Uncoupling proteins (UCP)2, UCP4 and UCP5 transport protons across the inner membrane of mitochondria in the central nervous system (CNS). Novel recombinant protein expression allowed expression of UCPs in Escherichia coli membranes. Functional neuronal UCPs formed multimers in membranes and interacted with various fatty acids (FAs) to transport protons. Self-association and unique ion transport properties of UCPs distinguish their physiological roles in the CNS.
Keywords: central nervous system (CNS), circular dichroism (CD) spectroscopy, fatty acid (FA), membrane protein folding, molecular association, proton transport, reconstitution, uncoupling proteins (UCPs)
Abstract
Mitochondrial inner membrane uncoupling proteins (UCPs) facilitate transmembrane (TM) proton flux and consequently reduce the membrane potential and ATP production. It has been proposed that the three neuronal human UCPs (UCP2, UCP4 and UCP5) in the central nervous system (CNS) play significant roles in reducing cellular oxidative stress. However, the structure and ion transport mechanism of these proteins remain relatively unexplored. Recently, we reported a novel expression system for obtaining functionally folded UCP1 in bacterial membranes and applied this system to obtain highly pure neuronal UCPs in high yields. In the present study, we report on the structure and function of the three neuronal UCP homologues. Reconstituted neuronal UCPs were dominantly helical in lipid membranes and transported protons in the presence of physiologically-relevant fatty acid (FA) activators. Under similar conditions, all neuronal UCPs also exhibited chloride transport activities that were partially inhibited by FAs. CD, fluorescence and MS measurements and semi-native gel electrophoresis collectively suggest that the reconstituted proteins self-associate in the lipid membranes. Based on SDS titration experiments and other evidence, a general molecular model for the monomeric, dimeric and tetrameric functional forms of UCPs in lipid membranes is proposed. In addition to their shared structural and ion transport features, neuronal UCPs differ in their conformations and proton transport activities (and possibly mechanism) in the presence of different FA activators. The differences in FA-activated UCP-mediated proton transport could serve as an essential factor in understanding and differentiating the physiological roles of UCP homologues in the CNS.
INTRODUCTION
Located in the inner mitochondrial membrane (IMM), uncoupling proteins (UCPs) dissipate the proton electrochemical gradient across the membrane, resulting in the reduction of ATP production through ATP synthase. Abundantly expressed in brown adipose tissue (BAT), human UCP1 mediates proton transport across the IMM and generates heat [1]. Expressed in the central nervous system (CNS), UCP2, UCP4 and UCP5 are referred to as neuronal UCPs [2]. UCP2 is also found in several other human tissues in addition to the CNS, including liver, endothelium and pancreatic tissues; and might be responsible for different physiological functions [2–4]. UCP4 and UCP5 are predominantly localized in neurons and neurosensory cells and their physiological roles are not clear [2,5–7]. Compared with the prototypic UCP1, the amino acid sequence identities of UCPs 2, 4, 5 are 59%, 34% and 30% respectively [8–11]. Despite these relatively low sequence identities to UCP1, both UCP4 and UCP5 possess comparable helical conformations and proton transport activities to other UCPs [11]. Neuronal UCP-mediated proton transport is activated by fatty acids (FAs) and inhibited by purine nucleotides (PNs) [11]. Recent studies relate the proton transport-mediated uncoupling activities of neuronal UCPs to possible protection against reactive oxygen species (ROS), produced in the mitochondria [4,12–17]. In addition to the proton transport function, all UCPs are also able to transport chloride and other small anions [9,11,18]. Protein sequence symmetry analysis and experimental results suggest that UCPs could also function as small substrate carriers using an H+-assisted mechanism [19,20]. The exact physiological role(s) of neuronal UCPs has not been fully established.
Examining the structure-function relationships of neuronal UCPs remains an intriguing approach for clarifying the physiological roles of these proteins in neurons. In our previous studies, using recombinant proteins expressed in bacterial systems, we obtained highly pure neuronal UCPs in inclusion bodies and reconstituted these proteins into mild detergents and liposomes for structural and functional studies [10,11,21]. Although this expression method produces proteins in high yield and allows for excellent purity, there is always a possibility of partial folding or misfolding of the reconstituted proteins that can influence the accurate analysis of their molecular functions. We recently reported a novel recombinant protein expression system using the auto-induction method and the small periplasmic leader sequence PelB (pectate lyase B) to direct UCP1 to the bacterial membrane, which resulted in the production of highly pure native-like folded UCP1, in high yields [22]. In that report, recombinant UCP1 extracted from the bacterial membranes and reconstituted in both detergents and liposomes adopted dominantly helical structures and displayed proton and chloride ion transport activity [22]. In the current study, we applied this novel expression method and report the expression of His-tagged recombinant versions of neuronal UCPs in Escherichia coli inner membranes for the first time. All three neuronal UCPs were extracted from bacterial membranes using the mild detergent octyl glucoside (OG) and reconstituted into liposomes. Similar to UCP1 [22], the neuronal UCPs showed evidence of self-association and displayed predominantly helical conformations in liposomes. The associated forms of neuronal UCPs also transported proton and chloride ions across lipid membranes. The effect of various FAs on the overall structure and the proton/chloride transport rates of neu-ronal UCPs led to new insights into specific functions of neuronal UCPs and possible differentiation of their physiological roles.
MATERIALS AND METHODS
UCP constructs
The human UCP2 cDNA clone (pET21a-UCP2) was a gift from M. Brand (MRC Dunn Human Nutrition Unit, Cambridge, U.K.). UCP4 cDNA was synthesized by GenScript Corp. and received in a pUC57 vector. UCP5 cDNA was purchased from A.T.C.C. and was received in a pCR-BluntII-TOPO vector. The neuronal UCP constructs were sub-cloned into the pET21d vector (Novagen) and modified to contain an N-terminal His-tag followed by the tobacco etch virus (TEV) protease recognition site (ENLYFQG), as previously described [10]. The His6-ENLYFQG-hUCPs coding sequences were sub-cloned into the pET26b(+) vector using standard molecular techniques such that the PelB leader sequence was in frame with the His-tag, TEV protease site and the protein. The constructs (pET26b-hUCP2, pET26b-hUCP4 and pET26b-hUCP5) were then transformed into E. coli BL21 CodonPlus (DE3)-RIPL for production of the recombinant fusion proteins containing an N-terminal PelB leader sequence fused to the respective neuronal UCP. To verify the identity of the clones, each of the recombinant cDNAs in the pET26b (+) expression vector was sequenced (TCAG, Sick Children's Hospital).
Chemicals
Egg yolk L-α-lecithin (Sigma) contained at least 60% (w/w) phosphatidylcholine. The remaining 40% was comprised mostly of phosphatidylethanolamine and other lipids. The phospholipid POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) was from Avanti Polar Lipids (Alabaster). C8E4 (octyltetraoxyethylene) was obtained from Bachem. DDM was from Calbiochem-EMD Biosciences. 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) and OG were from Affymetrix. The fluorescent probe SPQ [6-methoxy-N-(3-sulfopropyl)quinolinium; 99% purity] was from Biotium Inc.. All other chemicals were from Sigma.
Expression, membrane extraction and purification of neuronal UCPs
Recombinant versions of neuronal UCPs were overexpressed in E. coli BL21 CodonPlus (DE3)-RIPL using the auto-induction method, as previously described for UCP1 [22]. Briefly, the bacterial culture was grown in the auto-induction media [1% tryptone, 0.5% yeast extract, 1 mM MgSO4, 0.5% glycerol, 0.05% glucose, 0.2% lactose, 25 mM (NH4)2SO4, 50 mM KH2PO4, 50 mM Na2HPO4] at 22°C for 20 h before collection [23]. The bacterial cell pellets were collected at 8000 g for 10 min (4°C). Bacterial cells were suspended in extraction buffer (20 mM Tris/HCl, 500 mM NaCl, pH 8.0) and lysed using a Constant System cell disruption press (20 kPsi) in the presence of complete protease inhibitor cocktail (EDTA-free; Roche), DNase (0.5 mg/ml) and lysozyme (0.2 mg/ml). The cell lysate was subjected to centrifugation at 20000 g for 20 min (4°C) to remove inclusion bodies and unbroken cells. The supernatant was subsequently ultracentrifuged at 256631 g (TLA-100 rotor, Beckman Coulter) for 1 h (4°C) to yield the bacterial membranes in the pellet fraction and the cytosol in the supernatant. The enrichment of membranes in the pellet fractions was confirmed using the NADH oxidase assay [22].
The purification protocols of UCP2, UCP4 and UCP5 followed that of UCP1, as described previously [22]. In brief, the bacterial membranes were solubilized in binding buffer {20 mM Tris/HCl, 500 mM NaCl, 1mM [Tris (hydroxypropyl) phosphine], pH 8.0} containing 1% LDAO (lauryldimethylamine-oxide) detergent and 20 mM imidazole. This mixture was subjected to Ni-NTA (nitrilotriacetic acid) chromatography for purification under non-denaturing conditions. The columns were washed with 30 mM imidazole and eluted with 400 mM imidazole in 1% OG detergent. The pooled eluted fractions were desalted using Econo-Pac 10DG Columns (Bio-Rad). The final purity and concentration of purified proteins (∼0.3–0.5 mg protein/1 l culture) were analysed by SDS/PAGE and Bradford protein concentration assays respectively. The proteins were stored in desalting buffer (1% OG, 50 mM NaCl, 20 mM Tris/HCl, 1% glycerol, pH 8.0) at −80°C.
