Our study revealed that miR-148b was specifically down-regulated in hepatic cancer stem cells (HCSCs) and affected cell proliferation and metastasis in vitro and tumorigenicity in vivo by directly targeting to Neuropilin-1(NRP-1), a transmembrane co-receptor involved in metastasis, suggesting that enforced miR-148b expression might be an efficient therapeutic strategy to eradicate HCSCs and reduce metastasis.
Keywords: cancer stem cell, hepatocellular carcinoma, microRNA-148b, Neuropilin-1, side population cell
Abstract
The existence of cancer stem cells (CSCs) is considered as a direct reason for the failure of clinic treatment in hepatocellular carcinoma (HCC). Growing evidences have demonstrated that miRNAs play an important role in regulation of stem cell proliferation, differentiation and self-renewal and their aberrances cause the formation of CSCs and eventually result in carcinogenesis. We recently identified miRNA-148b as one of the miRNAs specifically down-regulated in side population (SP) cells of PLC/PRF/5 cell line. However, it remains elusive how miRNA-148b regulates CSC properties in HCC. In the present study, we observed that overexpression or knockdown of miR-148b through lentiviral transfection could affect the proportion of SP cells as well as CSC-related gene expression in HCC cell lines. In addition, miR-148b blocking could stimulate cell proliferation, enhance chemosensitivity, as well as increase cell metastasis and angiogenesis in vitro. More importantly, miR-148b could significantly suppress tumorigenicity in vivo. Further studies revealed that Neuropilin-1 (NRP1), a transmembrane co-receptor involved in tumour initiation, metastasis and angiogenesis, might be the direct target of miRNA-148b. Taking together, our findings define that miR-148b might play a critical role in maintenance of SP cells with CSC properties by targeting NRP1 in HCC. It is the potential to develop a new strategy specifically targeting hepatic CSCs (HCSCs) through restoration of miR-148b expression in future therapy.
INTRODUCTION
Hepatocellular carcinoma (HCC) is known as the fifth most common cancer and the third cause of cancer-related mortality in the world. The existence of hepatic cancer stem cells (HCSCs) has been considered as the direct reason of clinical tumour relapse and therapeutic failure [1]. Increasing evidences have suggested that many miRNAs play critical roles in controlling the self-renewal and differentiation of stem cell and some of them were proved to express abnormally in HCSCs and regulate the key biological characteristics of HCSCs [2]. It suggests a new clue to conquer HCC by directly targeting and eradicating HCSCs through related-miRNA regulation [3]. However, it remains unclear the detailed mechanism about how miRNAs regulate the formation of HCSCs and maintain the properties of HCSCs.
Based on cancer stem cell (CSC) hypothesis, tumours consist of heterogeneous cell populations, in which only a small population called CSCs has the capability to drive tumour initiation [4]. Traditional surgery, chemotherapy and radiotherapy could eliminate the bulk of cancerous cells but hard to eradicate CSCs that maintain a high capacity for self-renewal, chemoresistance and pluripotency, leading HCC patients to suffer frequent recurrence or metastasis. Therefore, it is in urgent need of novel treatment strategies, which could target CSCs directly. Several functional assays have been developed to isolate CSCs, including side population (SP) approach, surface biomarker-based isolation, sphere formation and ALDEFLUOR-approach [1]. SP cells those can actively efflux Hoechst 33342 dye through ATP-binding cassette proteins (ABC) transporters have been proved to harbour CSC properties early in 2006 [5]. Compared with non-SP (NSP) cells, SP cells showed enhanced proliferation, chemoresistance and tumorigenicity in HuH-7 and PLC/PRF/5 cell lines, indicating that SP cells isolation is an effective method to identify HCSCs [5].
miRNAs are non-coding RNAs of ~19–23 nts that are crucial for many biological processes through regulating gene expression. Several miRNAs, which regulate differentiation of normal stem cells, are also involved in carcinogenesis. Ji et al. [6,7] revealed that the miR-181 family are highly expressed in epithelial cell adhesion molecule (EpCAM)+AFP+ cells of HCC and maintained the CSC characteristics through targeting caudal type homeobox 2 (CDX2), GATA binding protein 6 (GATA6) and nemo-like kinase (NLK), which are essential for hepatic cell differentiation through regulating Wnt/β-catenin signal pathway. miR-21 was also detected to overexpress in SP cells of HCC and increase the ability of migration and invasion via degradation of tumour suppressors phosphatase and tensin homolog (PTEN), reversion-inducing-cysteine-rich protein with kazal motifs (RECK) and programmed cell death 4 (PDCD4) [8]. Besides these oncogenic miRNAs, more miRNAs were found to be specifically down-regulated in CSC of HCC as tumour suppressors, such as miR-142-3p, miR-150, miR-145, miR-612, miR-200a and miR-200c. They contribute to cell proliferation, drug resistance, cell migration and invasion, angiogenesis and tumorigenesis both in vitro and in vivo [9–14]. To target and suppress HCSCs by regulating these miRNAs may shed light on the development of efficient therapeutic strategies against HCC. However, it remains a substantial need for more thorough investigation on the role of miRNAs in maintenance of CSC-like properties and regulation of tumour initiation and progression.
In previous study, we performed the quantitative real-time (qRT)-PCR profile and detected 27 miRNAs aberrantly expressed in SP cells of PLC/PRF/5 cell line, including miR-148b, miR-9*, miR-194 and so on [15]. We noticed that miR-148b was one of the most down-regulated miRNAs, suggesting that it might function as a key regulator in SP cells. Coincidentally, Liu et al. [16] also detected differently miRNAs expression profile between SP and NSP F344 rat HCC cell and found out 10 under-expressed miRNAs in SP cells, which contained miR-148b*. Moreover, accumulating evidences support that miR-148b acts as a tumour-suppressor in various tumour types, such as breast cancer, lung cancer, pancreatic cancer, gastric cancer, colorectal cancer and non-Hodgkin's lymphoma [17–22]. A clinical research recently revealed that miR-148b was significantly decreased in HCC and involved in tumour invasion and progression, based on the qRT-PCR data from 156 cases of HCC and 36 cases of normal control specimens [23]. Zhang et al. [24] also confirmed that miR-148b was significantly down-regulated in human HCC tissues and correlated with larger tumour size, more tumour number, metastasis and worse prognosis. Overexpression of miR-148b inhibited cell proliferation, tumorigenicity and cell invasion in HepG2 by targeting the gene wingless-type MMTV integration site family, member 1 (WNT1) [24]. Nevertheless, how miR-148b regulates the formation and maintenance of SP cells with CSC properties in HCC is still poorly characterized.
In the present study, we proved that miR-148b was down-regulated in SP of PLC/PRF/5 and HuH-7 cell lines. Gain-of-function and loss-of-function studies via lentivirus transfection could change SP population and relative CSC properties, including cell proliferation, drug-resistance, cell migration and invasion, angiogenesis and tumorigenicity. Dual luciferase assay revealed that Neuropilin-1 (NRP1), a membrane-bound co-receptor to tyrosine kinase receptors for vascular endothelial growth factor (VEGF), transforming growth factor-β1 (TGF-β1), hepatocyte growth factor (HGF), the platelet-derived growth factor homodimer (PDGF-BB) and Semaphorin-3 (SEMA3) family members [25], was the direct target of miR-148b. In conclusion, we suggested that miR-148b might play a critical role in maintaining SP cells with CSC properties by regulating NRP1 in HCC and enforced miR-148b expression might be an efficient therapeutic strategy to eradicate HCSCs.
MATERIALS AND METHODS
Cell culture
Human liver cancer cell lines PLC/PRF/5 and HuH-7 were purchased from the Cell Resource Center, Shanghai Institute of Biochemistry and Cell Biology at the Chinese Academy of Sciences. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco) containing 10% FBS and incubated at 37°C in a humidified environment containing 5% CO2.
