Methylseleninic acid (MSA), as a potent second-generation selenium compound, could activate KLF4/miR-200a/Keap1/Nrf2 pathway in oesophageal squamous cell carcinoma cells.
Keywords: kelch-like ECH-associated protein 1 (Keap1), Krüpple-like factor 4 (KLF4), micro-ribonucleic acid-200a (miR-200a), methylseleninic acid (MSA), nuclear factor E2-related factor 2 (Nrf2), oesophageal squamous cell carcinoma (ESCC)
Abstract
Oesophageal squamous cell carcinoma (ESCC) occurs at a very high rates in certain regions of China. There are increasing evidences demonstrating that selenium could act as a potential anti-oesophageal cancer agent, but the precise mechanisms involved are still not completely understood. Methylseleninic acid (MSA), as a potent second-generation selenium compound, is a promising chemopreventive agent. Previous studies demonstrated that the kelch-like ECH-associated protein 1 (Keap1)/nuclear factor E2-related factor 2 (Nrf2) system plays a critical role in cancer prevention, but little is known about its association with MSA in ESCC cells. In the present study, we observed that MSA treatment significantly down-regulated Keap1, induced nuclear accumulation of Nrf2 and enhance the antioxidant response element (ARE) promoter activity in ESCC cells. MSA could also significantly induce miR-200a expression and inhibit Keap1 directly. Antagomir-200a could attenuate MSA treatment-induced Keap1 down-regulation in ESCC cells. Moreover, MSA-induced miR-200a expression was dependent on the mediation of Krüpple-like factor 4 (KLF4). These results reaffirm the potential role of MSA as a chemopreventive agent via the regulation of KLF4/miR-200a/Keap1/Nrf2 axis in ESCC cells.
INTRODUCTION
Selenium is an essential trace element with a lot of physiological functions in humans and animals. Many studies in animals and humans clinical trials have indicated that selenium is a promising chemopreventive agent for several major types of cancer, including oesophageal squamous cell carcinoma (ESCC) [1–4]. For instance, supplementation with selenium-containing compounds was associated with a significantly lower cancer mortality rate [4]. Selenomethionine had a protective effect against mild oesophageal squamous dysplasia at baseline [5]. However, the exact mechanism of selenium in the prevention of oesophageal carcinogenesis is still unclear.
Recent studies have shown that certain chemopreventive agents could active nuclear factor E2-related factor 2 (Nrf2) either by oxidative or covalent modification of its inhibitor kelch-like ECH-associated protein 1 (Keap1) or by phosphorylation of Nrf2 [6–8]. Nrf2, a redox sensitive transcription factor, is a master regulator of intracellular antioxidants and phase II detoxification enzymes by the transcriptional activation of many antioxidant response element (ARE)-containing genes [9,10]. Under homoeostatic conditions, Nrf2 is generally localized in the cytoplasm, where it is bound to Keap1. Once under oxidative stress or chemopreventive compounds, Keap1 acts as a molecular sensor and undergoes chemical modifications in a series of reactive cysteine residues, allowing the release of Nrf2, which escapes from degradation and translocates to the nucleus [11]. Previous study indicated that the Keap1/Nrf2 system regulates an important defensive mechanism against upper aerodigestive tract carcinogenesis, including cancers of the tongue and the oesophagus [12]. And, some selenium compounds have been shown to increase Nrf2 in prostate cancer [13]. The latest study reported that methylseleninic acid (MSA) treatment could up-regulate Nrf2 mRNA level in prostate tumour tissues [14]. However, the relationship between MSA and Nrf2 activation in ESCC has not been elucidated.
miRNAs as a type of endogenous non-coding RNA, mediate post-transcriptional gene regulation and participate in nearly all biological processes [15]. So far, a regulatory role for miRNAs in controlling Keap1 gene expression has been characterized in several studies [16–18]. Eades et al. [16] showed that miR-200a could target Keap1 directly and histone deacetylase (HDAC) inhibitor SAHA (suberanilohydroxamic acid, vorinostat) could induce re-expression of miR-200a in breast cancer cell lines. Furthermore, our previous study demonstrated that MSA could inhibit HDAC activity in ESCC cells [19]. All of these prompted us to hypothesize that MSA might regulate miR-200a to modulate Keap1/Nrf2 pathway in ESCC cells. To test this issue, we detected the expression level of miR-200a in ESCC cells with or without MSA treatment, respectively. We found that MSA treatment resulted in an increase in miR-200a expression and activation of Keap1/Nrf2 pathway. We also found that MSA treatment significantly increased Krüpple-like factor 4 (KLF4) binding to the miR-200a promoter region in ESCC cells.