Semi-native PAGE
To provide semi-native conditions, the content of SDS was significantly reduced compared with traditional SDS/PAGE [22]. Following the protocol from our previous study, 2 mM SDS was included in the electrophoresis running buffer to ensure the migration of proteins. Prior to electrophoretic experiments, each protein sample was mixed with the sample buffer containing 62.5 mM Tris/HCl, pH 6.8, 0.002% Bromophenol Blue and 10% glycerol. The gels were run at 110 V for 75 min and stained with Coomassie Brilliant Blue R-250.
Identification of proteins by western blots and MS analysis
The identity of overexpressed His6-tagged UCP2, UCP4 and UCP5 was confirmed by both western blot and MS analyses. In the western blot, 10–50 μg of total proteins were run on a SDS/PAGE (12% gel) and transferred (110 min, 15 V) to nitrocellulose membranes using the semi-dry technique. The membranes were stained with Amido Black to confirm the efficiency of transfer and were blocked overnight at 4°C in TBS-buffer containing 5% skim milk and 0.05% Tween-20. Mouse IgG2b anti-histidine antibody (Millipore) was used as the primary antibody (1:500 dilution). Rabbit peroxidase-conjugated antibody raised against mouse (Rockland) was used as the secondary antibody (1:4000 dilution). Immunodetection was achieved by chemiluminescence using ECL Western Blot reagent (GE Healthcare) and an image was captured using a Bio-Rad VersaDoc Imaging System.
To further confirm the identity of recombinant protein in bands of different molecular masses, purified protein bands resolved on electrophoretic gels were excised, trypsin-digested and analysed by LC-MS/MS in a data-dependent acquisition, as described previously [22]. Trypsin-digested protein fractions were separated using a NanoAcquity (Waters) LC-MS/MS equipped with a 25-cm × 75-μm inner diameter C18 column. MS analysis was performed on a QExactive (Thermo Scientific) using higher energy collision dissociation. De novo sequencing of fractionated proteins was performed with Peak Studio 6.0 Software using the Uniprot Swiss database for Homo sapiens, with a false discovery rate of <1%.
Reconstitution of UCPs into liposomes
Reconstitution of UCPs into liposomes followed the procedures in previously used protocols with minor modifications [11,22]. Briefly, stock lipids in chloroform were dried under vacuum (8-12 h) and resuspended in reconstitution buffers. The multilamellar liposomes were solubilized in C8E4 detergent [(detergent/lipid=2.5/1 (w/w)]. In the ion transport assays, the internal reconstitution buffer was supplemented with 3 mM fluorescent probe SPQ. Purified proteins were added to the transparent lipid/detergent mixed micelles. The spontaneous formation of liposomes/proteoliposomes was achieved following the detergent removal by SM-2 Biobeads (Bio-Rad). In ion transport assays, the external probe SPQ was removed using a coarse Sephadex G25-300 (GE Healthcare) spin column.
CD and fluorescence spectroscopic measurements
Far- and near- UV CD spectra were measured in 0.1 and 0.5 cm path length quartz cells respectively, on an Aviv 215 spectropolarimeter (Aviv Biomedical) at 1-nm resolution. Proteins’ ellipticities are exhibited as mean residue ellipiticity [θ] and the reported spectra are an average of at least eight measurements. Secondary structure content of proteins was estimated using the deconvolution program CDSSTR and the analysis was based on a set of 48 reference proteins and performed on the Dichroweb web site [24,25]. Thermal denaturation of neuronal UCPs in detergents was analysed by monitoring the decrease in protein ellipiticity at 222 nm, in the range of 15°C–90°C.
Steady-state fluorescence measurements were performed using a Cary Eclipse spectrophotometer (Varian). Excitation and emission bandwidth slits for all measurements were 5 nm and a scan speed of 600 nm/min was used throughout the experiments (25°C).
Ion transport measurements
Proton and chloride transport measurements were performed as previously described [11,22]. Following reconstitution, liposomes/proteoliposomes were formed with entrapped SPQ fluorescent probes. SPQ is quenched by anions. In each ion transport assay, 40 μl of liposomes/proteoliposomes were incubated in 1.96 ml of external buffer. In the proton transport assays, the internal medium consisted of TES buffer (20 mM), TEA2SO4 (TEA, tetraethylammonium; 54 mM) and EDTA (0.7 mM). The external medium in this case contained K2SO4 (54 mM) instead of TEA2SO4. In the chloride transport assays, the internal medium consisted of 110 mM sodium phosphate and 0.2 mM EDTA, whereas the external buffer contained 10 mM sodium phosphate 200 mM KCl and 0.2 mM EDTA. All media were kept constant at pH 7.2. The ion fluxes in both assays were initiated by the influx of K+ from the external media mediated by the K+-ionophore valinomycin. In the chloride transport assays, chloride influx quenched the SPQ and caused a decrease in its fluorescence signal. In the proton transport assays, FAs activated UCP-mediated proton efflux, resulting in deprotonation of TES buffer. TES anion quenched SPQ's fluorescence. All ion transport data were corrected by subtraction of the non-specific ion leakage and calibrated for the SPQ fluorescence response and internal volume of proteoliposomes. The final protein concentrations in proteoliposomes were quantified using a modified Lowry concentration assay, as described previously [11].
Sequence alignment and structural modelling of neuronal UCPs
Protein primary sequence alignment of UCPs and ADP/ATP carrier (AAC) were performed using Clustal Omega [26]. Protein sequence alignment analysis was viewed through Jalview program [27]. Protein helical contents were estimated by Distill program [28]. The 3D structural model of UCP2 (PDB ID: 2LCK) was viewed using Pymol (http://www.pymol.org) [30].
RESULTS
Expression of neuronal UCPs in bacterial membranes and their folding in detergents and liposomes
The incorporation of the PelB leader sequence into the N-terminus of the neuronal UCPs combined with the application of the auto-induction method resulted in all three neuronal UCPs (UCP2, UCP4 and UCP5) being targeted to the bacterial membranes. Bacterial cells were fractionated using differential velocity centrifugation and the identity of the neuronal UCPs in the E. coli membrane fraction was verified using Western blot analysis probed with anti-histidine antibody (Figure 1, inset) and mass spectrometric analysis (Supplementary Table S1). An NADH oxidase activity assay was used to confirm that the membrane fraction containing the UCPs was in fact enriched in bacterial membranes (results not shown).
Figure 1. Expression of neuronal UCPs in bacterial membranes and folding of the proteins in the detergent OG.
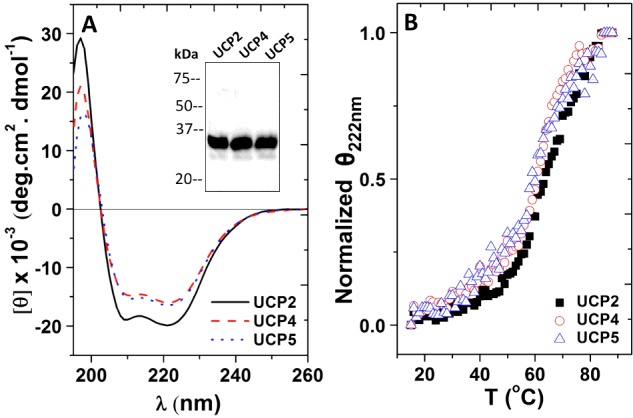
(A) Averaged far-UV CD spectra of 3–5 μM neuronal UCPs in buffer containing 1% OG. Inset shows the western blot analysis of neuronal UCPs in the membrane fraction isolated from E. coli probed with mouse IgG2b anti-histidine antibody, detected by chemiluminescence using a horseradish peroxidase-linked secondary antibody. Molecular mass markers (kDa) are indicated to the left. (B) Comparative thermal denaturation of neuronal UCPs in 1% OG. The molar ellipticity of UCPs at 222 nm was normalized and Tm for each UCP was calculated using Hill equation to fit the data
Neuronal UCPs were extracted from the bacterial membranes using the mild detergent OG and purified using nickel affinity chromatography, prior to being reconstituted into liposomes. The folding of neuronal UCPs in OG was examined using CD spectroscopy (Figure 1A; Table 1). As shown in the far-UV CD spectra, neuronal UCPs in 1% OG exhibited typical spectral footprints of helical conformations, with double negative maximum ellipticities at ∼222 and ∼208 nm and a positive maximum ellipticity of ∼195 nm (Figure 1A). Deconvolution of these CD spectra revealed 63%, 53% and 54% helical contents for UCP2, UCP4 and UCP5 respectively (Table 1). These helical contents were significantly higher than those obtained previously using recombinant UCPs refolded from inclusion bodies [11]. It is also important to note that the estimated helicities of neuronal UCPs extracted from the membranes are close to the protein helicities calculated based on the primary amino acid sequences (Table 1). To further characterize the stability of neuronal UCPs in OG, thermal denaturation experiments were performed in which the protein ellipticity at 222 nm was monitored in the 15°C–90°C temperature range (Figure 1B). All three proteins displayed relatively stable conformations that underwent a two-state unfolding process, with melting points in the range of 64°C–69°C (Figure 1B; Table 2).
Table 1. Estimated secondary structure composition of neuronal UCPs in detergents*, in lipid vesicles* and from primary amino acid sequence prediction†.
| Protein | Protein environment | α-Helix | β-Strand | Turn | Random | NRMSD |
|---|---|---|---|---|---|---|
| OG micelles | 63 | 5 | 12 | 19 | 0.014 | |
| UCP2 | POPC vesicles | 85 | 3 | 2 | 8 | 0.002 |
| Distill prediction | 69 | – | – | – | – | |
| OG micelles | 53 | 7 | 16 | 24 | 0.015 | |
| UCP4 | POPC vesicles | 80 | 6 | 4 | 11 | 0.009 |
| Distill prediction | 70 | – | – | – | – | |
| OG micelles | 54 | 9 | 17 | 20 | 0.009 | |
| UCP5 | POPC vesicles | 85 | 4 | 3 | 7 | 0.009 |
| Distill prediction | 75 | – | – | – | – |
*Deconvolution of CD spectra was performed using the CDSSTR program on the Dichroweb website (see ’Materials and Methods’). The values represent the percentage of secondary structure composition.NRMSD, normalized RMSD, denotes the best fit between the calculated and experimental CD spectra.