SP cell detection and isolation by flow cytometry
The cells were harvested, washed by PBS and suspended at 1×106 cells/ml in Hank's balanced salt solution supplemented with 3% FBS and 10 mM Hepes. Cells were incubated at 37°C for 90 min with 15 μg/ml Hoechst 33342 (Sigma) alone or in the presence of 50 μM verapamil (Sigma). During the incubation, the tubes were shaken up and down every 20 min to mix the cells with the solution. The cells were washed by Hank's solution for twice, added with 1 μg/ml propidium iodide (Sigma) and then filtered through a 70-μm cell strainer (BD). SP cell analysis and sorting were performed using MoFlo XDP (Beckman Coulter). Hoechst 33342 was excited with the UV laser at 350 nm and fluorescence emission was measured with 450 (Hoechst blue) and 570 (Hoechst red) optical filters. Propidium iodide labelling was measured for the discrimination of dead cells.
miRNA interference
Lentiviral miR-148b-Ubi-EGFP-multiple cloning sites (MCS) (miR-148b) and its control Ubi-EGFP-MCS (miR-NC), miR-148b-H1-MCS-cytomegalovirus (CMV)-EGFP (Inh-148b) and its control H1-MCS-CMV-EGFP (Inh-NC) were constructed by Genechem Company and transfected to the PLC/PRF/5 and HuH-7 cells according to the manufacturer's instructions, in the presence of virus at a multiplicity of infection of 10.
RNA extraction and quantitative real-time PCR
Total RNA from cultured cells was extracted using TRIzol reagent (Invitrogen). qRT-PCR assays were carried out to detect mRNA expression using the GoScript Reverse Transcription System (Promega) and QuantiFast SYBR Green PCR Kit (Qiagen) according to the manufacturer's instructions. miRNA from cultured cells was extracted using miRNeasy Mini Kit (Qiagen). qRT-PCR assays for miRNA detection were carried out using miScript II RT Kit (Qiagen) and miScript SYBR Green PCR Kit (Qiagen) according to the manufacturer's instructions. The primers for miR-148b and the internal control U6 were purchased from Qiagen. The primers for gene cluster of differentiation 133 (CD133), EpCAM, ATP-binding cassette, sub-family G (WHITE), member 2 (ABCG2), matrix metallopeptidase 9 (MMP9), tissue inhibitor of metalloproteinase-1 (TIMP-1), BMI1 proto-oncogene (BMI 1), snail family zinc finger 1 (SNAI 1), twist-related protein 1 (TWIST 1), cadherin 1 (CDH 1), CDH 2, NRP1 and control β-actin are shown in Table 1.
Table 1. Primer for the genes used in the present study.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| CD133 | 5′-CCTATAGAACAATCACTGAGC-3′ | 5′-CAGAGGAAGTATTGTTTGTG-3′ |
| EpCAM | 5′-GCTGGTGTTATTGCTGTTA-3′ | 5′-CTTCTCATACTTTGCCATTC-3′ |
| ABCG2 | 5′-CTGTGAGGCCTATAATAACC-3′ | 5′-GAGTGGCTTATCCTGCTT-3′ |
| MMP9 | 5′-CCTGGAGACCTGAGAAC-3′ | 5′-CAGGGACAGTTGCTTCT-3′ |
| TIMP-1 | 5′-ACTCTTGCACATCACTACCT-3′ | 5′-AAACACTGTGCATTCCTC-3′ |
| BMI 1 | 5′-CATTTTCTGCTGAACGACT-3′ | 5′-AGTGATCTTGATTCTCGTTG-3′ |
| SNAI 1 | 5′-AGGTGTGACTAACTATGCAA-3′ | 5′-CATTACTCACAGTCCCTTTT-3′ |
| TWIST 1 | 5′-ACCTAGATGTCATTGTTTCC-3′ | 5′-CTGTCTCGCTTTCTCTTTTA-3′ |
| CDH 1 | 5′-CTGGTTAGTGATGCAGTTAG-3′ | 5′-GAACCTTCTGATGCTAAAGT-3′ |
| CDH 2 | 5′- GGTTTGCCAGTGTGACT-3′ | 5′-GATAAGCAGGATGATGATG-3′ |
| NRP1 | 5′-ACAGCTTCTTCCCAGTATAG-3′ | 5′-GTATCCACTCTCGGTAGGA-3′ |
| β-Actin | 5′-GTTACACCCTTTCTTGACAA-3′ | 5′-CTTCACCGTTCCAGTTTT-3′ |
Soft agar colony formation assay
Make 0.6% agar–10% FBS DMEM bottom layer and 0.3% agar–10% FBS DMEM top layer with 5000 cells in each well of six-well plate. After 14 days incubation, colonies were fixed with methanol and stained with 0.1% Crystal Violet solution for 15 min and photographed.
Cell migration and invasion assay
Cell migration and invasion experiment was assessed using the Millipore 24-well Millicell Chamber of pore size 8 mm (Millipore). For the migration assay, a total of 1×105 cells in DMEM without FBS were added into the upper chamber of the insert. For the invasion assay, 2×105 cells in DMEM without FBS were added into the upper chamber pre-coated with Matrigel (Sigma). DMEM with 30% FBS was in the lower chamber. After incubated for 24 h, chamber was fixed and stained with 0.5% crystal violet for 30 min and the non-invading cells were removed with cotton swabs. The number of invasive cells on the lower surface of the chamber membrane was then counted under a microscope at a magnification of ×400 in five random fields.
Luciferase assay
PLC/PRF/5 cells were seeded in 96-well plates. After 24 h, 100 ng of pmirGLO-NRP1 3′-UTR [wild-type (WT)] or pmirGLO-mutant NRP1 3′-UTR [mutant-type (MUT)] were co-transfected into cells with miR-148b or miR-NC by Lipofectamine 2000. Forty-eight hours later, Firefly and Renilla luciferase activities were measured with a Dual-Luciferase Reporter System (Promega). The Renilla luciferase activities were used as an internal control for transfection efficiency. All experiments were repeated for three times.
Western blot
Cells were harvested, washed and lysed by lysis buffer. Proteins were separated by SDS/PAGE (8% gel) and transferred to nitrocellulose membrane. After blocking in 5% non-fat milk for 1 h, the membranes were incubated with the following primary antibodies:rabbit anti-NRP1 monoclonal antibody (1:1000, Cell Signaling Technology), mouse anti-α tubulin monoclonal antibody (1:1000, Abcam). The proteins were visualized with enhanced chemiluminescence reagents (Pierce).
Xenograft tumour model
Every group contains 10 male BALB/c nude mice of 6-week-old. Group 1 were injected with 5×106 control miR-NC or miR-148b PLC/PRF/5 cells in 100 μl of PBS into either shoulder, whereas group 2 were injected with 5×106 control Inh-NC or Inh-148b PLC/PRF/5 cells in 100 μl of PBS into either shoulder. Tumour volumes were measured with slide caliper every three days until scarification after 6 weeks. The tumours formed in BALB/c nude mice were fixed in formalin and embedded in paraffin. The sections were subjected to hematoxylin and eosin (H&E) staining or stained with anti-NRP1 antibody.
Statistical analysis
All results are expressed as mean±S.D. Comparisons between groups were conducted with the unpaired t test. The relationship between miR-148b and NRP1 (CST) expression was explored by Spearman correlation. Values were considered to be significantly different at P<0.05. All statistical analysis was conducted using SPSS 13.0 software.