MATERIAL AND METHODS
Cell culture, RNA isolation and quantitative real-time PCR
KYSE150, KYSE180, KYSE410 and KYSE510 were cultured in RPMI-1640 medium supplemented by 10% FBS at 5% CO2. Total RNA was isolated from cultured cells using TRIzol Reagent (Invitrogen) and reverse-transcribed to cDNA with M-MLV Reverse Transcriptase (Promega). Real-time PCR was performed on the StepOne Plus Real-Time PCR System (Applied Biosystems) with Power SYBR Green PCR Master Mix (Applied Biosystems), according to the manufacturer's protocol. The sequences of the PCR primers that were used to detect KLF4 and β-actin were reported previously [19].
Plasmids, transfection and reagents
pGL3-ARE and pcDNA3-HA-Nrf2 were generous gifts from Professor Xiaoming Yang. The 3′-UTR of Keap1 was amplified using the following primers: 5′-TCATACTAGTGGCACTTTTGTTTCTTGGGC-3′ and 5′-GCATTAAGCTTCAGGGTGAAAGACACTAG-3′ and cloned into pMiR-Report vector (Ambion) digested with HindIII and Spe I. We also generated three bases mutation in the predicated target site for miR-200a by using a QuickChange site-specific mutagenesis kit (Stratagene). All constructs were sequenced in Sangon Company. Transfection of plasmids was performed in 70%–80% confluent cells using Lipofectamine 2000 Reagent (Invitrogen) according to manufacturer's protocol.
MSA was purchased from Sigma–Aldrich (Sigma–Aldrich Inc.). Pre-miR miR-200a precursor and Pre-miR negative control were purchased from Ambion. Antagomir-200a was synthesized from Ribobio. KLF4 siRNA and scramble control were purchased from OriGene (OriGene Technologies). Transfections of Pre-miR miR-200a precursor, Pre-miR negative control, KLF4 siRNA and scramble control were performed by using siPORT NeoFX Transfection Agent (Life Technologies) according to manufacturer's protocol.
miRNA-specific quantitative real-time RT-PCR
For miRNA analysis from cultured cells, miRNA was isolated using a mirVana RNA isolation kit (Ambion). Reverse transcription and real-time PCR were performed as described [20] by using miRNA-specific quantitative real-time PCR (Applied Biosystems). The small RNA U6 was used as an internal control for normalization. Real-time PCR was performed using a StepOne Plus Detection System and fold changes in gene expression were calculated using the 2−△△Ct method [21]. The mean miRNA level from three real-time quantitative PCR experiments was calculated for each case.
Western blot analysis
Cells were harvested at indicated time points and lysed in RIPA buffer (Sigma). Western blot analysis was performed with the use of conventional protocols as described previously [22]. Nuclear and cytoplasmic proteins were extracted in accordance with the manufacturer's instructions (Pierce Biotechnology). The antibodies and dilutions used included anti-β-actin (AC-15; 1:2000; Sigma), anti-Keap1 (D1G10; 1:1000; Cell Signaling Technology), anti-Nrf2 (D1Z9C; 1:1000; Cell Signaling Technology), anti-LaminB (M-20; 1:1000; Santa Cruz), anti-GFP (A00185.01; 1:1000; Santa Cruz). After extensively washed, the membranes were incubated with anti-mouse or anti-rabbit IgG-horseradish peroxidase conjugate antibody (Zhongshan Company) for 1 h at room temperature and developed with a Luminol chemiluminescence detection kit (Santa Cruz). Membranes were reprobed for β-actin antibodies for normalization and accurate quantification. Protein expression level was quantified by using a Gel EDAS 293 analysis system (Cold Spring USA Corporation) and Gel-Pro Analyzer 3.1 software (Media Cybernetics).
Reporter assay
To measure the transcriptional activity of Nrf2, reporter assays were performed using the ARE promoter reporter construct. Cells were transfected in 24-well plates with pGL3-ARE and/or pcDNA3-HA-Nrf2. The total amount of transfected DNA was kept constant by adding pCL3-basic and pcDNA3-HA plasmids. Transfection efficiencies were estimated using co-transfected pEGFP-C1 (20 ng). The protein level of GFP was detected by western blotting. After transfection, cells were treated with or without MSA for 48 h. Then, Firefly luciferase activity was determined using the Dual-luciferase reporter assay system (Promega). All results were expressed as means ± S.D. for independent triplicate cultures.