†Helical contents of neuronal UCPs were predicted using the Distill prediction program, as described in the ‘Materials and Methods’.
Table 2. Stability of neuronal UCPs in the OG detergent and liposomes.
| Dissociation constants: SDS titration of reconstituted UCPs* (mM SDS) | |||||
|---|---|---|---|---|---|
| Protein | OG detergent Tm† | CD K1/2 | Fl Ka1/2 | Fl Kb1/2 | Δλmax |
| UCP2 | 69.3±1.1 | 1.1±0.1 | 0.61±0.1 | 7.1±0.3 | 5–7 |
| UCP4 | 64.5±1.9 | 1.0±0.1 | 0.60±0.1 | 9.8±0.3 | 7–8 |
| UCP5 | 67.7±2.6 | 1.0±0.1 | 0.60±0.1 | 8.9±0.2 | 7–8 |
†Tm (melting temperature) of UCPs in the detergent OG was calculated using the Hill equation to fit the data of the thermal denaturation curve depicted in Figure 1(B).
*Dissociation constants were calculated using the Hill equation to fit the data of SDS dissociation experiments for reconstituted UCPs in liposomes. The CD K1/2 values were obtained from fitting the θ208/θ222 compared with [SDS] curve, which represented the SDS concentrations at 50% of protein dissociation (Figures 2A–2F). The Fl Ka1/2 values were obtained from fitting the maximum Trp fluorescence intensity of UCPs compared with [SDS] at a low SDS concentration range (0–10 mM; Figures 3A–3F). The Fl Kb1/2 values were obtained from fitting the maximum Trp fluorescence intensity of UCPs compared with [SDS] at a high SDS concentration range (10–100 mM; Figures 3A–3F). The Δλmax indicates the range of SDS concentrations where Trp's λmax started to red-shift (Figures 3G–3I).
Abbreviation: Fl, fluorescence.
Self-association of neuronal UCPs in liposomes
Following isolation in detergent, the recombinant neuronal UCPs were reconstituted into liposomes to allow for characterization in a more native-like membrane environment. CD spectra of UCPs in liposomes displayed characteristics typical of α-helices, with the π ➔ π* exciton split bands at ∼192 and 208 nm and the n ➔ π* transition at ∼222 nm (Figures 2A–2C). Compared with their conformation in detergent, each of the neuronal UCPs in liposomes displayed a marked enhancement in their helical content, which could indicate the important role of lipids in the folding of these proteins. Despite their high helical content, the neuronal UCPs exhibited atypical far-UV CD spectra for helical proteins. Specifically, the parallel π ➔ π* band of neuronal UCPs' far-UV CD spectra at 208–210 nm exhibited a shoulder-like negative maximum with lower ellipticity than the negative maximum at 222 nm (Figure 2). The θ208/θ222 ratios of the CD spectra, in both OG detergent and liposomes, were less than 1 (0.60, 0.63 and 0.61 for UCP2, UCP4 and UCP5 respectively). Such ellipticity ratios have been observed in the spectra of coiled coil and helical bundle motifs and as a result of protein association [22,31–34]. Using complementary electrophoresis and MS methods, the atypical helical conformation of the neuronal UCPs can mostly probably be attributed to a predominant population of UCP multimers (tetramers) in liposomes with molecular mass of ∼ 140–150 kDa (Figures 2G–2I; Supplementary Table S1). This is the first direct observation for the self-association of neuronal UCPs in vitro. A similar phenomenon was reported previously for UCP1 [22].
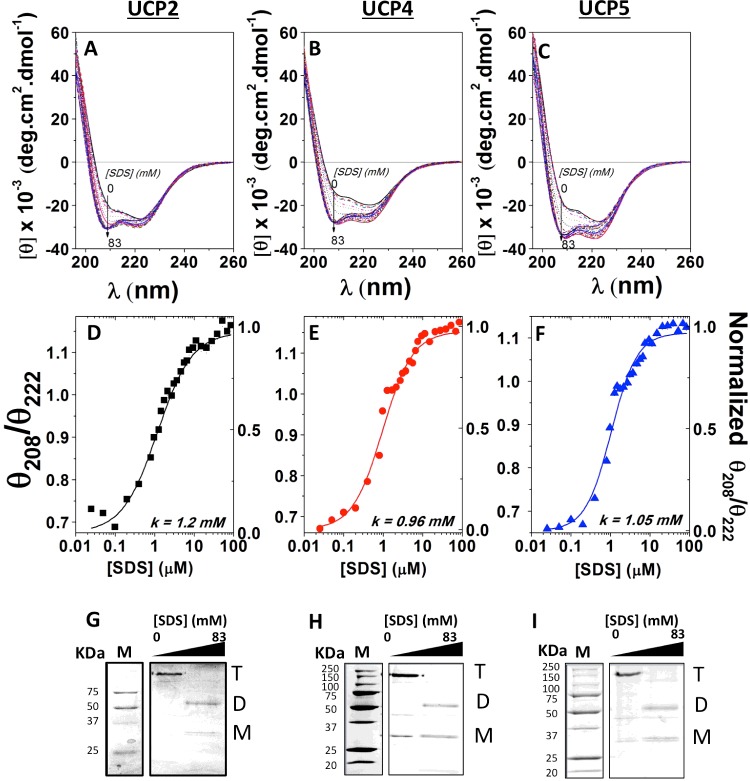
Figure 2. Titration of neuronal UCPs in POPC liposomes with SDS, monitored by CD spectroscopy and semi-native PAGE.
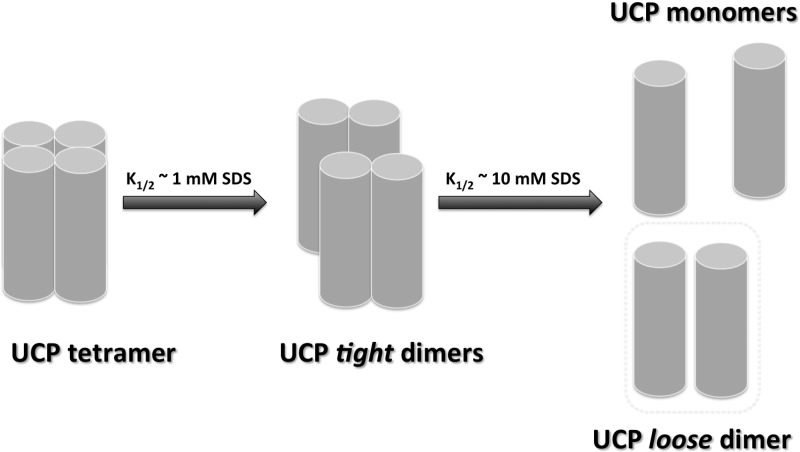
(A, B) and C) show the far-UV CD spectra of reconstituted UCP2, UCP4 and UCP5 respectively in POPC liposomes titrated with SDS (0–83 mM). Corresponding plot of θ208/θ222 compared with [SDS] reveals a cooperative dissociation of UCP2 (D), UCP4 (E) and UCP5 (F) tetramers in the presence of SDS. The θ208/θ222 ratio was normalized and K1/2 ([SDS] at 50% protein dissociation) was calculated using the Hill fitting. The K1/2 values of UCP2, UCP4 and UCP5 are 1.2 mM SDS, 0.96 mM SDS and 1.05 mM SDS respectively. The corresponding semi-native PAGE gels (12%) show reconstituted UCP2 (G), UCP4 (H) and UCP5 (I) in POPC liposomes titrated with SDS, stained with Coomassie Brilliant Blue. UCP tetramers (T) were the predominant species detected in POPC liposomes when treated with loading buffer lacking SDS. As the SDS concentration increased beyond the critical micelle concentration of SDS (∼2 mM), transition towards predominantly UCP dimers (D) was observed. At 83 mM SDS (∼40× CMC of SDS), UCP tetramers were completely absent, UCP dimers became prominent and a small concentration of monomers (M) was present.