RESULTS
SP cells of HCC cell lines harbour cancer stem cell-like properties
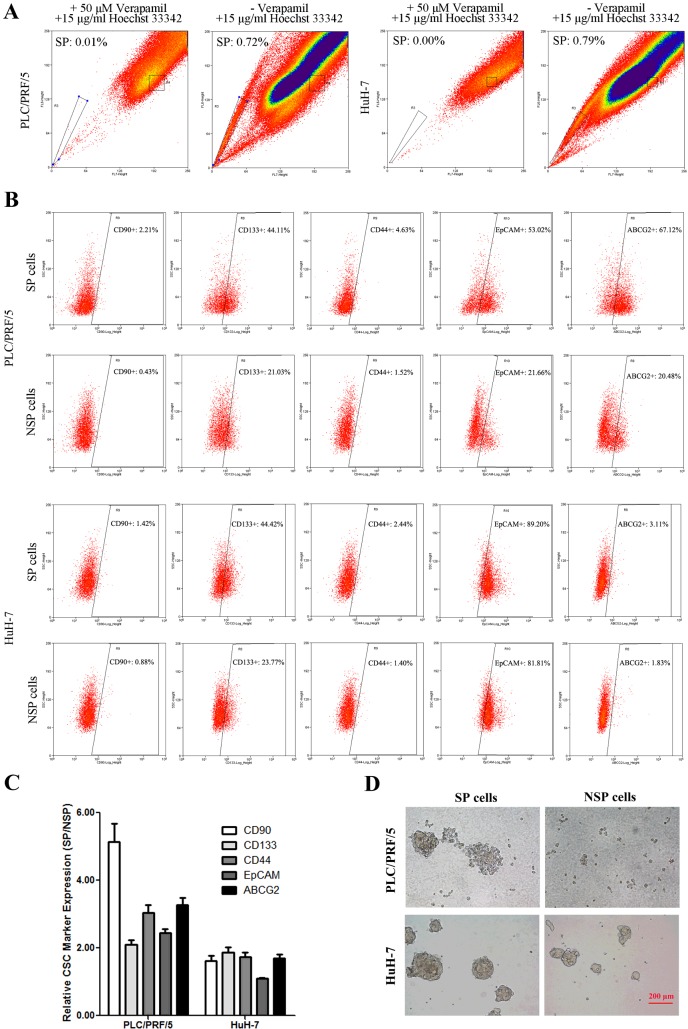
SP cell assay based on actively efflux of Hoechst 33342 dye was reported to be an effective method to identify CSCs in HCC [5]. To investigate the characteristics of SP cells in HCC, we first detected and isolated SP and NSP cells from PLC/PRF/5 and HuH-7 cell lines by flow cytometry. As the data showed, the percentage of SP was 0.74±0.08% in PLC/PRF/5 and 0.77±0.09% in HuH-7 cells in the presence of Hoechst 33342, whereas the SP cells were diminished in both cell lines in the presence of the calcium channel blocker verapamil and Hoechst 33342 (Figure 1A). We further tested the expression of HCC-related CSC markers, including CD90, CD133, CD44, EpCAM and ABCG2, on the surface of SP and NSP cells by flow cytometry. The percentage of PLC/PRF/5 cell positive for CD90, CD133, CD44, EpCAM and ABCG2 was 2.21%, 44.11%, 4.63%, 53.02% and 67.12% in SP and 0.43%, 21.03%, 1.52%, 21.66% and 20.48% in NSP respectively (Figures 1B and 1C). Similarly, the percentage of HuH-7 cell positive for CD133, CD44, CD90, EpCAM and ABCG2 was 1.42%, 44.42%, 2.44%, 89.20% and 3.11% in SP and 0.88%, 23.77%, 1.40%, 81.81% and 1.83% in NSP respectively (Figures 1B and 1C). The results indicated that all of the five CSC markers were expressed much higher on the surface of SP compared with NSP cells in HCC cell lines. Additionally, sphere formation assay data showed SP cells could form much larger and more spheres than NSP cells after one-week culture in serum-free medium (Figure 1D), confirming that our sorted SP cells harbour CSC-like properties.
Figure 1. SP cell isolation and CSC-related properties analysis.
(A) PLC/PRF/5 and HuH-7 cells were incubated with 15 μg/ml Hoechst 33342 alone or in the presence of 50 μM verapamil and analysed by flow cytometry. (B) SP and NSP cells were immunostained with CSC-related marker CD90, CD133, CD44, EpCAM and ABCG2 and analysed by flow cytometry. (C) The relative CSC-related marker expressed on the surface of SP and NSP cells were showed by the histogram. (D) SP and NSP cells of PLC/PRF/5 and HuH-7 were cultured in serum-free medium for 2 weeks and observed under microscopes. Bar=200 μm.
miR-148b was significantly down-regulated in SP cells in HCC cell lines
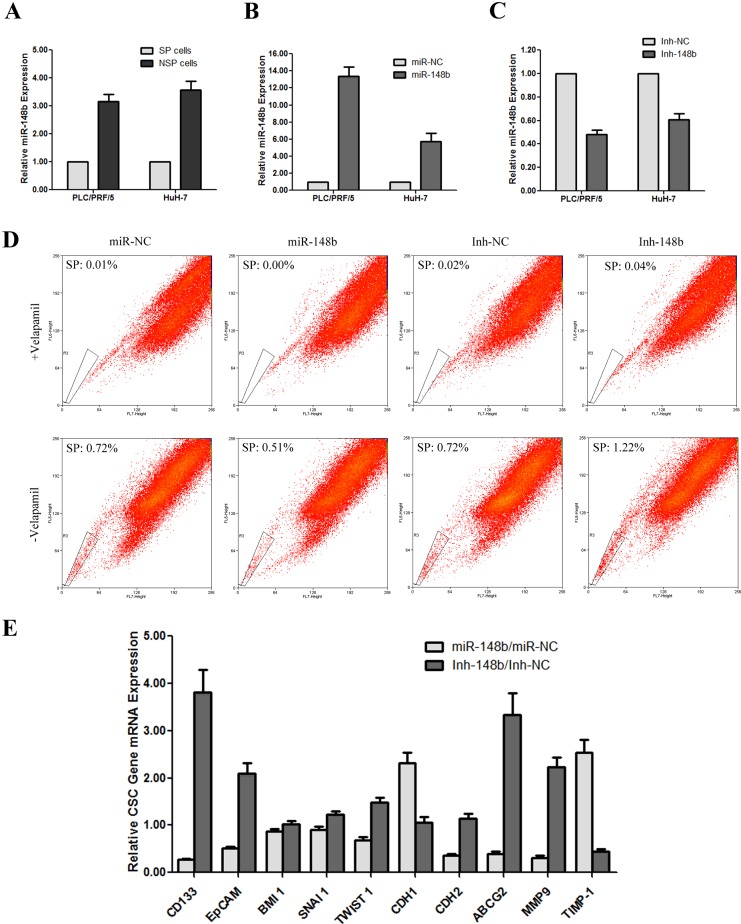
In previous study, we identified miR-148b as one of the 27 differentially expressed miRNAs between SP and NSP cells from PLC/PRF/5 [15]. In the present study, to re-verify the result in both PLC/PRF/5 and HuH-7 cell lines, we sorted SP and NSP cells separately by MoFlo XDP and analysed their miR-148b expression by qRT-PCR. As the results showed, the fold change of SP/NSP was −3.16±0.26 in PLC/PRF/5 and −3.58±0.30 in HuH-7 cells respectively (Figure 2A), similar with our previous study (SP/NSP=−3.14).
Figure 2. miR-148b was down-regulated in SP cells and affected the proportion of SP and the CSC-related gene expression in HCC cell lines.
(A) miR-148b was down-regulated in SP cells of both PLC/PRF/5 and HuH-7 cell lines. (B) qRT-PCR analysis of miR-148b overexpression cells (miR-148b) and its control cells (miR-NC) transfected by lentivirus in both PLC/PRF/5 and HuH-7 cell lines. (C) qRT-PCR analysis of miR-148b knockdown cells (Inh-148b) and its control cells (Inh-NC) transfected by lentivirus in both PLC/PRF/5 and HuH-7 cell lines. (D) miR-148b affected the proportion of SP cells in PLC/PRF/5. E. miR-148b affected the CSC-related gene expression in PLC/PRF/5.
miR-148b expression affects the percentage of SP cells and the CSC-related gene expression in PLC/PRF/5
In order to further investigate the role of miR-148b, we established miR-148b stable overexpressing and inhibiting cell lines with lentivirus transfection. As shown is Figure 2(B), miR-148b-Ubi-EGFP-MCS (miR-148b) transfection increased the expression of miR-148b by 13.36- and 5.7-fold in PLC/PRF/5 and HuH-7 respectively. Conversely, miR-148b-H1-MCS-CMV-EGFP transfection (Inh-148b) reduced the expression of miR-148b by 2.08- and 1.64-fold in PLC/PRF/5 and HuH-7 respectively (Figure 2C).
Through SP cells detection by flow cytometry, we found that the fraction of SP in PLC/PRF/5 was increased from 0.70±0.08% to 1.24±0.11% when miR-148b was knocked-down, whereas the percentage of SP cells was decreased from 0.72±0.06% to 0.50±0.03% when miR-148b was overexpressed (Figure 2D), indicating that the expression level of miR-148b was tightly related with SP population maintenance in HCC.
