact, it means that high HMGA1 gene or its pseudogene expression allows an increase in HMGA2 and EZH2 protein levels then contributing to cancer progression.14 Such a mechanism is likely to occur in ATC where the overexpression of EZH2 has been detected in ATC but not in the undifferentiated thyroid carcinomas.25,26
Interestingly, Kumar et al. have recently demonstrated that the HMGA2 3′ UTR contains 7 conserved seed sequences for let-7, which has been previously demonstrated to constrain lung cancer development. They identified 6 ceRNA targets that are regulated by hmga2 in a let7-dependent manner: Transforming growth factor β receptor III (tgfbr3), Angiopoietin-related protein 2 (Angptl2), Fibronectin Type III Domain Containing Protein 3 (Fndc3), Ski-like protein (Skil) and Hmga1.27 Then, on the basis of these results also HMGA1, through its 3’UTR, may function as decoy for HMGA2 and other cancer-related gene expression. Therefore, a synergism between members of HMGA protein family might be envisaged based not only on common functions but also on the ability of the HMGA mRNAs to act as decoy for miRNAs able to target themselves.
In conclusion, the results published by Esposito et al. indicate that also the expression of the HMGA1 pseudogenes contribute to carcinogenesis and, together with the paper by Kumar et al., reveal another mechanism by which HMGA gene family has a critical role in cancer progression based on the ability to regulate gene expression also as non-coding RNAs (Fig. 2). Therefore, these new reports make even more important the HMGA gene family in cancer diagnosis and prognosis, and as potential target for an innovative cancer therapy.
Figure 2.
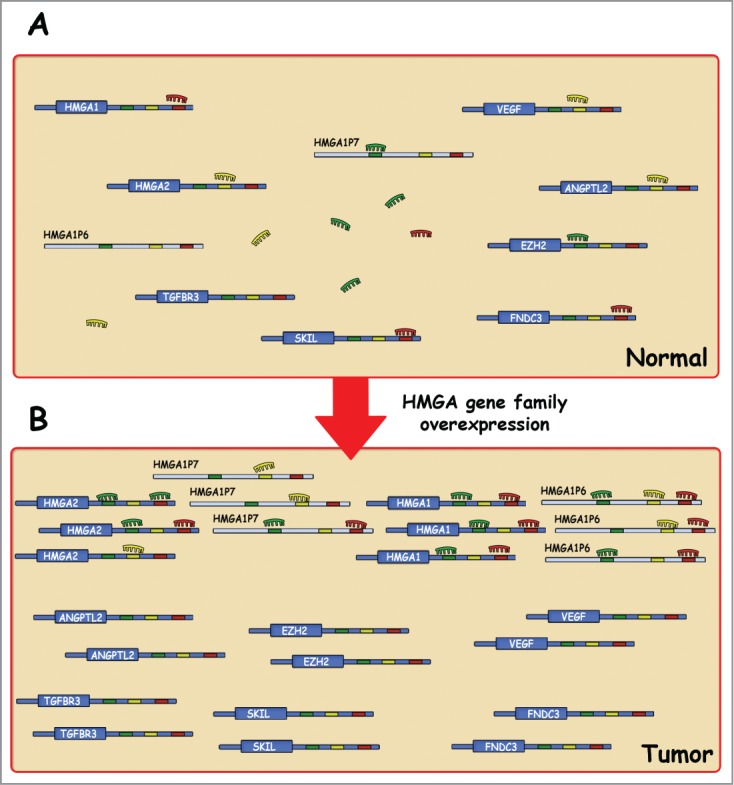
A ceRNA model for the HMGA gene family. HMGA RNA transcripts have seed sequences for the same miRNAs shared with other transcripts (A). As proposed by Esposito and Kumar, in malignancies, the HMGA gene family overexpression increases cellular concentrations of particular seed sequences, resulting in the derepression of several cancer-related gene transcripts that contain the same seed sequences (B).