SDS titration of associated neuronal UCPs
To characterize the stability of the associated forms of neuronal UCPs, the proteoliposomes were titrated with SDS (Figures 2A–2F and 3). It was expected that with increasing SDS concentration a gradual dissociation of the multimers would occur until their complete dissociation at high SDS concentrations [22]. During SDS titrations, overall and local conformational changes of the proteins were monitored by CD and fluorescence spectroscopies respectively (Figures 2A–2F and 3). Figures 2(A)–2(C) show the far-UV CD spectra of the titrations of reconstituted neuronal UCP tetramers with SDS. The θ208/θ222 ratios in the spectra of each protein were plotted against SDS concentrations to monitor the gradual transformation of the proteins from their associated to monomeric forms (Figures 2D–2F). The far-UV CD spectra of reconstituted UCPs displayed characteristics of associated conformations, with a negative maximum at 222 nm and a negative shoulder at ∼210 nm (θ208/θ222 < 1; Figures 2A–2C). As the SDS concentration increased from 0 to 2 mM, the CD spectral shape of UCPs exhibited a drastic change. The negative ellipticities at 210 and 222 nm were both enhanced and the negative shoulder at 210 blue-shifted towards 208 nm and gradually became a distinct minimum (Figures 2A–2C). Concurrent with this transformation, the θ208/θ222 ratios increased and approached the value of 1 (Figures 2D–2F). These results suggest a transition of reconstituted UCPs towards a less constrained multimeric conformation with weaker intermolecular interactions and more independent monomers. Addition of SDS above 2 mM (2–83 mM) did not affect the θ208 ellipticity, whereas the θ222 negative ellipticity further decreased (Figures 2A–2C). Above 50 mM SDS, the θ208/θ222 was greater than 1 and remained unchanged, suggesting a stable conformation of dissociated proteins (Figures 2D–2F). As discussed in a previous study, high concentrations of SDS could also induce the denaturation of protein tertiary structures [22]. Plotting the θ208/θ222 ratio as a function of [SDS] (mM) revealed a two-state dissociation path for neuronal UCPs reconstituted in POPC liposomes, with the dissociation constant K1/2 representing the SDS concentration at which 50% of the protein population was dissociated (Figures 2D–2F). The K1/2 values of UCP2, UCP4 and UCP5 were determined to be approximately 1 mM SDS (Table 2). Semi-native PAGE analysis of UCPs complemented the CD results, showing that addition of SDS to proteoliposomes resulted in the gradual loss of tetramers and concurrent enrichment of dimers and monomers (Figures 2G–2I). At the end titration point of 83 mM SDS, the UCP tetramers were completely replaced by UCP dimers and monomers (Figures 2G–2I; Supplementary Table S1).
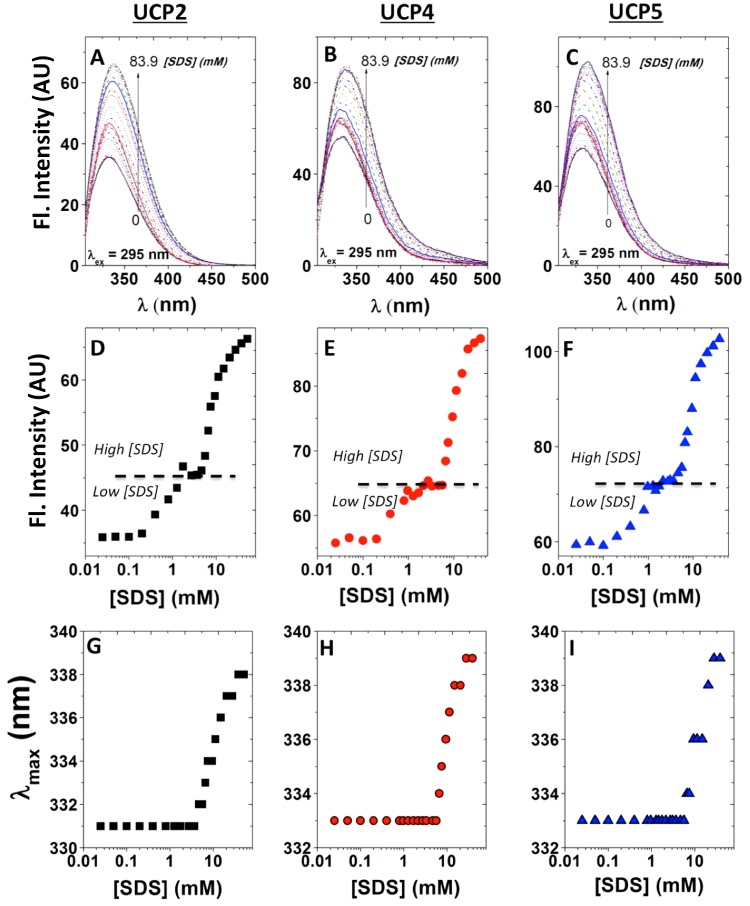
Figure 3. Dissociation of neuronal UCPs in POPC liposomes by SDS, monitored by Trp fluorescence.
The Trp fluorescence spectra of UCP2 (A), UCP4 (B) and UCP5 (C) (λex=295 nm) were monitored as SDS was added to the proteoliposomes ([SDS]=0–83.9 mM). The corresponding plots of maximum Trp intensity reveal a three-state dissociation process for neuronal UCP tetramers in lipid vesicles, as shown for UCP2 (D), UCP4 (E) and UCP5 (F). In the low [SDS] region (0–10 mM), UCP tetramers are suggested to transition toward tight dimeric units. In the high [SDS] region (10–100 mM), UCP tight dimers could be further dissociated into their loose dimeric and monomeric forms. The K1/2 for the first (Ka1/2) and second (Kb1/2) transitions of all neuronal UCPs were obtained and summarized in Table 2. In addition, corresponding plots of the maximum emission wavelength (λmax) compared with [SDS] for UCP2 (G), UCP4 (H) and UCP5 (I) reveal no significant change in the maximum Trp emission wavelength up to ∼10 mM SDS, above which the fluorophores became more exposed to the polar environments and exhibited a red shift in its maximum emission wavelength.
Dissociation/unfolding of UCPs by SDS was also monitored using fluorescence spectroscopy (Figures 3A–3C). The total Trp fluorescence of all neuronal UCPs (λex=295 nm) increased and the maximum emission (λmax) gradually red-shifted in the presence of increasing concentrations of SDS (Figures 3A–3C). Plotting the fluorescence intensity compared with [SDS] revealed a three-state transition curve for each of the neuronal UCPs (Figures 3D–3F). Signs of this three-state dissociation/transition were also apparent (but less resolved) in the ellipticity ratio compared with [SDS] plots of UCPs 4 and 5 (and less for UCP2; Figures 2D–2F). The first transition for all neuronal UCPs occurred at an SDS concentration of ∼0.6 mM, which aligned with the conformational dissociation point (∼1 mM) observed using CD (Figures 2D–2E). The second transition occurred at higher SDS concentrations (∼7–10 mM), which could be interpreted as the point at which further dissociation of UCP dimers to monomers and partial unfolding of monomers occurs (Figures 3D–3F; Table 2). Examination of the λmax of Trp compared with [SDS] revealed that Trp residues experienced a red shift towards higher wavelengths (335–339 nm) at SDS concentrations between ∼5–8 mM, signifying the exposure of Trp residues to more polar environments (Figures 3G–3I). This shift in λmax of Trp residues occurred at [SDS] close to the second transition point observed for UCP fluorescence intensities (Figures 3D–3F; Table 2).
In summary, the results from CD and fluorescence spectroscopy measurements, semi-native PAGE and MS suggest that neuronal UCPs are able to self-associate into tetramers in lipid membranes under our experimental conditions. These results also show two distinct transition states for neuronal UCP tetramer dissociation during SDS titration experiments. The first transition occurs at low SDS concentrations (∼1 mM), where UCP tetramers are dissociated into their corresponding dimeric units. Conformational analysis of neuronal UCPs revealed a transition from highly associated to less-constrained helices (Figure 2). At the low SDS concentration range, Trp residues remain in relatively less polar environments, as shown by the λmax values of ∼331–333 nm (Figure 3). The second transition occurred at higher SDS concentrations (∼10 mM), where neuronal UCP dimers were further dissociated into a mixture of loose dimers and monomers. During this transition, neuronal UCPs exhibited small overall conformational changes and their Trp residues were exposed to more polar environments (λmax ∼ 338–339 nm).
Effects of FAs on the proton and chloride transports mediated by neuronal UCPs
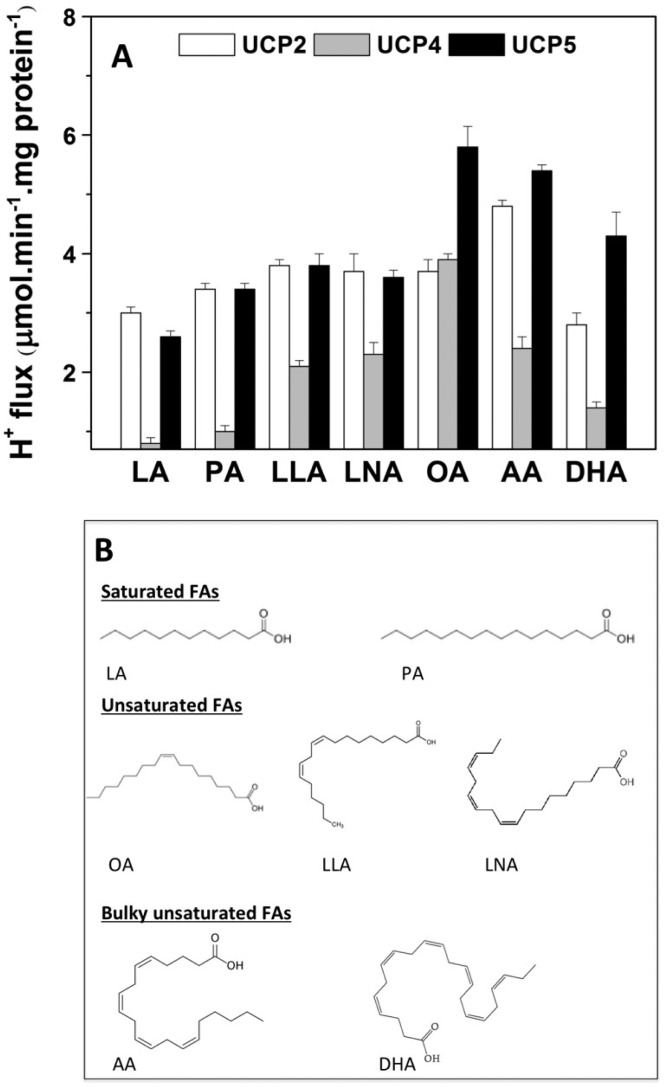
In an effort to characterize the FA-activation mechanism of the UCP-mediated proton transport pathway, we assessed the effects of various FAs on UCP-mediated ion transport rates (Figures 4 and 5). In addition, CD spectroscopy was used to examine the FA–UCP interactions (Supplementary Figures S1 and S2). In the present study, seven different FAs were used, which varied in their hydrophobicity, structure and hydrocarbon chain length lauric acid (LA), palmitic acid (PA), oleic acid (OA), linoleic acid (LLA), linolenic acid (LNA), arachidonic acid (AA) and docohexaneoic acid (DHA; Figure 4B).