We further examined whether miR-148b could affect the expression of CSC-related genes by qRT-PCR. As shown in Figure 2(E), miR-148b inhibition lead to a significant expression change of CD133 (increased 3.81 times), EpCAM (increased 2.10 times), ABCG2 (increased 3.33 times), MMP9 (increased 2.22 times) and TIMP-1 (decreased 2.28 times). Unexpected, the level of BMI 1, SNAI 1, TWIST 1, CDH1 and CDH2 kept almost unchanged, compared with the control Inh-NC cells. On the other hand, miR-148b overexpression did not affect the expression of BMI 1, SNAI 1 and TWIST 1 at all, but showed expression change of the other genes, including CD133 (decreased 3.74 times), EpCAM (decreased 1.95 times) and ABCG2 (decreased 2.53 times), MMP9 (decreased 3.25 times), TIMP-1 (increased 2.54 times), CDH1 (increased 2.32 times) and CDH2 (decreased 2.78 times), relative to control miR-NC cells. These findings suggested that the expression of miR-148b might inhibit the CSC properties and metastasis ability of cells in HCC.
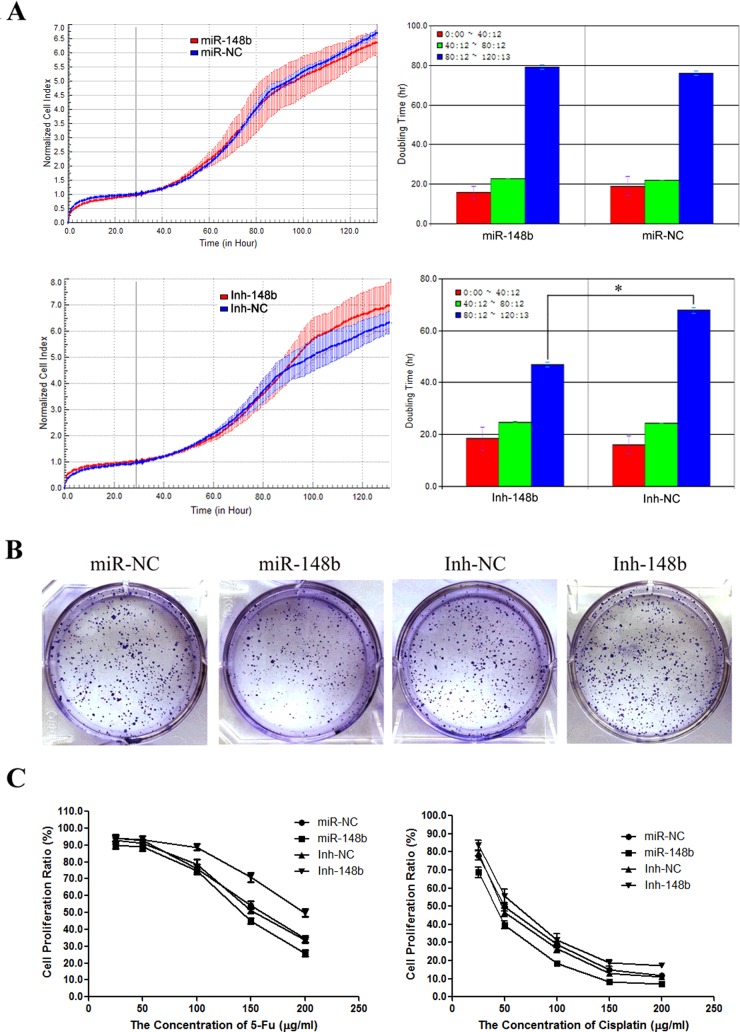
miR-148b inhibits cell proliferation and enhances chemosensitivity in PLC/PRF/5
Using real-time cellular analysis (RTCA), we found that the doubling time of Inh-148b cells from 80 to 120 h (46.93±1.01 h) was significantly shortened compared with that of control Inh-NC cells (67.80±1.08 h), whereas the doubling time of Inh-148b cells from 0 to 40 h and 40 to 80 h was not different from that of control Inh-NC cells in PLC/PRF/5 (18.39±4.45 compared with 16.04±3.28 and 24.85±0.13 compared with 24.43±0.03 respectively; Figure 3A). The data suggested that the knockdown of miR-148b could stimulate cells to keep growth even when culture environment changes to poor and unavailable. However, miR-148b overexpression seemed not to affect the cell proliferation as the doubling time of miR-148b and control miR-NC cells showed almost no difference during the whole culture time.
Figure 3. miR-148b inhibition stimulated cell proliferation and enhanced chemosensitivity.
(A) The detection of cell doubling time of miR-148b, miR-NC, Inh-148b and Inh-NC PLC/PRF/5 cells by RTCA. (B) Soft agar colony formation by miR-148b, miR-NC, Inh-148b and Inh-NC PLC/PRF/5 cells. (C) Chemosensitivity of miR-148b, miR-NC, Inh-148b and Inh-NC PLC/PRF/5 cells to 5-FU and cisplatin.
We further performed soft agar colony formation assay and observed that Inh-148b cells displayed more colonies compared with control Inh-NC cells; conversely, miR-148b cells displayed fewer colonies than miR-NC cells (Figure 3B). The finding indicated that the miR-148b blocking significantly stimulated cell proliferation.
To examine the effect of miR-148b on chemosensitivity of cells in HCC, we treated miR-148b, miR-NC, Inh-148b and Inh-NC PLC/PRF/5 cells with gradient concentrations of 5-Fu or cisplatin, which were two of the most commonly used traditional chemotherapy agents in clinic for HCC and tested the alternation of cell proliferation by WST-1 assay. As expected, the IC50 of Inh-148b was significantly increased under the treatment of 5-Fu (202.7 μg/ml compared with 154.8 μg/ml) or cisplatin (61.49 μg/ml compared with 50.60 μg/ml), whereas the IC50 of miR-148b was decreased under the treatment of 5-Fu (139.7 μg/ml compared with. 157.7 μg/ml) or cisplatin (39.57 μg/ml compared with 52.69 μg/ml; Figure 3C).
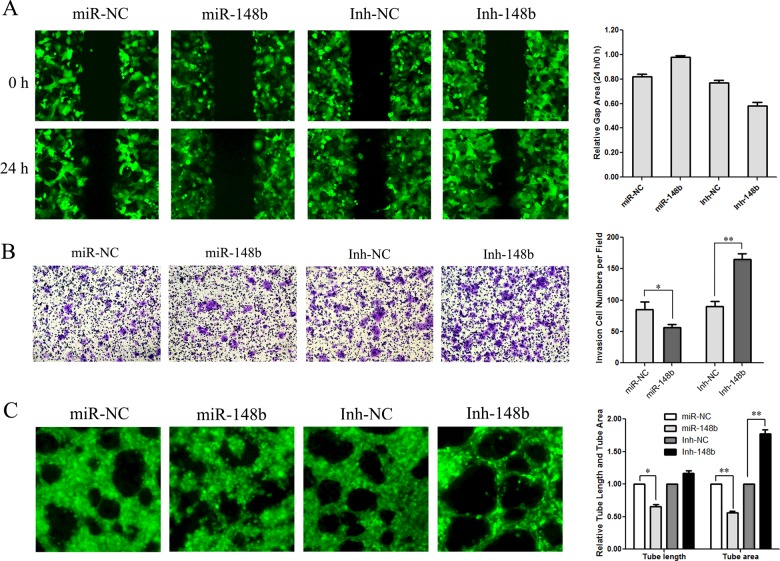
miR-148b inhibits migration, invasion and angiogenesis of PLC/PRF/5 cells
We further performed scratch wound healing assay and transwell assay to test the cell migration and invasion ability in miR-148b gain-of-function or loss-of-function cells. The scratch wound healing assay results showed that 24 h after the wound was created, the gap area of Inh-148b cells reduced to 58.1±2.9% whereas that of control Inh-NC cells only reduced to 76.8±2.3%; conversely, the gap area of miR-148b cells after 24 h almost had no change (Figure 4A), confirming that miR-148b expression abolished the cell migration ability. The transwell assay results also showed that compared with Inh-NC cells, Inh-148b raised the cell invasion ability by 1.84±0.11-fold, whereas miR-148b decreased the ability of invasion by 1.50±0.07-fold (Figure 4B), indicating that miR-148b could suppress metastasis in HCC. Moreover, the effect of miR-148b on the formation of capillary tubes in matrigel was also assessed. As the data showed miR-148b PLC/PRF/5 cells reduced the capillary tube length by 1.53±0.08-fold and area by 1.78±0.07-fold; in contrast, blocking miR-148b increased both the tube length by 1.16±0.04-fold and the tube area by 1.77±0.06-fold (Figure 4C), suggesting that miR-148b could inhibit the capillary-like structure formation. Collectively, our results indicated that miR-148b could suppress migration, invasion and angiogenesis in PLC/PRF/5.