To demonstrate the miR-200a directly regulates Keap1 expression by binding to its 3′-UTR, reporter assays were performed using Keap1 3′-UTR-wt and Keap1 3′-UTR-mut. Cells in 24-well plates were transfected with the indicated plasmids (300 ng) and the internal control plasmid pRL-SV40 (2 ng). After transfection, cells were treated with or without MSA for 24 h. Then, luciferase activity was determined as mentioned above. All results were expressed as means ± S.D. for independent triplicate cultures.
ChIP
Using Chromatin immunoprecipitation Kit & Shearing Kit (Active Motif), ten millions of KYSE150 cells were cross-linked and lysed and chromatin was sheared to 200–700 bp fragments. Sheared chromatin–DNA mixture was incubated with 4 μg of KLF4 (H-180, Santa Cruz), negative control IgG antibody per reaction overnight at 4°C. Cross-links were reversed and protein was removed by digestion with proteinase K. Purified DNA was used for RT-PCR. Using the following primers: miR-200a promoter (244-bp,–1048—-804) forward: 5′-GCTCACCCTTGCAGGTCTCC-3′ and reverse: 5′CCCGAAACCCAGCCGCATC-3′.
Statistical analysis
SPSS for Windows (SPSS Inc.) was used for statistical analysis. A Student's two-tailed non-paired ttest was used to determine significant differences between treatment and control values in all the experiments. Values of P<0.05 were considered statistically significant.
RESULTS
MSA could activate Keap1/Nrf2 pathway in ESCC cells
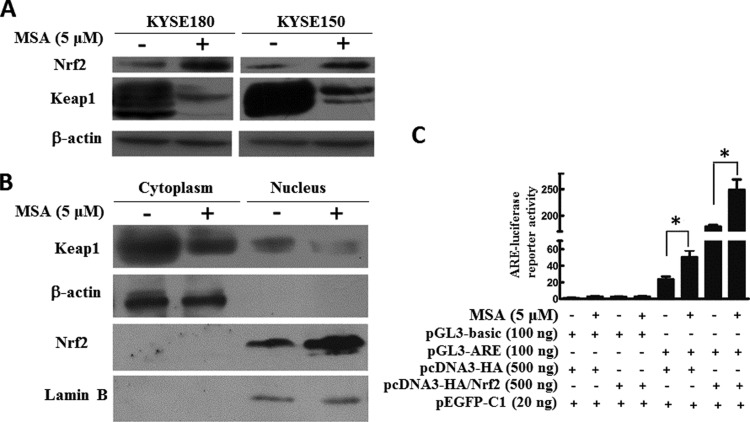
We firstly examined the impact of MSA on endogenous Keap1 expression in ESCC cell lines. In both KYSE180 and KYSE150, MSA treatment significantly down-regulated Keap1 and up-regulated Nrf2 at protein level compared with the control cells without MSA treatment respectively (Figure 1A). We further examined Nrf2 and Keap1 expression in cytoplasm and nuclear extracts with western blotting. Upon MSA treatment, we found an elevated expression level of Nrf2 located in the nucleus and a decrease expression level of Keap1 both in the cytoplasm and in the nucleus, in comparison with the nuclear protein loading control (lamin B) and the cytoplasm loading control (β-actin; Figure 1B). Meanwhile, the luciferase reporter ARE had been used to characterize Nrf2 transcriptional activity in KYSE150 cells with or without MSA treatment. As shown in Figure 1(C), co-transfection with Nrf2 could enhance the ARE promoter activity much higher with MSA treatment than without.
Figure 1. MSA activated Keap1/Nrf2 pathway in ESCC cells.
(A) KYSE180 and KYSE150 were treated with MSA (5 μM) for 24 h. Then, total cell lysates were prepared. Western blotting was performed to examine Keap1 and Nrf2. β-Actin was used as a loading control. (B) KYSE150 cells were treated with MSA (5 μM) for 24 h. Then, protein lysates were prepared. Keap1 and Nrf2 were detected in the cytoplasmic and nuclear extracts by western blot. β-Actin was used as a loading control of cytoplasmic protein and lamin B was shown as a loading control of nuclear protein. (C) KYSE150 cells were transfected with 100 ng of pGL3-ARE or pGL3-basic and 500 ng of pcDNA3-HA or pcDNA3-HA/Nrf2 plasmids with or without MSA treatment respectively. The pEGFP-C1 (20 ng) plasmid was co-transfected to normalize transfection efficiency. The luciferase activities were measured 48 h after transfection. Means ± S.D., n=3, *P<0.05.