Figure 4. Effect of FAs on the proton transport activity of associated neuronal UCPs in liposomes.
(A) Average corrected proton transport rates mediated by neuronal UCPs in L-α-lecithin vesicles. FAs were used at the concentration of 50 μM to activate the proton transport by UCPs. The reported rates are the means of 5–10 independent measurements±the S.E.M.s. The phospholipid concentration was ∼20 mg/ml; the final protein content in the liposomes was 2–5 μg/mg of lipid. The final proton transport rates were corrected for the total protein content in the liposomes. (B) Structure of FAs tested in the present study.
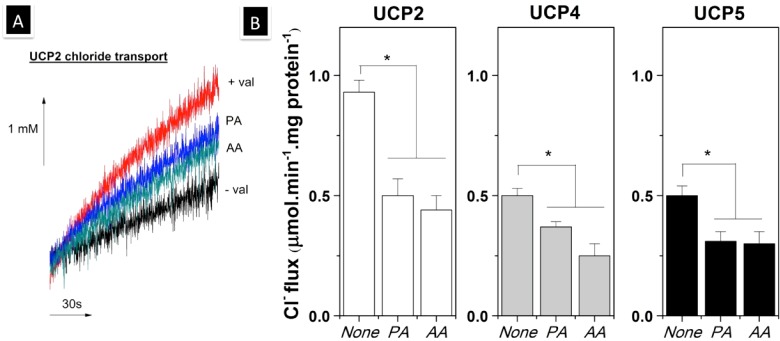
Figure 5. Effect of FAs on the chloride transport activity of associated neuronal UCPs in liposomes.
(A) Chloride influx mediated by UCP2 in the absence (+val) and presence of PA (PA) or AA (AA). The non-UCP2 specific flux was shown for comparison (−val). (B) Average corrected chloride transport rates mediated by neuronal UCPs in L-α-lecithin vesicles. FAs were used at the concentration of 50 μM to examine their effect on the chloride transport by UCPs. The reported rates are the means of 5–10 independent measurements±the S.E.M.s. The phospholipid concentration was ∼20 mg/ml; the final protein content in the liposomes was 2–5 μg/mg of lipid. The final chloride transport rates were corrected for the total protein content in the liposomes. A one-way ANOVA statistical test was performed to determine the statistical significance of data, and P-values were obtained. P<0.05 when comparing the chloride transport mediated by neuronal UCPs in the absence and presence of FAs (*).
Results from CD spectroscopy and proton transport experiments showed that all FAs interacted with UCPs and differentially activated their proton transport activities (Figure 4; Supplementary Figures S1 and S2). The UCP-mediated proton transport rates varied between 1–6 μmol·min−1·mg protein−1 [or 0.5–3 protons per second for each UCP protein], depending on the specific UCP and FA (Figure 4). A comparative proton transport rate in all neuronal UCPs shows that with the exception of DHA, unsaturated FAs are generally stronger proton transport activators for neuronal UCPs than saturated FAs (LA and PA; Figure 4). As revealed by far- and near-UV CD spectra, minor conformational changes in UCPs imply that, among the tested FAs, LA had the weakest interaction with these proteins (Supplementary Figures S1 and S2). In general, based on the enhanced negative double maxima ellipticities at the 208 and 222 nm wavelengths in the far-UV CD spectra, it is apparent that FA–UCP interactions induce less densely packed protein conformations (Supplementary Figure S1). Near-UV CD spectra of neuronal UCPs also show an enhanced overall signal intensity of UCPs in the presence of FAs (except for LA; Supplementary Figure S2). Among neuronal UCPs, UCP4 exhibited the lowest proton transport rates (except in the case of OA activation; Figure 4). In the presence of the same FA activator, UCP4-mediated proton transport rates were ∼2–3 fold lower than those of UCP2 and UCP5 (Figure 4). However, in the presence of OA, UCP4 exhibited its highest proton transport rate at ∼4 μmol·min−1·mg protein−1, similar to that of UCP2 (Figure 4). OA was also the strongest proton transport activator for UCP5 (Figure 4). On the other hand, AA was the most potent proton transport activator for UCP2 (Figure 4). Using planar lipid bilayer systems, it was also shown that AA is a strong FA activator for the proton transport activity of UCP2 [35].
In addition to proton transport, chloride transport was also detected for reconstituted neuronal UCPs (Figure 5). Overall, all neuronal UCPs transported chloride ions at lower rates than protons (Figures 4 and 5). UCP2 transported chlorides at a higher rate (∼1 μmol·min−1·mg·protein−1) compared with UCP4 and UCP5 (∼ 0.5 μmol·min−1·mg·protein−1; Figure 5). To further examine the ion transport mechanism of UCPs, we also examined the effects of FAs (PA and AA) on the chloride transport activity of neuronal UCPs in proteoliposomes. At the concentration of 50 μM, both PA and AA partially inhibited chloride transport by all neuronal UCPs (Figure 5). AA, a more potent UCP-proton transport activator, displayed a slightly stronger inhibitory effect on the chloride transport of all UCPs (Figures 4 and 5). Consequently, our experimental results suggest that FAs interact with UCPs and differentially modulate their proton (activated) and chloride (inhibited/reduced) transport activities.
DISCUSSION
Novel recombinant expression for studying the structure and function of neuronal UCPs
Molecular and structural studies of UCPs have been challenging due to difficulties in obtaining sufficient yield of functional proteins reconstituted in lipid membranes [36]. While facilitating the production of large quantities of pure proteins, bacterial expression systems also pose challenges for expression of heterologous proteins, especially membrane proteins such as UCPs [9–11,36,37]. Previous expression of mammalian UCPs in E. coli resulted in the production of protein inclusion bodies [10,11]. Once extracted from inclusion bodies and reconstituted in model membranes, the proteins might be only partially folded into their functional forms [10,11]. To improve the folding quality of the neuronal UCPs, we utilized the PelB leader sequence to target the proteins to bacterial membranes, as we have done previously for UCP1 [22]. This novel expression system proved to be successful for neuronal UCPs as well (Figure 1A, inset). The neuronal UCPs that were targeted to the bacterial membranes were folded, highly helical and stable in both detergents and liposomes (Figures 1 and 2; Table 1). In addition, all reconstituted neuronal UCPs displayed putative UCP ion transport functions (Figures 4 and 5). It is important to note that UCP5, when folded from inclusion bodies in a previous study, exhibited chloride transport in L-α-PC (phosphatidylcholine) liposomes only when cardiolipin (CL) was included with the membrane lipid [11]. In the current study, reconstituted UCP5 transported chloride in L-α-PC liposomes in the absence of added CL (Figure 5). Based on our previous studies, the presence of CL can enhance the ion transport activity of UCP5 as well as other UCPs [11,22]. Given that E. coli membranes contain ∼5% CL, UCP5 localized to the bacterial membranes could associate with CL and the intact UCP5–CL complex could be maintained and carried into liposomes during the purification and reconstitution processes [38]. It is also possible that UCP5 extracted from bacterial membranes possesses a different conformation from that of UCP5 extracted from inclusion bodies and therefore facilitated chloride transport. Consequently, the fusion of the PelB leader sequence, together with the use of mild auto-induction expression method, allows for the preparation of pure neuronal UCPs in high yield that are suitable for structural and functional studies.
Understanding the self-association of neuronal UCPs in lipid membranes
Self-association of mitochondrial carrier family (MCF) proteins has been reported previously in several studies [22,39–41]. As with our previous study on UCP1 [22], the predominant molecular form of neuronal UCPs in liposomes, observed in the current study, was tetrameric (Figure 1A, inset; Supplementary Table S1). Tetrameric UCPs were capable of transporting both protons and chloride ions across lipid bilayers (Figures 4 and 5). The effect of SDS titration on the tetrameric forms of neuronal UCPs, as revealed by their electrophoretic and mass spectrometric analyses, further confirms our previously proposed molecular model for UCP1 multimerization (Figure 6) [22]. In this molecular model, neuronal UCP tetramers can be easily dissociated into loosely-bound dimeric units at low SDS concentrations (0.6–1 mM; Figures 2, 3 and 6). Within each UCP dimer, monomers interact strongly and the dimeric unit cannot be completely dissociated to monomers even at a high concentration of SDS (∼83 mM; Figure 6). Other studies in the past also reported the existence of UCP1 dimers [39,42] and the dimeric unit of UCPs (UCPs 1, 2, 4 and 5) could be physiologically significant [22]. The dimerization of other mitochondrial carriers has been reported to be important for the metabolite transport function of these proteins [43–45]. In our previous study, neuronal UCPs transported protons and chloride in their monomeric forms [11]. The results of the current study imply that UCPs can also transport protons (FA-activated) in multimeric forms.
Figure 6. Dissociation model of neuronal UCP tetramers.
Neuronal UCP tetramers are composed of two loosely associated homo-dimers. The tetramers can be dissociated into homo-dimers at low SDS concentrations ([SDS] ∼ CMC). UCP tetramers were dissociated at ∼1 mM SDS to form dimers. UCP monomers interact and tightly associate with each other through direct protein–protein interactions and/or protein interactions mediated by lipids, to form a tight dimer. These tight dimers can be partially unfolded/dissociated at higher concentrations of SDS to form loose dimers and/or monomers. Loose dimers may be fully separated to monomers at much higher concentrations of SDS.