Figure 4. miR-148b affected cell migration, invasion and angiogenesis in vitro.
(A) miR-148b affected scratch wound healing. (B) miR-148b affected cell invasion by transwell assay. (C) miR-148b affected the tube formation in matrigel.
miR-148b suppresses tumorigenicity of PLC/PRF/5 in vivo
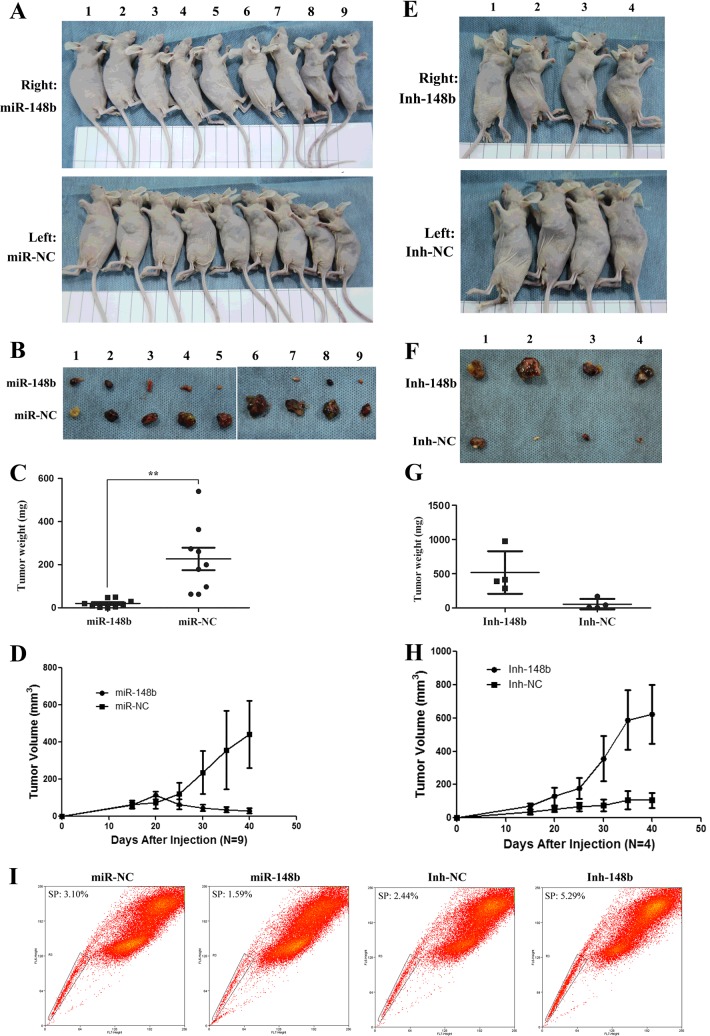
To further confirm the function of miR-148b in tumorigenicity, an in vivo tumour xenograft model was used. Ten nude mice with 6-week-old were injected with miR-148b (right side) and control miR-NC (left side) PLC/PRF/5 cells in both shoulders (Group 1) and other ten nude mice were injected with Inh-148b and control Inh-NC PLC/PRF/5 cells in both shoulders too (Group 2). After 6 weeks, nine mice were left in Group 1, whereas only four mice were left in Group 2; the rest died because of non-controllable factors. As shown in Figures 5(A)–5(D), the tumours formed by miR-148b cells injection were much smaller than those formed by miR-NC cells injection (20.1±18.7 mg compared with 226.7±155.6 mg (P<0.05), as well as the tumour growth rate of miR-148b cells was much slower than that of miR-NC after injection. On the other hand, the mean tumour weight of mice injected with Inh-148b cells was remarkably higher than that injected with control Inh-NC cells (518.4±310.7 mg compared with 56.1±76.9 mg; Figures 5E–5H) and the tumour growth rate of Inh-148b was much faster than that of Inh-NC after injection. The results strongly suggested that miR-148b could inhibit the tumorigenicity of PLC/PRF/5 in vivo. Moreover, through analysing the SP percentage of tumour formed by injection of miR-148b or Inh-148b PLC/PRF/5 cells, we found that the tumour formed by Inh-148b cells contained 5.29% SP cells, whereas the compared tumour formed by Inh-NC cells only contained 2.44% SP cells; conversely, miR-148b tumour only contained 1.59% SP cells, which was much less than miR-NC tumour contained SP cells (3.10%; Figure 5I), indicating that miR-148b could stimulate the SP cell formation in vivo.
Figure 5. miR-148b suppressed the tumorigenicity in vivo.
(A) Six-week-old male nude mice were injected by 5×106 control miR-NC or miR-148b PLC/PRF/5 cells in either shoulder (Group 1). Six weeks later, the mice were killed. (B) The tumours were showed. (C) The weight of tumours was analysed by statistics. (D) The growth of tumour volume after injection. (E) Six-week-old male nude mice were injected by 5×106 control Inh-NC or Inh-148b PLC/PRF/5 cells in either shoulder (Group 2). Six weeks later, the mice were killed. (F) The tumours were showed. (G) The weight of tumours was analysed by statistics. (H) The growth of tumour volume after injection. (I) The SP fraction percentage of tumour formed in mice was analysed by flow cytometry. The weight and volume of miR-NC and Inh-NC tumours were different partly because the sample sizes for the two nude mice groups were different (10 nude mice for Group 1; only four nude mice for Group 2 and the other six mice died because of too heavy tumour burden) and partly because the growth of the miR-NC or Inh-NC tumour was affected by the tumour of miR-148b or Inh-148b on the other side shoulder of the same mouse. Especially the huge tumours of Inh-148b competed and obtained much more nutrition and resources and lead the tumours of Inh-NC on the other side shoulders to be smaller than expectation.
miR-148b targets to NRP1 directly
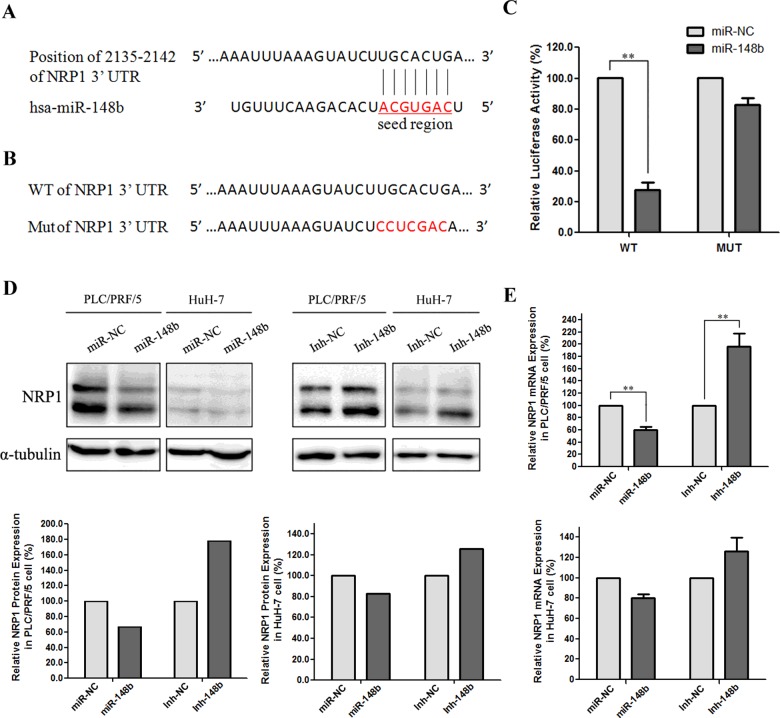
To explore the function of miR-148b, we used three computational algorithms, including TargetScan, miRNA and miRDB (a microRNA target predietion and functional annotation database), to identify the target genes of miR-148b. Among the predicted targets, we focus on gene NRP1 since it was reported to be involved in induction of epithelial-to-mesenchymal transition (EMT) and maintenance of CSC phenotype [25]. According to the prediction by the seed complementarity, a potential miR-148b-binding site was found at nt 2135–2142 of NRP1 3′-UTR. The sequence alignment of miR-148b and the 3′-UTR of NRP1 transcripts was shown in Figure 6(A). To verify the direct interaction between miR-148b and NRP1, we cloned 3′-UTR sequences of NRP1 that contain predicted target site (WT) and seed-region-mutated sequences (MUT) of miR-148b into the pmirGLO vector respectively (Figure 6B). The results showed that co-transfection of miR-148b lentivirus significantly decreased the firefly luciferase activity of the reporter with WT 3′-UTR of NRP1, but not that of the mutant reporter, which indicates that miR-148b could directly target the 3′-UTR of NRP1 (Figure 6C). Western blot and qRT-PCR analysis confirmed that miR-148b caused the decrease in NRP1 expression on the level of both mRNA and protein in PLC/PRF/5 and HuH-7 cells (Figures 6D and 6E).