In all, these findings demonstrated that up-regulation of Nrf2 in the nucleus caused by MSA plays its transcriptional role in ESCC cells.
MSA activates Keap1/Nrf2 pathway via up-regulating miR-200a
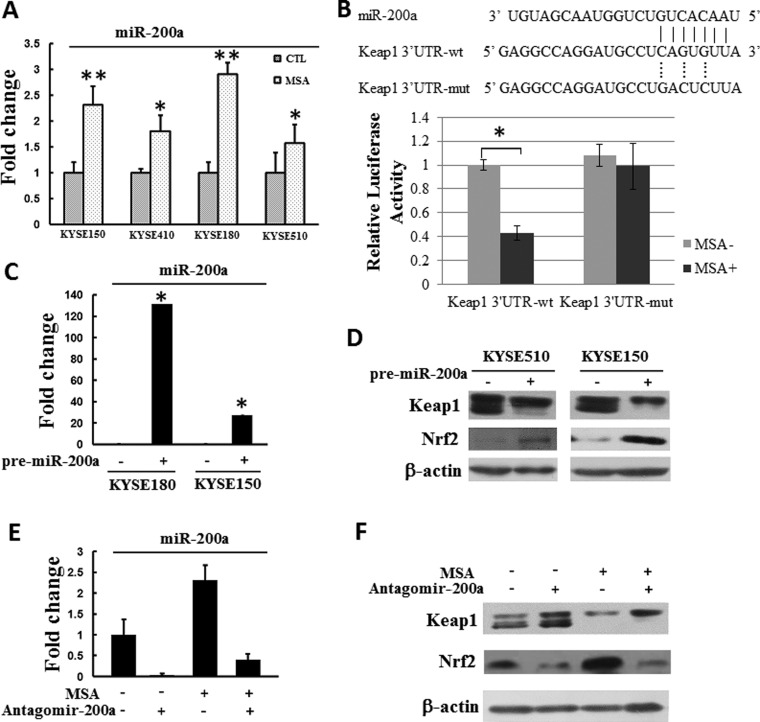
Previous study has showed that Keap1 is a direct target of miR-200a [16,18]. In order to test if MSA inhibited Keap1 through miR-200a at the post-transcriptional level, KYSE 150, KYSE 410, KYSE 180 and KYSE 510 cells were treated with MSA (5 μM) for 24 h, the level of miR-200a was detected by real-time PCR afterwards. As shown in Figure 2(A), MSA could significantly induce miR-200a expression in ESCC cells.
Figure 2. MSA activates Keap1/Nrf2 pathway via up-regulating miR-200a.
(A). KYSE150, KYSE 410, KYSE 180 and KYSE510 were treated with or without MSA (5 μM) for 24 h, then the expression level of miR-200a was detected by real-time PCR. U6 was used as an internal control. The level of miR-200a in each cell lines without MSA treatment was designated as unit 1 respectively. Means ± S.D., n=3, *P<0.05, **P<0.01. (B) Predicted duplex formation between human Keap1 3′-UTR and miR-200a. Luciferase activity of Keap1 3′-UTR wild-type (Keap1 3′-UTR-wt) or mutant (Keap1 3′-UTR-mut) reporter gene in KYSE150 cells with or without MSA treatment (5 μM, 24 h) were detected. Mean ± S.D. (n=3), *P<0.05. (C) KYSE180 and KYSE150 cells were transfected with either 30 nM Pre-miR miR-200a precursor or a Pre-miR negative control miRNA precursor. miRNA was extracted 24 h after transfection. Real-time PCR was performed to examine miR-200a level. The level of miR-200a in the corresponding cells that transfected with the negative control was designated as unit 1 respectively. Means ± S.D., n=3. (D) KYSE510 and KYSE150 cells were transfected with miR-200a precursor (30 nM) or pre-miR negative control (30 nM) respectively. Keap1 and Nrf2 protein levels were detected 24 h after transfection. β-Actin was used as a loading control. (E) miR-200a level in KYSE150 cells after the administration of MSA (5 μM) and/or antagomir-200a (20 μM) was measured by real-time PCR. U6 was used as the endogenous control. The level of miR-200a without MSA and antagomir-200a treatment was designated as unit 1. Means ± S.D., n=3. (F) The levels of Keap1 and Nrf2 in KYSE150 cells treated with MSA (5 μM) following antagomir-200a (20 μM) transfection were detected by western blot. β-Actin was used as a loading control.