The self-association of neuronal UCPs and other UCP homologues can occur via protein–protein interaction or protein—lipid–protein interaction. Amino acid sequence analysis revealed the existence of conserved GxxxG motifs in all UCPs [46]. These motifs are commonly found in TM (transmembrane) domains that are involved in helix–helix associations; their presence in the TM domains of UCPs provides indirect support for the likelihood of UCP self-association, discussed above (Table 3) [46]. In addition, sequence alignment of UCPs demonstrated many conserved amino acid sequences that could participate in protein self-association (Table 3). In our previous study of UCP2 mutants, two positively charged residues located in TM2 domain at the protein/IMS (intermembrane space) interface (Arg96 and Lys104) were shown to participate in UCP2 protein–protein interactions [47]. These amino acids are conserved and involved in formation of a salt-bridge network of UCPs in the protein/IMS interface, which have been suggested to play an important role in regulating the substrate-binding and transport of UCPs (Supplementary Figure S3) [19]. In addition to protein–protein interaction, UCP association may also occur through protein–lipid–protein interaction. In our previous study, the mitochondrial lipid CL was shown to influence UCP1 self-association and enhancing its proton transport activity [22]. Therefore, given its abundance in the brain mitochondrial membranes, CL could play an essential role in the molecular association and ion transport activity of neuronal UCPs.
Table 3. Similarities and differences in expression profiles, amino acid sequences and common motifs of neuronal UCPs*.
| Conserved/semi-conserved sequences | |||
|---|---|---|---|
| UCP2 | UCP4 | UCP5 | |
| MCF conserved sequences [48] | Px[D/E]xx[K/R]x[R/K]-20-30 aa-[D/E]Gxxxx[W/Y/F]-[K/R]G | ||
| Inter-helical association motif [46] | GxxxG | ||
| Proton transport residue [54] | Asp28 | Glu34 | Glu55 |
| Charged residues in chloride-binding region [55] | Asp35, Lys38, Arg88, Asp138, Lys141, Lys239 and Asp236 | Asp41, Lys44, Arg97, Asp149, Lys152, Lys253 and Asp250 | Asp60, Lys66, Arg113, Asp163, Lys166, Lys260 and Asp257 |
| PN-binding residues [9] | Arg88, Arg185, Arg279 | Arg97, Arg199, Arg299 | Arg113, Arg206, Arg305 |
| Unique biochemical/biophysical properties | |||
| UCP2 | UCP4 | UCP5 | |
| Expression profile | Expression in various tissues and organs (brain, muscle, liver, heart, kidney, pancreas) | Primarily expressed in the brain and the CNS (neurons, astrocytes, Purkinje cells) | Primarily expressed in the brain and to a lesser extent in testis, uterus, kidney, lung, stomach |
| Sequence identity to UCP1 | 59% | 34% | 30% |
| Proton transport rate (μmol·min−1·mg·protein−1) | ∼2–5 (strongest activator: AA) | ∼1–4 (strongest activator: OA) | ∼2–6 (strongest activator: OA) |
| Chloride transport rate (μmol·min−1·mg·protein−1) | ∼1 | ∼0.5 | ∼0.5 |
| Interaction with CL [11] | CL induced higher UCP2-mediated H+ flux | CL had no effect on UCP4-mediated H+ flux | CL induced higher UCP5-mediated H+ flux |
FA binding and ion transport of neuronal UCPs
Information about the structure and physiological function of brain-specific UCP4 and UCP5 is scarce as compared with other UCPs. With the goal of demystifying the biological function(s) of UCPs in the CNS and characterizing their molecular physiology using a biophysical approach, the current study reports new findings on the structures and ion transport activities of neuronal UCPs, especially UCPs 4 and 5. The data underscore the common structural and functional features of all neuronal UCPs (Table 3). On the other hand, each UCP exhibits variations in its ion (proton and chloride) transport and FA-binding properties, which could be relevant to the specific physiological roles of these proteins in the CNS (Table 3).
All three neuronal UCPs display self-associated helical conformations that transport proton (FA-activated) and chloride (FA-inhibited) ions across lipid membranes (Figures 1–5; Table 3). As members of the MCF, neuronal UCPs share a conserved motif Px[D/E]xx[K/R]x[R/K]-20–30 aa-[D/E]Gxxxx[W/Y/F]-[K/R]G spanning from the odd-numbered TMs (1, 3 and 5) to the even-numbered TMs (2, 4 and 6) [4,8,48]. This signature motif may play an important role in forming general funnel-shaped channel-like structures spanning the inner mitochondrial membranes (Table 3) [30,48]. In addition, neuronal UCPs share UCP-specific conserved amino acids, including the salt-bridge networks at the IMS/protein and mitochondrial matrix/protein interfaces, the GxxxG motifs involved in inter-helical association, the conserved amino acid residues in proton and chloride transport and the conserved arginine residues in PN-binding sites (Table 3). Consequently, the conserved amino acid sequences and motifs in UCPs can translate to their similarity in self-association and ion transport function.
Despite similarity in their general structures and functions, neuronal UCPs displayed distinct proton and chloride transport activities in the presence of various FA activators. For instance, UCP2 transported protons at the highest rate in the presence of AA, whereas UCP4 and UCP5 achieved their maximum proton transport rates in the presence of OA (Figure 4; Table 3). These differences could be related to the specificity in binding of FAs to UCPs. Whereas a specific site for FA–UCP interaction has not been identified, the results from the present study can shed light on understanding the differences in FA-binding regions of neuronal UCPs. Since FAs with longer hydrophobic chains (C16 and above) activated the proton transport of neuronal UCPs more strongly than shorter chain FAs (C12 and below), it is plausible that specific FA-binding regions, with a threshold hydrophobicity, exists in UCPs. These FA-binding hydrophobic regions might also possess a specific geometry that can better accommodate certain shapes of FAs over the others; for example, long chain compared with short chain or bulky compared with linear FAs. It has been also proposed that stable hydrophobic binding between FAs and UCPs is required for proton transport function of the proteins [49]. Therefore, long chain FAs could stay associated with UCPs due to the specific hydrophobic interactions above the threshold hydrophobicity and might also act as a shuttle to translocate protons from the IMM to the mitochondrial matrix. In contrast, in the case of short chain FAs, hydrophobic interactions with UCPs are close to or below threshold hydrophobicity, resulting in weak interactions that lead to FA anions being dissociated from UCPs before becoming protonated. In addition to the possibility of the existence of hydrophobic regions with threshold hydrophobicity, the geometry of FAs might also play a role in their specific interaction with UCPs. For example, with one double bond, the C18-chain OA has a high conformational flexibility (Figure 4). On the other hand, the C20-chain AA possesses a much lower conformational flexibility and a more bulky (and compact) shape due to the presence of four, configurationally restricted, conjugated double bonds (Figure 4). Therefore, the FA-binding hydrophobic region of UCP2 could be broader than those of UCPs 4 and 5 to better accommodate bulky, long-chain FAs, such as AA, rather than more linear, long-chain FAs, such as OA. Overall, in addition to the hydrophobic nature and specific geometry of the FA-binding sites, the configuration of FAs can also influence the specificity of FA binding to UCPs.
Neuronal UCPs: ion transport mechanism and specific physiological roles
At the moment, two main questions remain unanswered regarding the proton transport mechanism of UCPs and their physiological functions in the mitochondria. Our data suggest that the specific hydrophobic interaction between FAs and UCPs plays an important role in the proton flux mediated by the proteins. As detected through far- and near-UV CD spectra, all FAs interacted with UCPs and induced conformational changes in the proteins. Moreover, compared with short chain FAs, long chain FAs (≥ C16) induced drastic conformational changes and higher proton transport rates in neuronal UCPs (Figures S1 and S2). Similar results were obtained for proton transport in a patch-clamp study of UCP1 in BAT mitochondria [49]. In comparison with their proton transport activity, all neuronal UCPs transported chloride at much lower rates. Interestingly, the chloride transport by all neuronal UCPs was partially inhibited by FAs such as PA and AA. Therefore, FAs could have dual roles in modulating the function of UCPs, as an activator of proton transport and as an inhibitor of chloride transport. It is also possible that both transport pathways might share one or more key amino acids. Identifying these amino acid residues could be essential for elucidating the ion transport mechanism of UCPs, in general, and their proton transport mechanism in particular.