Figure 6. NRP1 was the direct target of miR-148b.
(A) The interaction of miR-148b and the 3′-UTR of NRP1. (B) The mutation of miR-148b-binding site of NRP1 3′-UTR. (C) Luciferase reporter assays with co-transfection of WT or MUT 3′-UTR of NRP1 and miR-148b or miR-NC as indicated. Firefly luciferase activity was normalized by Renilla luciferase activity. (D) Western blot analysis of NRP1 protein level in miR-148b overexpressed or knocked-down cells of PLC/PRF/5 and HuH-7. E. qRT-PCR analysis of NRP1 mRNA expression in miR-148b overexpressed or knocked-down cells of PLC/PRF/5 and HuH-7.
NRP1 increases expression in SP cells of HCCs
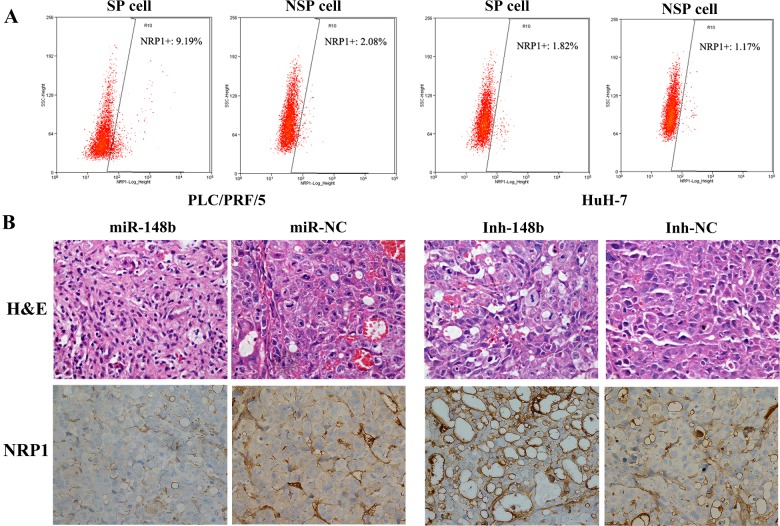
We further investigated the NRP1 expression in SP cells of PLC/PRF/5 and HuH-7 and found that the expression of NRP1 in SP cells was much higher than that in NSP cells of PLC/PRF/5 (9.29% compared with 2.08%) and of HuH-7(1.82% compared with 1.17%; Figure 7A), indicating that NRP1 played a vital role in SP cells of HCC. Moreover, through H&E staining and NRP1 immunohistochemistry, we found that the tumour formed by Inh-148b cells injection showed more tube formation and NRP1 overexpression, as well as excessive cell division and increased nucleo-cytoplasmic ratio, whereas the tumour formed by miR-148b cells injection showed the opposite effects (Figure 7B), indicating that miR-148b might regulate the tumorigenesis by inhibit the expression of NRP1 and further reduce the formation of SP cells with CSC-like properties.
Figure 7. NRP1 was specifically overexpressed in SP cells of PLC/PRF/5 and HuH-7 and under the regulation of miR-148b.
(A) NRP1 was specifically overexpressed in SP cells of PLC/PRF/5 and HuH-7. (B) H&E staining and anti-NRP1 immunohistochemistry of tumour formed in mice.
DISCUSSION
In previous study, we have detected miR-148b as one of the most down-regulated miRNAs in SP sub-population of PLC/PRF/5 cell line based on the miRNA expression profile screening [15]. In the present study, we further investigated the potential role of miR-148b in maintaining SP properties and regulating the progression of tumour initiation and development in HCC. Our data showed that miR-148b could inhibit the formation of SP cells, as well as affect cell proliferation, drug resistance, metastasis and angiogenisis by directly targeting to NRP1 in HCC. Additionally, in vivo experiment confirmed that miR-148b could significantly suppress the tumour growth, suggesting the potential application of miR-148b in future clinic therapy of HCC.
Accumulating evidences support that miR-148b acts as a tumour-suppressor and down-regulation of miR-148b is a frequent event in various tumour types, including liver, gastric, colorectal, pancreatic, breast and lung cancers [17–19,21–24]. Moreover, miR-148b is considered as a major co-ordinator of malignancy or an independent prognostic marker since its expression is closely associated with tumour invasion and progression in both breast cancer and liver cancer [19,23,24]. However, the possible role of miR-148b in HCSC has not been elucidated. In the present study, we re-verified that miR-148b was down-regulated in SP cells of both PLC/PRF/5 and HuH-7 cell lines and further found that the expression level of miR-148b could affect the fraction of SP cells significantly as blocking miR-148b increased SP percentage, whereas overexpressing miR-148b decreased SP percentage both in vitro and in vivo. These results suggest that miR-148b might serve as a potential suppressor in HCC through controlling the SP sub-population, which possess self-renewal and tumour initial ability. Through qRT-PCR, we observed that miR-148b inhibition could lead to the up-regulation of CD133, EpCAM, ABCG2 and MMP9 and the down-regulation of TIMP-1, whereas overexpressing miR-148b in cells could lead to the almost opposite results. Among these genes, CD133 and EpCAM are proved to be the most important markers for CSC properties in HCC, ABCG2 is especially overexpressed in SP cells as one of the ABC [1]. In addition, MMP9 and TIMP-1 are thought to be involved in tumour metastasis [26]. Our data supported that miR-148b might not only play a critical role in maintenance of SP cells with CSC properties, but also associate with tumour metastasis. By coincident, it was reported that the proportion of SP cells was positively related to metastatic potential of parent cells [27]. However, to our surprise, knockdown of miR-148b resulted in no remarkable changes in the expression of EMT-related genes, such as BMI 1, SNAI 1, TWIST 1, CDH 1 and CDH 2. Although it was reported that BMI 1 was up-regulated in SP cells in HCC [27], we proposed that miR-148b might regulate the maintenance of SP cells in HCC through the distinguished pathway from BMI 1.
Given that CSCs often display higher tolerance to poor environment and chemotherapeutic agents, we tested the influence of miR-148b on cell proliferation and drug resistance in the present study and confirmed that miR-148b blocking could keep the cell growth rate and increase resistance to cisplatin and 5-FU, the traditional chemotherapeutic drugs used as first-line therapy for HCC. In consonance with our results, Song et al. [16,17] proved that miR-148b could inhibit cell proliferation of both gastric cancer and colorectal cancer and Wu et al. [20] revealed that miR-148b could enhance the radiosensitivity of non-Hodgkin's lymphoma. Since cell migration and invasion was one of the most important characteristics of CSC, we further investigation confirmed that miR-148b inhibition strongly stimulated cell migration and invasion, whereas miR-148b overexpression lead cell to reduce the metastasis ability. Meanwhile, we also found that miR-148b inhibition enhance the angiogenesis through tube formation assay. In the previous study, miR-148b has been found to promote tumour metastasis in pancreatic cancer and breast cancer [19,21]. However, it was the first time to discover the function of miR-148b in metastasis and angiogenesis in HCC.