References
- 1. Furuno M, Pang KC, Ninomiya N, Fukuda S, Frith MC, Bult C, Kai C, Kawai J, Carninci P, Hayashizaki Y, et al. Clusters of Internally Primed Transcripts Reveal Novel Long Noncoding RNAs. PLoS Genet 2006; 2:e37; PMID:16683026; http://dx.doi.org/ 10.1371/journal.pgen.0020037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009; 136:629-41; PMID:19239885; http://dx.doi.org/ 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 3. Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 2001; 293: 1146-50; PMID:11498593; http://dx.doi.org/ 10.1126/science.1064023 [DOI] [PubMed] [Google Scholar]
- 4. Zhang Y, Li J, Kong L, Gao G, Liu QR, Wei L. NATsDB: Natural Antisense Transcripts DataBase. Nucl Acids Res 2007; 35:D156-61; PMID:17082204; http://dx.doi.org/ 10.1093/nar/gkl782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci U S A 2003; 100:9779-84; PMID:12902540; http://dx.doi.org/ 10.1073/pnas.1630797100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol 2004; 2:e363; PMID:15502875; http://dx.doi.org/ 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet 2005; 37:495-500; PMID:15806104; http://dx.doi.org/ 10.1038/ng1536 [DOI] [PubMed] [Google Scholar]
- 8. Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol 2013; 11:59; PMID:23721193; http://dx.doi.org/ 10.1186/1741-7007-11-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mighell AJ, Smith NR, Robinson PA, Markham AF. Vertebrate pseudogenes. FEBS Lett 2000; 468:109-14; PMID:10692568; http://dx.doi.org/ 10.1016/S0014-5793(00)01199-6 [DOI] [PubMed] [Google Scholar]
- 10. Harrison PM, Hegyi H, Balasubramanian S, Luscombe NM, Bertone P, Echols N, Johnson T, Gerstein M. Molecular fossils in the human genome: Identification and analysis of the pseudogenes in chromosomes 21 and 22. Genome Res 2002; 12:272-80; PMID:11827946; http://dx.doi.org/ 10.1101/gr.207102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Balakirev ES, Ayala FJ. Pseudogenes: Are they “junk” or functional DNA? Annu Rev Genet 2003; 37:123-51; PMID:14616058; http://dx.doi.org/ 10.1146/annurev.genet.37.040103.103949 [DOI] [PubMed] [Google Scholar]
- 12. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010; 465:1033-38; PMID:20577206; http://dx.doi.org/ 10.1038/nature09144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang ZD, Frankish A, Hunt T, Harrow J, Gerstein M. Identification and analysis of unitary pseudogenes: historic and contemporary gene losses in humans and other primates. Genome Biol 2010; 11:R26; PMID:20210993; http://dx.doi.org/ 10.1186/gb-2010-11-3-r26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Esposito F, De Martino M, Petti MG, Forzati F, Tornincasa M, Federico A, Arra C, Pierantoni GM, Fusco A. HMGA1 pseudogenes as candidate proto-oncogenic competitive endogenous RNAs. Oncotarget 2014; 5:8344-54; PMID:25268743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fusco A, Fedele M. Roles of the HMGA proteins in cancer. Nat Rev Cancer 2007; 7:899-910; PMID:18004397; http://dx.doi.org/ 10.1038/nrc2271 [DOI] [PubMed] [Google Scholar]
- 16. Chiappetta G, Avantaggiato V, Visconti R, Fedele M, Battista S, Trapasso F, Merciai BM, Fidanza V, Giancotti V, Santoro M, et al. High level expression of the HMGA1 gene during embryonic development. Oncogene 1996; 13:2439-46; PMID:8957086 [PubMed] [Google Scholar]