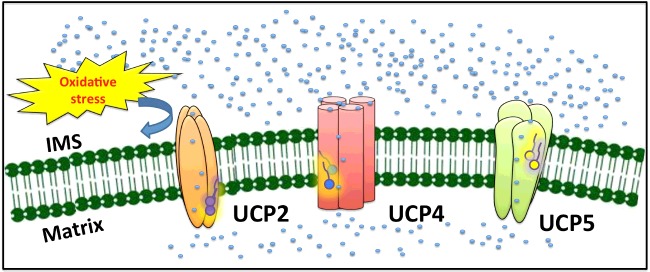
The proton transport function of UCPs in the CNS reduces the electrochemical potential across the IMM, which in turn leads to the reduction of ROS production [4,12–14,50]. However, the question regarding the mechanism of UCP activation to transport protons is not fully answered. As discussed earlier, long chain FAs and their oxidized derivatives can interact with UCPs through hydrophobic interactions and activate proton transport [16,47,49]. A hypothesis for the activation mechanism of proton transport in UCPs suggests the oxidative stress-induced lipolysis pathway [4]. Specifically, under oxidative stress, lipolysis (via mitochondrial phospholipase A2) in the mitochondrial membranes can release free FAs (and their oxidized derivatives). These substrates then interact with neuronal UCPs and activate their proton transport pathway in a feedback mechanism (Figure 7) [51,52]. In other words, oxidative stress generates ROS products and free FAs and their oxidized forms as substrates for binding to UCPs and activating their proton transport function. Proton transport in turn leads to the reduction of ROS production and lipolytic activity of phospholipase A2. Once a lower threshold level in FA concentration is reached, UCPs’ proton transport function becomes deactivated, which results in overproduction of ROS species and the cycle is repeated. We observed long-chain FAs, such as OA and AA, to be potent activators of the proton transport activity of neuronal UCPs (Figure 4). These FAs are abundantly found in the CNS and could play a potential role in regulating the function of UCPs in these tissues [53]. The variation in potencies of FAs for activating proton transport in each UCP could imply the existence of distinct and specific hydrophobic FA-binding domains in each protein. As discussed previously, AA was the most potent proton transport activator for UCP2, whereas OA was the most potent proton transport activator for UCP4 and UCP5 (Figure 4; Table 3). In addition, UCP2 transported chloride ∼2-fold faster than UCPs 4 and 5 (Figure 5; Table 3). The difference between UCP2 compared with UCP4 and UCP5 is also reflected in their sequence homology and tissue expression profiles (Table 3). UCP2 is not only found in CNS tissues but is also expressed in liver, pancreas, muscle and other human tissues (Table 3). On this basis, the specific FA activation of UCPs’ proton transport could be linked to the tissue in which each protein is expressed and the availability of FAs in those tissues. Analysis of the FA components of the brain mitochondrial lipids reveals a higher content of OA over AA and DHA (53). Therefore, based on the relative availability of FAs in the brain tissues, it can be speculated that in these tissues, UCP4 and UCP5 are more specifically activated (and, therefore, more capable in transporting protons) than UCP2.
Figure 7. Proposed model of stress-induced proton transport mechanism of neuronal UCPs.
Under oxidative stress, lipolysis can be activated through enzymes such as mitochondrial phospholipase A2. The lipolysis of mitochondrial phospholipid releases free FAs and their oxidized derivatives (e.g., FAOOH), which can directly interact with UCPs and activate the proton transport. Each neuronal UCP might possess a specific FA-binding site. The FA-binding sites of neuronal UCPs could have an optimal binding affinity for FAs that are available in specific expression tissues. The proton transport mediated by neuronal UCPs can help reduce the membrane potential across the IMM and consequently lower ROS production.
Among neuronal UCPs, UCP4 transported protons and chloride at the lowest rates (Figures 4 and 5). In addition, UCP4 is the only UCP whose ion transport activities were unaffected by CL, a mitochondria-specific lipid that is localized at the cristae membrane of the IMM [11]. These unique biophysical properties of UCP4 could be related to its location within the IMM and its physiological roles. A recent microscopy study revealed that UCP4 is preferentially localized in the IMM inner boundary membrane section, whereas ATP synthase is more centrally located in the cristae membrane section [17]. This local separation of proteins precludes a significant role for UCP4 in uncoupling phosphorylation from proton pumping in the ETC (electron transport chain), but is not in conflict with the original view about the role of UCP4 in modulating excessive proton gradient that is associated with ROS production [17].
In addition to a proton transport-mediated uncoupling effect, UCPs in neurons may indirectly achieve their protective roles. In a recent study, a carrier/exchanger function has been proposed for UCP2 in transporting C4 metabolites out of the mitochondria in exchange for phosphate and proton ions, thereby regulating glucose and glutamine oxidation and, indirectly, reducing ROS production [20]. It is therefore possible that UCP2 possesses different functional modes (i.e. ion transporter and C4 substrate carrier/exchanger) under different physiological conditions. In analogy, despite common structural and functional features shared among neuronal UCPs, UCP4 and UCP5 could also possess other functional modes in addition to their ion transport activities.
Acknowledgments
We are grateful to Ms Tu Hoang for her technical assistance in purifying neuronal UCPs.
Abbreviations
- AA
arachidonic acid
- AAC
ADP/ATP carrier
- BAT
brown adipose tissue
- CMC
critical micelle concentration
- CNS
central nervous system
- CL
cardiolipin
- C8E4
octyltetraoxyethylene
- DHA
docohexanenoic acid
- FA
fatty acid
- IMM
inner mitochondrial membrane
- IMS
intermembrane space
- LA
lauric acid
- LDAO
lauryldimethylamine-oxide
- MCF
mitochondrial carrier family
- OA
oleic acid
- OG
octyl glucoside
- PA
palmitic acid
- PC
phosphatidylcholine
- PN
purine nt
- ROS
reactive oxygen species
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- SPQ
6-methoxy-N-(3-sulfopropyl)quinolinium
- TEA
tetraethylammonium
- TEV
tobacco etch virus
- TM
transmembrane
- UCP
uncoupling protein
AUTHOR CONTRIBUTION
Tuan Hoang performed molecular cloning, protein expression and purification, reconstitution, spectroscopic and ion transport experiments and analysis for all neuronal UCPs and assisted in drafting the article. Miljan Kuljanin performed MS experiment and analysis. Matthew Smith provided technical advice and expertize in molecular cloning and expression of UCPs and helped plan and draft the article. Masoud Jelokhani-Niaraki conceived the study and participated in data analysis and drafting of the article. All authors have read and approved the final manuscript.
FUNDING
This work was supported by the Canada Foundation for Innovation [grant numbers CFI6786 (to M.J.-N.) and CFI11292 (to M.D.S.)]; the Natural Sciences and Engineering Research Council of Canada [grant numbers DG250119 (to M.J.-N.) and DG312143 (to M.D.S.)]; the NSERC Alexander Graham Bell Doctoral scholarship [grant number CGSD3-409084-2011 (to T.H.)]; and the Ontario Graduate Scholarship (to T.H.).
References
- 1.Nicholls D.G., Bernson V.S., Heaton G.M. The identification of the component in the inner membrane of brown adipose tissue mitochondria responsible for regulating energy dissipation. Exp. Suppl. 1978;32:89–93. doi: 10.1007/978-3-0348-5559-4_9. [DOI] [PubMed] [Google Scholar]
- 2.Andrews Z.B., Diano S., Horvath T.L. Mitochondrial uncoupling proteins in the CNS: in support of function and survival. Nat. Rev. Neurosci. 2005;6:829–840. doi: 10.1038/nrn1767. [DOI] [PubMed] [Google Scholar]
- 3.Chan C.B., De Leo D., Joseph J.W., McQuaid T.S., Ha X.F., Xu F., Tsushima R.G., Pennefather P.S., Salapatek A.M., Wheeler M.B. Increased uncoupling protein-2 levels in beta-cells are associated with impaired glucose-stimulated insulin secretion: mechanism of action. Diabetes. 2001;50:1302–1310. doi: 10.2337/diabetes.50.6.1302. [DOI] [PubMed] [Google Scholar]
- 4.Echtay K.S. Mitochondrial uncoupling proteins—what is their physiological role? Free Radic. Biol. Med. 2007;43:1351–1371. doi: 10.1016/j.freeradbiomed.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Mao W., Yu X.X., Zhong A., Li W., Brush J., Sherwood S.W., Adams S.H., Pan G. UCP4, a novel brain-specific mitochondrial protein that reduces membrane potential in mammalian cells. FEBS Lett. 1999;443:326–330. doi: 10.1016/S0014-5793(98)01713-X. [DOI] [PubMed] [Google Scholar]
- 6.Sanchis D., Fleury C., Chomiki N., Goubern M., Huang Q., Neverova M., Grégoire F., Easlick J., Raimbault S., Lévi-Meyrueis C., et al. BMCP1, a novel mitochondrial carrier with high expression in the central nervous system of humans and rodents, and respiration uncoupling activity in recombinant yeast. J. Biol. Chem. 1998;273:34611–34615. doi: 10.1074/jbc.273.51.34611. [DOI] [PubMed] [Google Scholar]
- 7.Yu X.X., Mao W., Zhong A., Schow P., Brush J., Sherwood S.W., Adams S.H., Pan G. Characterization of novel UCP5/BMCP1 isoforms and differential regulation of UCP4 and UCP5 expression through dietary or temperature manipulation. FASEB J. 2000;14:1611–1618. doi: 10.1096/fj.14.11.1611. [DOI] [PubMed] [Google Scholar]
- 8.Krauss S., Zhang C., Lowell B.B. The mitochondrial uncoupling-protein homologues. Nat. Rev. Mol. Cell Biol. 2005;6:248–261. doi: 10.1038/nrm1592. [DOI] [PubMed] [Google Scholar]