Several genes, such as cholecystokinin B receptor (CCKBR), AMP-activated protein kinase α1 (AMPK α1), carcinoembryonic antigen (CEA) and DNA methyl transferase 3B (DNMT3B), are validated as miR-148b target genes and function in regulation of tumour cell growth, tumorigenesis, metastasis and DNA hypomethylation in previous reviews [17,21,22,28]. In our study, NRP1 was confirmed to be a novel direct target of miR-148b for the first time. NRP1 is a transmembrane co-receptor for VEGF, TGF-β1, HGF, PDGF-BB and SEMA3 family members. Although it was not essential for receptor signalling, it could enhance response of tyrosine kinase receptors to their ligands [25]. NRP1 was reported to be overexpressed in several human tumour types, including HCC and responsible for tumour initiation and cancer progression, especially for angiogenesis [29,30]. Our data showed that NRP1 is preferentially expressed in SP cells of PLC/PRF/5 and HuH-7, indicating that NRP1 plays a critical role in cancer stem-like cells in HCC. In fact, a link between NRP1 and CSCs has been established in several other cancers, such as breast cancer, skin tumour and glioblastoma multiforme [31–33]. Both in skin tumour and in glioblastoma multiforme, NRP1 was proved to promote the cell self-renewal, stemness and tumorigenicity through VEGF-VEGFR2-NRP1 axis; whereas in breast cancer, nuclear factor of kappa light polypeptide gene enhancer in B cells (NF-kappa B) pathway activation is also involved [31–33]. Although the contribution of NRP1 to regulating the CSC properties of SP cells with needs further investigation, it appears to be a good target for anti-CSC therapy in HCC. Both anti-NRP1 monoclonal antibodies (mAbs) and cell-penetrating peptides of NRP1 have been tested in cancer treatments and shown efficacy in certain cancers [34–36]. Based on our data, raising the miR-148b level of patients might be another good therapeutic strategy to treat HCC since the in vivo experiment confirmed that miR-148b could strongly suppress tumorigenicity, inhibit the tissue NRP1 expression and block the angiogenesis.
In conclusion, our data suggest that miR-148b is down-regulated in SP of HCC cell lines PLC/PRF/5 and HuH-7 and affects the CSC properties, including cell proliferation, drug-resistance, metastasis and tumorigenesis. miR-148b might act as a CSC suppressor by directly targeting to NRP1 in HCC. Enforced expression of miR-148b by nanoparticle conjugates or other methods might supply an effective tool to conquer HCC [37].
Acknowledgments
We thank Dr Wansong Lin (Fujian Provincial Cancer Hospital, P.R. China) for the pmirGLO vector as a generous gift. We also thank Professor Chuanmao Zhang (College of Life Sciences, Peking University, P.R. China) for the kind suggestion and support.
Abbreviations
- ABC
ATP-binding cassette proteins
- ABCG2
ATP-binding cassette, sub-family G (WHITE), member 2
- AMPK α1
AMP-activated protein kinase α1
- BMI 1
BMI1 proto-oncogene
- CCKBR
cholecystokinin B receptor
- CD
cluster of differentiation
- CDH
cadherin
- CDX2
caudal type homeobox 2
- CEA
carcinoembryonic antigen
- CMV
cytomegalovirus
- CSC
cancer stem cell
- DMEM
Dulbecco’s modified Eagle’s medium
- DNMT3B
DNA methyltransferase 3B
- EMT
epithelial-to-mesenchymal transition
- EpCAM
epithelial cell adhesion molecule
- GATA6
GATA binding protein 6
- HCC
hepatocellular carcinoma
- HCSC
hepatic cancer stem cell
- H&E
hematoxylin and eosin
- HGF
hepatocyte growth factor
- mAbs
monoclonal antibodies
- MCS
multiple cloning sites
- MMP9
matrix metallopeptidase 9
- MUT
mutant-type
- NF-kappa B
nuclear factor of kappa light polypeptide gene enhancer in B cells
- NLK
nemo-like kinase
- NRP1
Neuropilin-1
- NSP
non-SP
- PDCD4
programmed cell death 4
- PDGF-BB
the platelet-derived growth factor homodimer
- PTEN
phosphatase and tensin homolog
- qRT
quantitative real-time
- RECK
reversion-inducing-cysteine-rich protein with kazal motifs
- RTCA
real-time cellular analysis
- SEMA3
Semaphorin-3
- SNAI 1
snail family zinc finger 1
- SP
side population
- TGF-β1
transforming growth factor-β1
- TIMP-1
tissue inhibitor of metalloproteinase-1
- TWIST 1
twist-related protein 1
- VEGF
vascular endothelial growth factor
- WNT1
wingless-type MMTV integration site family, member 1
- WT
wild-type
AUTHOR CONTRIBUTION
Qinying Liu conceived and designed the project, performed experiments, wrote the paper and provided financial support. Yangmei Xu conceived and designed the project, performed experiments and wrote the paper. Shenghong Wei, Wei Gao, Tong Zhou performed experiments and analysed data. Li Chen, Zhen Wang and Mingang Ying designed the project and analysed data. Qiuhong Zheng supervised the project and provided financial support.
FUNDING
This work was supported by the Chinese National Natural Science Fund [grant numbers 31201034, 81201969 and 21102015]; the Joint Project of Ministry of Sanitation and Ministry of Education in Fujian Province [grant number WKJ-FJ-14]; the Natural Science Foundation of Fujian Province [grant numbers 2012J05138 and 2014J01301]; and the Key Project of Department of Science and Technology in Fujian Province [grant number 2012Y0017].
References
- 1.Pang R.W., Poon R.T. Cancer stem cell as a potential therapeutic target in hepatocellular carcinoma. Curr. Cancer Drug Targets. 2012;12:1081–1094. doi: 10.2174/156800912803987995. [DOI] [PubMed] [Google Scholar]
- 2.Leal J.A., Lleonart M.E. MicroRNAs and cancer stem cells: therapeutic approaches and future perspectives. Cancer Lett. 2012;338:174–183. doi: 10.1016/j.canlet.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 3.Qi W., Liang W., Jiang H., Miuyee Waye M. The function of miRNA in hepatic cancer stem cell. Biomed. Res. Int. 2014;2013:358902. doi: 10.1155/2013/358902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zimmerman A.L., Wu S. MicroRNAs, cancer and cancer stem cells. Cancer Lett. 2010;300:10–19. doi: 10.1016/j.canlet.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiba T., Kita K., Zheng Y.W., Yokosuka O., Saisho H., Iwama A., Nakauchi H., Taniguchi H. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. Hepatology. 2006;44:240–251. doi: 10.1002/hep.21227. [DOI] [PubMed] [Google Scholar]
- 6.Ji J., Yamashita T., Wang X.W. Wnt/beta-catenin signaling activates microRNA-181 expression in hepatocellular carcinoma. Cell Biosci. 2011;1:4. doi: 10.1186/2045-3701-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji J., Yamashita T., Budhu A., Forgues M., Jia H.L., Li C., Deng C., Wauthier E., Reid L.M., Ye Q.H., et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472–480. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou L., Yang Z.X., Song W.J., Li Q.J., Yang F., Wang D.S., Zhang N., Dou K.F. MicroRNA-21 regulates the migration and invasion of a stem-like population in hepatocellular carcinoma. Int. J. Oncol. 2013;43:661–669. doi: 10.3892/ijo.2013.1965. [DOI] [PubMed] [Google Scholar]
- 9.Chai S., Tong M., Ng K.Y., Kwan P.S., Chan Y.P., Fung T.M., Lee T.K., Wong N., Xie D., Yuan Y.F., et al. Regulatory role of miR-142-3p on the functional hepatic cancer stem cell marker CD133. Oncotarget. 2014;5:5725–5735. doi: 10.18632/oncotarget.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia Y., Liu H., Zhuang Q., Xu S., Yang Z., Li J., Lou J., Zhang W. Tumorigenicity of cancer stem-like cells derived from hepatocarcinoma is regulated by microRNA-145. Oncol. Rep. 2012;27:1865–1872. doi: 10.3892/or.2012.1701. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J., Luo N., Luo Y., Peng Z., Zhang T., Li S. microRNA-150 inhibits human CD133-positive liver cancer stem cells through negative regulation of the transcription factor c-Myb. Int. J. Oncol. 2011;40:747–756. doi: 10.3892/ijo.2011.1242. [DOI] [PubMed] [Google Scholar]