In order to test whether miR-200a could directly regulate Keap1 by binding to its 3′-UTR in ESCC cells, the Keap1 3′-UTR was cloned into the pMiR-Report vector, downstream the luciferase gene (pMiR-Report/Keap1 3′-UTR-wt, abbreviated to Keap1 3′-UTR-wt). For control group, a mutated Keap1 3′-UTR with three bases mutations from the site of perfect complementarity was cloned (pMiR-Report/Keap1 3′-UTR-mut, abbreviated to Keap1 3′-UTR-mut). As shown in Figure 2(B), transfection of Keap1 3′-UTR-wt following with MSA treatment led to a decrease in the luciferase activity in KYSE 150 cells. In contrast, MSA treatment did not influence the luciferase activity of Keap1 3′-UTR-mut, demonstrating that mutation of the miR-200a-binding site in the Keap1 3′-UTR abolished the inhibitory effect of miR-200a to regulate its expression. In addition, transfection of Pre-miR miR-200a precursor could effectively express miR-200a (Figure 2C) and increased expression of miR-200a led to a reduced production of Keap1 at protein level (Figure 2D). Furthermore, we also treated KYSE150 cells with antagomir-200a and re-examined the impact of MSA treatment. The results showed that antagomir-200a could attenuate MSA treatment-induced Keap1 down-regulation and Nrf2 up-regulation (Figures 2E and 2F).
In summary, these results showed that Keap1 was a direct target of miR-200a in ESCC cells. Also, MSA could up-regulate the expression of miR-200a.
MSA-induced miR-200a expression depended on the mediation of KLF4
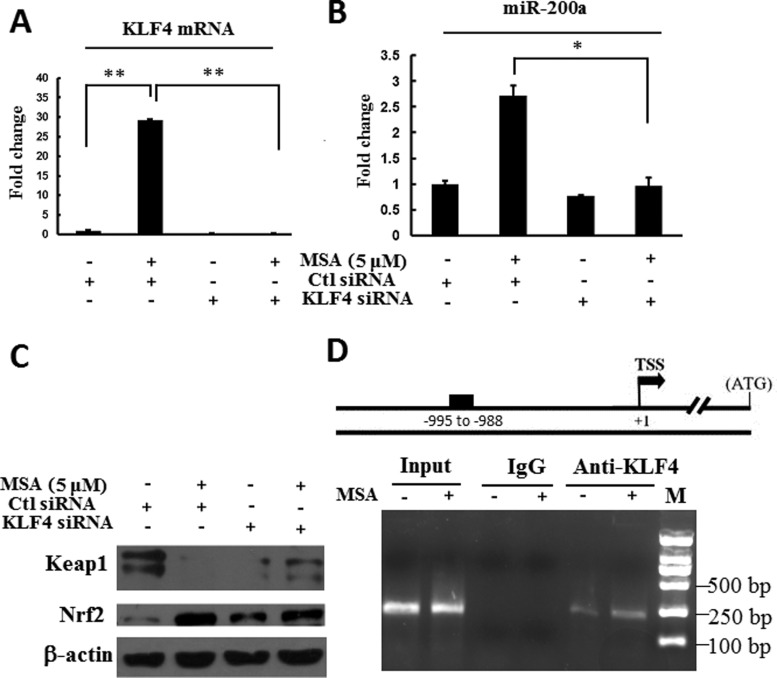
Our previous study reported that KLF4 could be up-regulated by MSA in ESCC cells [19]. We examined the genomic sequences in the 5′-direction of the mir-200a gene. Intriguingly, a putative binding site at the ∼1 kb upstream of the mir-200a was observed for KLF4 transcription factor. To determine whether KLF4 could up-regulate miR-200a in human ESCC cells, quantitative real-time PCR was performed. As shown in Figures 3(A) and 3(B), MSA could up-regulate the mRNA level of KLF4. When KLF4 expression was inhibited by using KLF4 siRNA, MSA could not induce miR-200a expression anymore. The results of western blotting showed that knockdown of KLF4 could abolish MSA treatment-induced Keap1 down-regulation and Nrf2 up-regulation. Furthermore, ChIP assay showed that MSA could significantly increase the binding of KLF4 to the miR-200a promoter region in ESCC cells. All of these results indicated that MSA-induced miR-200a expression was dependent on the mediation of KLF4.
Figure 3. KLF4 mediated miR-200a up-regulation induced by MSA in ESCC cells.