- 17. Piscuoglio S, Zlobec I, Pallante P, Sepe R, Esposito F, Zimmermann A, Diamantis I, Terracciano L, Fusco A, Karamitopoulou E. HMGA1 and HMGA2 protein expression correlates with advanced tumour grade and lymph node metastasis in pancreatic adenocarcinoma. Histopathology 2012; 60:397-404; PMID:22276603; http://dx.doi.org/ 10.1111/j.1365-2559.2011.04121.x [DOI] [PubMed] [Google Scholar]
- 18. Pegoraro S, Ros G, Piazza S, Sommaggio R, Ciani Y, Rosato A, Sgarra R, Del Sal G, Manfioletti G. HMGA1 promotes metastatic processes in basal-like breast cancer regulating EMT and stemness. Oncotarget 2013; 4:1293-1308; PMID:23945276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Berlingieri MT, Manfioletti G, Santoro M, Bandiera A, Visconti R, Giancotti V, Fusco A. Inhibition of HMGI-C protein synthesis suppresses retrovirally induced neoplastic transformation of rat thyroid cells. Mol Cell Biol 1995; 15:1545-53; PMID:7862147 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20. Berlingieri MT, Pierantoni GM, Giancotti V, Santoro M, Fusco A. Thyroid cell transformation requires the expression of the HMG1 proteins. Oncogene 2002; 21:2971-80; PMID:12082527; http://dx.doi.org/ 10.1038/sj.onc.1205368 [DOI] [PubMed] [Google Scholar]
- 21. Scala S, Portella G, Fedele M, Chiappetta G, Fusco A. Adenovirus mediated suppression of the HMGI(Y) protein synthesis as a potential therapy of human malignant neoplasias. Proc Natl Acad Sci USA. 2000; 97:4256-61; http://dx.doi.org/ 10.1073/pnas.070029997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vallone D, Battista S, Pierantoni GM, Fedele M, Casalino L, Santoro M, Viglietto G, Fusco A, Verde P. Neoplastic transformation of rat thyroid cells requires the junB and fra-1 gene induction which is dependent on the HMGI-C gene product. EMBO J 1997; 16:5310-21; PMID:9311991; http://dx.doi.org/ 10.1093/emboj/16.17.5310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Palmieri D, Valentino T, De Martino I, Esposito F, Cappabianca P, Wierinckx A, Vitiello M, Lombardi G, Colao A, Trouillas J, et al. PIT1 upregulation by HMGA proteins has a role in pituitary tumorigenesis. Endocr Relat Cancer 2012; 19:123-35; PMID:22199144; http://dx.doi.org/ 10.1530/ERC-11-0135 [DOI] [PubMed] [Google Scholar]
- 24. Fedele M, Pentimalli F, Baldassarre G, Battista S, Klein-Szanto AJ, Kenyon L, Visone R, De Martino I, Ciarmiello A, Arra C, et al. Transgenic mice overexpressing the wild-type form of the HMGA1 gene develop mixed growth hormone/prolactin cell pituitary adenomas and natural killer cell lymphomas. Oncogene 2005; 24:3427-35; PMID:15735694; http://dx.doi.org/ 10.1038/sj.onc.1208501 [DOI] [PubMed] [Google Scholar]
- 25. Borbone E, Troncone G, Ferraro A, Jasencakova Z, Stojic L, Esposito F, Horning N, Fusco A, Orlando V. Enhancer of zeste homolog 2 overexpression has a role in the development of anaplastic thyroid carcinomas. J Clin Endocrinol Metab 2011; 96:1029-38; PMID:21289264; http://dx.doi.org/ 10.1210/jc.2010-1784 [DOI] [PubMed] [Google Scholar]
- 26. Esposito F, Tornincasa M, Pallante P, Federico A, Borbone E, Pierantoni GM, Fusco A. Down-regulation of the miR-25 and miR-30d contributes to the development of anaplastic thyroid carcinoma targeting the polycomb protein EZH2. J Clin Endocrinol Metab 2012; 97:E710-8; PMID:22399519; http://dx.doi.org/ 10.1210/jc.2011-3068 [DOI] [PubMed] [Google Scholar]
- 27. Kumar MS, Armenteros-Monterroso E, East P, Chakravorty P, Matthews N, Winslow MM. HMGA2 functions as a competing endogenous RNA to promote lung cancer progression. Nature 2014; 505:212-17; PMID:24305048; http://dx.doi.org/ 10.1038/nature12785 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]