- 12.Tang J., Tao Z.H., Wen D., Wan J.L., Liu D.L., Zhang S., Cui J.F., Sun H.C., Wang L., Zhou J., et al. MiR-612 suppresses the stemness of liver cancer via Wnt/beta-catenin signaling. Biochem. Biophys. Res. Commun. 2014;447:210–215. doi: 10.1016/j.bbrc.2014.03.135. [DOI] [PubMed] [Google Scholar]
- 13.Liu J., Ruan B., You N., Huang Q., Liu W., Dang Z., Xu W., Zhou T., Ji R., Cao Y., et al. Downregulation of miR-200a induces EMT phenotypes and CSC-like signatures through targeting the beta-catenin pathway in hepatic oval cells. PLoS One. 2013;8:e79409. doi: 10.1371/journal.pone.0079409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oishi N., Kumar M.R., Roessler S., Ji J., Forgues M., Budhu A., Zhao X., Andersen J.B., Ye Q.H., Jia H.L., et al. Transcriptomic profiling reveals hepatic stem-like gene signatures and interplay of miR-200c and epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma. Hepatology. 2012;56:1792–1803. doi: 10.1002/hep.25890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y., Xie Y., Wang X., Chen X., Liu Q., Ying M., Zheng Q. Identification of cancer stem cells from hepatocellular carcinoma cell lines and their related microRNAs. Oncol. Rep. 2013;30:2056–2062. doi: 10.3892/or.2013.2703. [DOI] [PubMed] [Google Scholar]
- 16.Liu W.H., Tao K.S., You N., Liu Z.C., Zhang H.T., Dou K.F. Differences in the properties and mirna expression profiles between side populations from hepatic cancer cells and normal liver cells. PLoS One. 2011;6:e23311. doi: 10.1371/journal.pone.0023311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song Y., Xu Y., Wang Z., Chen Y., Yue Z., Gao P., Xing C., Xu H. MicroRNA-148b suppresses cell growth by targeting cholecystokinin-2 receptor in colorectal cancer. Int. J. Cancer. 2011;131:1042–1051. doi: 10.1002/ijc.26485. [DOI] [PubMed] [Google Scholar]
- 18.Song Y.X., Yue Z.Y., Wang Z.N., Xu Y.Y., Luo Y., Xu H.M., Zhang X., Jiang L., Xing C.Z., Zhang Y. MicroRNA-148b is frequently down-regulated in gastric cancer and acts as a tumor suppressor by inhibiting cell proliferation. Mol. Cancer. 2011;10:1. doi: 10.1186/1476-4598-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cimino D., De Pitta C., Orso F., Zampini M., Casara S., Penna E., Quaglino E., Forni M., Damasco C., Pinatel E., et al. miR148b is a major coordinator of breast cancer progression in a relapse-associated microRNA signature by targeting ITGA5, ROCK1, PIK3CA, NRAS, and CSF1. FASEB J. 2012;27:1223–1235. doi: 10.1096/fj.12-214692. [DOI] [PubMed] [Google Scholar]
- 20.Wu Y., Liu G.L., Liu S.H., Wang C.X., Xu Y.L., Ying Y., Mao P. MicroRNA-148b enhances the radiosensitivity of non-Hodgkin's Lymphoma cells by promoting radiation-induced apoptosis. J. Radiat. Res. 2012;53:516–525. doi: 10.1093/jrr/rrs002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao G., Zhang J.G., Liu Y., Qin Q., Wang B., Tian K., Liu L., Li X., Niu Y., Deng S.C., et al. miR-148b functions as a tumor suppressor in pancreatic cancer by targeting AMPKalpha1. Mol. Cancer Ther. 2012;12:83–93. doi: 10.1158/1535-7163.MCT-12-0534-T. [DOI] [PubMed] [Google Scholar]
- 22.Liu G.L., Liu X., Lv X.B., Wang X.P., Fang X.S., Sang Y. miR-148b functions as a tumor suppressor in non-small cell lung cancer by targeting carcinoembryonic antigen (CEA) Int. J. Clin. Exp. Med. 2014;7:1990–1999. [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z., Zheng W., Hai J. MicroRNA-148b expression is decreased in hepatocellular carcinoma and associated with prognosis. Med. Oncol. 2014;31:984. doi: 10.1007/s12032-014-0984-6. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J.G., Shi Y., Hong D.F., Song M., Huang D., Wang C.Y., Zhao G. MiR-148b suppresses cell proliferation and invasion in hepatocellular carcinoma by targeting WNT1/beta-catenin pathway. Sci. Rep. 2015;5:8087. doi: 10.1038/srep08087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prud’homme G.J., Glinka Y. Neuropilins are multifunctional coreceptors involved in tumor initiation, growth, metastasis and immunity. Oncotarget. 2012;3:921–939. doi: 10.18632/oncotarget.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKenna G.J., Chen Y., Smith R.M., Meneghetti A., Ong C., McMaster R., Scudamore C.H., Chung S.W. A role for matrix metalloproteinases and tumor host interaction in hepatocellular carcinomas. Am. J. Surg. 2002;183:588–594. doi: 10.1016/S0002-9610(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 27.Chiba T., Miyagi S., Saraya A., Aoki R., Seki A., Morita Y., Yonemitsu Y., Yokosuka O., Taniguchi H., Nakauchi H., Iwama A. The polycomb gene product BMI1 contributes to the maintenance of tumor-initiating side population cells in hepatocellular carcinoma. Cancer Res. 2008;68:7742–7749. doi: 10.1158/0008-5472.CAN-07-5882. [DOI] [PubMed] [Google Scholar]
- 28.Sandhu R., Rivenbark A.G., Coleman W.B. Loss of post-transcriptional regulation of DNMT3b by microRNAs: a possible molecular mechanism for the hypermethylation defect observed in a subset of breast cancer cell lines. Int. J. Oncol. 2012;41:721–732. doi: 10.3892/ijo.2012.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berge M., Allanic D., Bonnin P., de Montrion C., Richard J., Suc M., Boivin J.F., Contreres J.O., Lockhart B.P., Pocard M., et al. Neuropilin-1 is upregulated in hepatocellular carcinoma and contributes to tumour growth and vascular remodelling. J. Hepatol. 2011;55:866–875. doi: 10.1016/j.jhep.2011.01.033. [DOI] [PubMed] [Google Scholar]
- 30.Chu W., Song X., Yang X., Ma L., Zhu J., He M., Wang Z., Wu Y. Neuropilin-1 promotes epithelial-to-mesenchymal transition by stimulating nuclear factor-kappa B and is associated with poor prognosis in human oral squamous cell carcinoma. PLoS One. 2014;9:e101931. doi: 10.1371/journal.pone.0101931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beck B., Driessens G., Goossens S., Youssef K.K., Kuchnio A., Caauwe A., Sotiropoulou P.A., Loges S., Lapouge G., Candi A., et al. A vascular niche and a VEGF-Nrp1 loop regulate the initiation and stemness of skin tumours. Nature. 2011;478:399–403. doi: 10.1038/nature10525. [DOI] [PubMed] [Google Scholar]
- 32.Glinka Y., Mohammed N., Subramaniam V., Jothy S., Prud’homme G.J. Neuropilin-1 is expressed by breast cancer stem-like cells and is linked to NF-kappaB activation and tumor sphere formation. Biochem. Biophys. Res. Commun. 2012;425:775–780. doi: 10.1016/j.bbrc.2012.07.151. [DOI] [PubMed] [Google Scholar]
- 33.Hamerlik P., Lathia J.D., Rasmussen R., Wu Q., Bartkova J., Lee M., Moudry P., Bartek J., Jr, Fischer W., Lukas J., et al. Autocrine VEGF-VEGFR2-Neuropilin-1 signaling promotes glioma stem-like cell viability and tumor growth. J. Exp. Med. 2012;209:507–520. doi: 10.1084/jem.20111424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang W.C., Dennis M.S., Stawicki S., Chanthery Y., Pan Q., Chen Y., Eigenbrot C., Yin J., Koch A.W., Wu X., et al. Function blocking antibodies to neuropilin-1 generated from a designed human synthetic antibody phage library. J. Mol. Biol. 2007;366:815–829. doi: 10.1016/j.jmb.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 35.Teesalu T., Sugahara K.N., Kotamraju V.R., Ruoslahti E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc. Natl. Acad. Sci. U.S.A. 2009;106:16157–16162. doi: 10.1073/pnas.0908201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth L., Agemy L., Kotamraju V.R., Braun G., Teesalu T., Sugahara K.N., Hamzah J., Ruoslahti E. Transtumoral targeting enabled by a novel neuropilin-binding peptide. Oncogene. 2011;31:3754–3763. doi: 10.1038/onc.2011.537. [DOI] [PubMed] [Google Scholar]
- 37.Qureshi A.T., Monroe W.T., Dasa V., Gimble J.M., Hayes D.J. miR-148b-nanoparticle conjugates for light mediated osteogenesis of human adipose stromal/stem cells. Biomaterials. 2013;34:7799–7810. doi: 10.1016/j.biomaterials.2013.07.004. [DOI] [PubMed] [Google Scholar]