KYSE180 cells were transfected with KLF4 siRNA or control siRNA for 48 h. Then, the cells were treated with or without MSA (5 μM) for additional 24 h. Total RNA was extracted. KLF4 mRNA (A) and miR-200a level (B) were determined by real-time PCR. The level of KLF4 or miR-200a in KYSE180 cells with control siRNA transfection and without MSA treatment was designated as unit 1 respectively. Means ± S.D., n=3. (C). The levels of Keap1 and Nrf2 in KYSE180 cells treated as indicated were detected by western blot. β-Actin was used as a loading control. (D). ChIP assay result showed the KLF4 that associated with miR-200a prompter. KYSE150 cells were treated with MSA (5 μM, 24 h). Then, the lysates were immunoprecipitated by KLF4 antibody and–1048—-804 region of miR-200a promoter was amplified with specific primers.
DISCUSSION
ESCC occurs at very high rates in certain regions of China [23]. There are increasing evidences demonstrating that selenium could act as a potential anti-oesophageal cancer agent, but the precise mechanisms involved are still not completely understood. Significant risk factors for ESCC include tobacco smoking and alcohol consumption [24]. It has been reported that carcinogenesis induced by tobacco and alcohol is mediated, at least in part, by oxidative stress [25]. Oxidative stress is a term used to denote the imbalance between the concentrations of reactive oxygen species (ROS), reactive nitrogen species (RNS) and the defence mechanisms of the body. There is ongoing oxidative stress in the human body. Such an imbalance plays a pivotal role in many pathological conditions, including cancer [25]. The Keap1/Nrf2 signal pathway has been considered to protect cells against carcinogenesis and attenuate cancer development via neutralization ROS or carcinogen [26,27]. MSA, as a potent second-generation selenium compound, is a known antioxidant and chemopreventive agent. Our previous study demonstrated that MSA could inhibit HDAC activity in ESCC cells [19]. In the present study, for the first time, our results indicated that MSA strongly inhibited Keap1 expression via miR-200a and induced Nrf2 protein expression and nucleus accumulation in ESCC cells. In agreement, the regulation of Keap1 expression by miR-200a has been previously studied in breast cancer [16]. Besides, there are evidences that HDAC inhibition could activate Nrf2 activation by inhibiting Keap1 expression and the decrease in Keap1 expression was an indirect effect [28]. Thus the miRNA inhibition might be an underlying mechanism to explain the phenotype. Furthermore, a recent study showed that Nrf2-knockdown mice were more susceptible to 4-nitroquinoline-1-oxide (4NQO)-induced tongue and oesophageal carcinogenesis than wild-type mice, suggesting that Keap1/Nrf2 system is an important defence mechanism for upper aerodigestive tract carcinogenesis [12]. These data support the idea that the chemopreventive property of MSA in oesophageal carcinogenesis may depend on the activation of Keap1/Nrf2 pathway. In vivo studies are needed to further assess the chemopreventive effect of MSA.
It was reported that KLF4 could be up-regulated by MSA in human prostate cancer and ESCC cells [19,29]. In our study, KLF4 regulation of miR-200a expression was examined in ESCC cells. Previous study has validated that HDAC4 could inhibit the expression of miR-200a and reduce the histone H3 acetylation level at the mir-200a promoter through a Sp1-dependent pathway in hepatocellular carcinoma [30]. Our earlier experimental evidences support the idea that MSA could enhance the acetylation of histone H3 in ESCC cells [19]. An interesting possibility is that MSA-induced histone H3 acetylation may directly bind to the mir-200a promoter region and thus increased miR-200a expression in ESCC cells. Therefore, the possible involvement of histone H3 acetylation in the regulation of miR-200a deserves further investigation.
In conclusion, our study not only demonstrated that MSA could activate Keap1/Nrf2 pathway via up-regulating miR-200a but also found a novel mechanism by which miR-200a expression was regulated in ESCC cells.
Acknowledgments
We thank Professor Xiaoming Yang for the pGL3-ARE and pcDNA3-HA-Nrf2 plasmids.
Abbreviations
- ARE
antioxidant response element
- ESCC
oesophageal squamous cell carcinoma
- HDAC
histone deacetylase
- Keap1
kelch-like ECH-associated protein 1
- KLF4
Krüpple-like factor 4
- MSA
methylseleninic acid
- mut
mutation
- Nrf2
nuclear factor E2-related factor 2
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- SAHA
suberanilohydroxamic acid
- Sp1
specificity protein 1
- wt
wild type
AUTHOR CONTRIBUTION
Mei Liu, Ningzhi Xu and Hongxia Zhu conceived and designed the experiments. Chenfei Hu, Mei Liu, Qing Xu, Lechuang Chen and Kai Ma performed the experiments. Chenfei Hu and Mei Liu analysed the data. Mei Liu, Ningzhi Xu and Hongxia Zhu contributed to the writing of the manuscript. All authors read and approved the final manuscript.
FUNDING
This work was supported by the National Natural Science Foundation [grant numbers 81472561, 81321091, 81302279, 81071713 and 81021061].
References
- 1.Duffield-Lillico A.J., Reid M.E., Turnbull B.W., Combs G.F., Jr, Slate E.H., Fischbach L.A., Marshall J.R., Clark L.C. Baseline characteristics and the effect of selenium supplementation on cancer incidence in a randomized clinical trial: a summary report of the Nutritional Prevention of Cancer Trial. Cancer Epidemiol. Biomarkers Prev. 2002;11:630–639. [PubMed] [Google Scholar]
- 2.Li G.X., Lee H.J., Wang Z., Hu H., Liao J.D., Watts J.C., Combs G.F., Jr., Lu J. Superior in vivo inhibitory efficacy of methylseleninic acid against human prostate cancer over selenomethionine or selenite. Carcinogenesis. 2008;29:1005–1012. doi: 10.1093/carcin/bgn007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L., Bonorden M.J., Li G.X., Lee H.J., Hu H., Zhang Y., Liao J.D., Cleary M.P., Lu J. Methyl-selenium compounds inhibit prostate carcinogenesis in the transgenic adenocarcinoma of mouse prostate model with survival benefit. Cancer Prev. Res. 2009;2:484–495. doi: 10.1158/1940-6207.CAPR-08-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mark S.D., Qiao Y.L., Dawsey S.M., Wu Y.P., Katki H., Gunter E.W., Fraumeni J.F., Jr., Blot W.J., Dong Z.W., Taylor P.R. Prospective study of serum selenium levels and incident esophageal and gastric cancers. J. Natl. Cancer Inst. 2000;92:1753–1763. doi: 10.1093/jnci/92.21.1753. [DOI] [PubMed] [Google Scholar]
- 5.Limburg P.J., Wei W., Ahnen D.J., Qiao Y., Hawk E.T., Wang G., Giffen C.A., Roth M.J., Lu N., Korn E.L., et al. Randomized, placebo-controlled, esophageal squamous cell cancer chemoprevention trial of selenomethionine and celecoxib. Gastroenterology. 2005;129:863–873. doi: 10.1053/j.gastro.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 6.Chun K.S., Kundu J., Kundu J.K., Surh Y.J. Targeting Nrf2-Keap1 signaling for chemoprevention of skin carcinogenesis with bioactive phytochemicals. Toxicol. Lett. 2014;229:73–84. doi: 10.1016/j.toxlet.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 7.Yang Y., Cai X., Yang J., Sun X., Hu C., Yan Z., Xu X., Lu W., Wang X., Cao P. Chemoprevention of dietary digitoflavone on colitis-associated colon tumorigenesis through inducing Nrf2 signaling pathway and inhibition of inflammation. Mol. Cancer. 2014;13:48. doi: 10.1186/1476-4598-13-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaramillo M.C., Zhang D.D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang D.D. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab. Rev. 2006;38:769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- 10.Venugopal R., Jaiswal A.K. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl. Acad. Sci. U.S.A. 1996;93:14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang D.D., Lo S.C., Cross J.V., Templeton D.J., Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohkoshi A., Suzuki T., Ono M., Kobayashi T., Yamamoto M. Roles of Keap1-Nrf2 system in upper aerodigestive tract carcinogenesis. Cancer Prev. Res. 2013;6:149–159. doi: 10.1158/1940-6207.CAPR-12-0401-T. [DOI] [PubMed] [Google Scholar]
- 13.Terazawa R., Garud D.R., Hamada N., Fujita Y., Itoh T., Nozawa Y., Nakane K., Deguchi T., Koketsu M., Ito M. Identification of organoselenium compounds that possess chemopreventive properties in human prostate cancer LNCaP cells. Bioorg. Med. Chem. 2010;18:7001–7008. doi: 10.1016/j.bmc.2010.08.019. [DOI] [PubMed] [Google Scholar]
- 14.Singh C.K., Ndiaye M.A., Siddiqui I.A., Nihal M., Havighurst T., Kim K., Zhong W., Mukhtar H., Ahmad N. Methaneseleninic acid and gamma-tocopherol combination inhibits prostate tumor growth in vivo in a xenograft mouse model. Oncotarget. 2014;5:3651–3661. doi: 10.18632/oncotarget.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Eades G., Yang M., Yao Y., Zhang Y., Zhou Q. miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells. J. Biol. Chem. 2011;286:40725–40733. doi: 10.1074/jbc.M111.275495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Jaarsveld M.T., Helleman J., Boersma A.W., van Kuijk P.F., van Ijcken W.F., Despierre E., Vergote I., Mathijssen R.H., Berns E.M., Verweij J., et al. miR-141 regulates KEAP1 and modulates cisplatin sensitivity in ovarian cancer cells. Oncogene. 2013;32:4284–4293. doi: 10.1038/onc.2012.433. [DOI] [PubMed] [Google Scholar]
- 18.Wei J., Zhang Y., Luo Y., Wang Z., Bi S., Song D., Dai Y., Wang T., Qiu L., Wen L., et al. Aldose reductase regulates miR-200a-3p/141-3p to coordinate Keap1-Nrf2, Tgfbeta1/2, and Zeb1/2 signaling in renal mesangial cells and the renal cortex of diabetic mice. Free Radic. Biol. Med. 2014;67:91–102. doi: 10.1016/j.freeradbiomed.2013.10.811. [DOI] [PubMed] [Google Scholar]
- 19.Hu C., Liu M., Zhang W., Xu Q., Ma K., Chen L., Wang Z., He S., Zhu H., Xu N. Upregulation of KLF4 by methylseleninic acid in human esophageal squamous cell carcinoma cells: modification of histone H3 acetylation through HAT/HDAC interplay. Mol. Carcinog. 2014;54:1051–1059. doi: 10.1002/mc.22174. [DOI] [PubMed] [Google Scholar]
- 20.Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Zhou C., Liu S., Zhou X., Xue L., Quan L., Lu N., Zhang G., Bai J., Wang Y., Liu Z., et al. Overexpression of human pituitary tumor transforming gene (hPTTG), is regulated by beta-catenin/TCF pathway in human esophageal squamous cell carcinoma. Int. J. Cancer. 2005;113:891–898. doi: 10.1002/ijc.20642. [DOI] [PubMed] [Google Scholar]
- 23.Ke L. Mortality and incidence trends from esophagus cancer in selected geographic areas of China circa. Int. J. Cancer. 2002;102:1970, 271–90. doi: 10.1002/ijc.10706. [DOI] [PubMed] [Google Scholar]
- 24.Engel L.S., Chow W.H., Vaughan T.L., Gammon M.D., Risch H.A., Stanford J.L., Schoenberg J.B., Mayne S.T., Dubrow R., Rotterdam H., et al. Population attributable risks of esophageal and gastric cancers. J. Natl. Cancer Inst. 2003;95:1404–1413. doi: 10.1093/jnci/djg047. [DOI] [PubMed] [Google Scholar]
- 25.Mena S., Ortega A, Estrela J.M. Oxidative stress in environmental-induced carcinogenesis. Mutat. Res. 2009;674:36–44. doi: 10.1016/j.mrgentox.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 26.Kwak M.K., Kensler T.W. Targeting NRF2 signaling for cancer chemoprevention. Toxicol. Appl. Pharmacol. 2010;244:66–76. doi: 10.1016/j.taap.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klaunig J.E., Kamendulis L.M, Hocevar B.A. Oxidative stress and oxidative damage in carcinogenesis. Toxicol. Pathol. 2010;38:96–109. doi: 10.1177/0192623309356453. [DOI] [PubMed] [Google Scholar]
- 28.Wang B., Zhu X., Kim Y., Li J., Huang S., Saleem S., Li R.C., Xu Y., Dore S., Cao W. Histone deacetylase inhibition activates transcription factor Nrf2 and protects against cerebral ischemic damage. Free Radic. Biol. Med. 2012;52:928–936. doi: 10.1016/j.freeradbiomed.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu S., Zhang H., Zhu L., Zhao L., Dong Y. Kruppel-like factor 4 is a novel mediator of selenium in growth inhibition. Mol. Cancer Res. 2008;6:306–313. doi: 10.1158/1541-7786.MCR-07-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan J.H., Yang F., Chen B.F., Lu Z., Huo X.S., Zhou W.P., Wang F., Sun S.H. The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology. 2011;54:2025–2035. doi: 10.1002/hep.24606. [DOI] [PubMed] [Google Scholar]