- 9.Klingenberg M., Echtay K.S. Uncoupling proteins: the issues from a biochemist point of view. Biochim. Biophys. Acta. 2001;1504:128–143. doi: 10.1016/S0005-2728(00)00242-5. [DOI] [PubMed] [Google Scholar]
- 10.Ivanova M.V., Hoang T., McSorley F.R., Krnac G., Smith M.D., Jelokhani-Niaraki M. A comparative study on conformation and ligand binding of the neuronal uncoupling proteins. Biochemistry. 2009;49:512–521. doi: 10.1021/bi901742g. [DOI] [PubMed] [Google Scholar]
- 11.Hoang T., Smith M.D., Jelokhani-Niaraki M. Toward understanding the mechanism of ion transport activity of neuronal uncoupling proteins UCP2, UCP4, and UCP5. Biochemistry. 2012;51:4004–4014. doi: 10.1021/bi3003378. [DOI] [PubMed] [Google Scholar]
- 12.Echtay K.S., Roussel D., St-Pierre J., Jekabsons M.B., Cadenas S., Stuart J.A., Harper J.A., Roebuck S.J., Morrison A., Pickering S., et al. Superoxide activates mitochondrial uncoupling proteins. Nature. 2002;415:96–99. doi: 10.1038/415096a. [DOI] [PubMed] [Google Scholar]
- 13.Chu A.C., Ho P.W., Kwok K.H., Ho J.W., Chan K., Liu H., Kung M.H., Ramsden D.B., Ho S. Mitochondrial UCP4 attenuates MPP+ - and dopamine-induced oxidative stress, mitochondrial depolarization, and ATP deficiency in neurons and is interlinked with UCP2 expression. Free Radic. Biol. Med. 2009;46:810–820. doi: 10.1016/j.freeradbiomed.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 14.Kwok K.H., Ho P.W., Chu A.C., Ho J.W., Liu H., Yiu D.C., Chan K., Kung M.H., Ramsden D.B., Ho S. Mitochondrial UCP5 is neuroprotective by preserving mitochondrial membrane potential, ATP levels, and reducing oxidative stress in MPP+ and dopamine toxicity. Free Radic. Biol. Med. 2010;49:1023–1035. doi: 10.1016/j.freeradbiomed.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 15.Smorodchenko A., Rupprecht A., Fuchs J., Gross J., Pohl E.E. Role of mitochondrial uncoupling protein 4 in rat inner ear. Mol. Cell. Neurosci. 2011;47:244–253. doi: 10.1016/j.mcn.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Malingriaux E.A., Rupprecht A., Gille L., Jovanovic O., Jezek P., Jaburek M., Pohl E.E. Fatty acids are key in 4-hydroxy-2-nonenal-mediated activation of uncoupling proteins 1 and 2. PLoS One. 2013;8:e77786. doi: 10.1371/journal.pone.0077786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klotzsch E., Smorodchenko A., Löfler L., Moldzio R., Parkinson E., Schütz G.J., Pohl E.E. Superresolution microscopy reveals spatial separation of UCP4 and F0F1-ATP synthase in neuronal mitochondria. Proc. Natl. Acad. Sci. U.S.A. 2015;112:130–135. doi: 10.1073/pnas.1415261112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamaguchi H., Jelokhani-Niaraki M., Kodama H. Second transmembrane domain of human uncoupling protein 2 is essential for its anion channel formation. FEBS Lett. 2004;577:299–304. doi: 10.1016/j.febslet.2004.09.070. [DOI] [PubMed] [Google Scholar]
- 19.Robinson A.J., Overy C., Kunji E.R.S. The mechanism of transport by mitochondrial carriers based on analysis of symmetry. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17766–17771. doi: 10.1073/pnas.0809580105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vozza A., Parisi G., De Leonardis F., Lasorsa F.M., Castegna A., Amorese D., Marmo R., Calcagnile V.M., Palmieri L., Ricquier D., et al. UCP2 transports C4 metabolites out of mitochondria, regulating glucose and glutamine oxidation. Proc. Natl. Acad. Sci. U.S.A. 2014;111:960–965. doi: 10.1073/pnas.1317400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jelokhani-Niaraki M., Ivanova M.V., McIntyre B.L., Newman C.L., McSorley F.R., Young E.K., Smith M.D. A CD study of uncoupling protein-1 and its transmembrane and matrix-loop domains. Biochem. J. 2008;411:593. doi: 10.1042/BJ20071326. [DOI] [PubMed] [Google Scholar]
- 22.Hoang T., Smith M.D., Jelokhani-Niaraki M. Expression, folding and proton transport activity of human uncoupling protein-1 (UCP1) in lipid membranes: evidence for associated functional forms. J. Biol. Chem. 2013;288:36244–36258. doi: 10.1074/jbc.M113.509935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Studier F.W. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 24.Whitmore L., Wallace B.A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lees J.G., Miles A.J., Wien F., Wallace B.A. A reference database for circular dichroism spectroscopy covering fold and secondary structure space. Bioinformatics. 2006;22:1955–1962. doi: 10.1093/bioinformatics/btl327. [DOI] [PubMed] [Google Scholar]
- 26.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baú D., Martin A.J.M., Mooney C., Vullo A., Walsh I., Pollastri G. Distill: a suite of web servers for the prediction of one-, two- and three-dimensional structural features of proteins. BMC Bioinformatics. 2006;7:402. doi: 10.1186/1471-2105-7-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reference deleted.
- 30.Berardi M.J., Shih W.M., Harrison S.C., Chou J.J. Mitochondrial uncoupling protein 2 structure determined by NMR molecular fragment searching. Nature. 2011;476:109–113. doi: 10.1038/nature10257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenfield N.J., Hitchcock-DeGregori S.E. Conformational intermediates in the folding of a coiled-coil model peptide of the N-terminus of tropomyosin and alpha alpha-tropomyosin. Protein Sci. 1993;2:1263–1273. doi: 10.1002/pro.5560020809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper T.M., Woody R.W. The effect of conformation on the CD of interacting helices: a theoretical study of tropomyosin. Biopolymers. 1990;30:657–676. doi: 10.1002/bip.360300703. [DOI] [PubMed] [Google Scholar]
- 33.Salom D., Hill B.R., Lear J.D., DeGrado W.F. pH-dependent tetramerization and amantadine binding of the transmembrane helix of M2 from the influenza A virus. Biochemistry. 2000;39:14160–14170. doi: 10.1021/bi001799u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campbell J.H., Hoang T., Jelokhani-Niaraki M., Smith M.D. Folding and self-association of atTic20 in lipid membranes: implications for understanding protein transport across the inner envelope membrane of chloroplasts. BMC Biochem. 2014;15:462. doi: 10.1186/s12858-014-0029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beck V., Jaburek M., Demina T., Rupprecht A., Porter R.K., Jezek P., Pohl E.E. Polyunsaturated fatty acids activate human uncoupling proteins 1 and 2 in planar lipid bilayers. FASEB J. 2007;21:1137–1144. doi: 10.1096/fj.06-7489com. [DOI] [PubMed] [Google Scholar]
- 36.He Y., Wang K., Yan N. The recombinant expression systems for structure determination of eukaryotic membrane proteins. Protein Cell. 2014;5:658–672. doi: 10.1007/s13238-014-0086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jabůrek M., Garlid K.D. Reconstitution of recombinant uncoupling proteins UCP1, -2, and -3 have similar affinities for ATP and are unaffected by coenzyme Q10. J. Biol. Chem. 2003;278:25825–25831. doi: 10.1074/jbc.M302126200. [DOI] [PubMed] [Google Scholar]
- 38.Renner L.D., Weibel D.B. Cardiolipin microdomains localize to negatively curved regions of Escherichia coli membranes. Proc. Natl. Acad. Sci. U.S.A. 2011;108:6264–6269. doi: 10.1073/pnas.1015757108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin C.S., Hackenberg H., Klingenberg E.M. The uncoupling protein from brown adipose tissue mitochondria is a dimer - a hydrodynamic study. FEBS Lett. 1980;113:304–306. doi: 10.1016/0014-5793(80)80614-4. [DOI] [PubMed] [Google Scholar]
- 40.Schroers A., Burkovski A., Wohlrab H., Krämer R. The phosphate carrier from yeast mitochondria. Dimerization is a prerequisite for function. J. Biol. Chem. 1998;273:14269–14276. doi: 10.1074/jbc.273.23.14269. [DOI] [PubMed] [Google Scholar]
- 41.Kotaria R., Mayor J.A., Walters D.E., Kaplan R.S. Oligomeric state of wild-type and cysteine-less yeast mitochondrial citrate transport proteins. J. Bioenerg. Biomembr. 1999;31:543–549. doi: 10.1023/A:1005460810527. [DOI] [PubMed] [Google Scholar]
- 42.Klingenberg M., Appel M. The uncoupling protein dimer can form a disulfide cross-link between the mobile C-terminal SH groups. Eur. J. Biochem. 1989;180:123–131. doi: 10.1111/j.1432-1033.1989.tb14622.x. [DOI] [PubMed] [Google Scholar]
- 43.Zara V., Ferramosca A., Palmisano I., Palmieri F., Rassow J. Biogenesis of rat mitochondrial citrate carrier (CIC): the N-terminal presequence facilitates the solubility of the preprotein but does not act as a targeting signal. J. Mol. Biol. 2003;325:399–408. doi: 10.1016/S0022-2836(02)01236-6. [DOI] [PubMed] [Google Scholar]
- 44.Palmisano A., Zara V., Hönlinger A., Vozza A., Dekker P.J., Pfanner N., Palmieri F. Targeting and assembly of the oxoglutarate carrier: general principles for biogenesis of carrier proteins of the mitochondrial inner membrane. Biochem. J. 1998;333(Pt 1):151–158. doi: 10.1042/bj3330151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Capobianco L., Ferramosca A., Zara V. The mitochondrial tricarboxylate carrier of silver eel: dimeric structure and cytosolic exposure of both N- and C-termini. J. Prot. Chem. 2002;21:515–521. doi: 10.1023/A:1022473504904. [DOI] [PubMed] [Google Scholar]
- 46.Russ W.P., Engelman D.M. The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 47.Hoang T., Matovic T., Parker J., Smith M.D., Jelokhani-Niaraki M. The role of positively charged residues of the second transmembrane domain in the ion transport activity and conformation of human uncoupling protein-2. Biochemistry. 2015;54:2303–2313. doi: 10.1021/acs.biochem.5b00177. [DOI] [PubMed] [Google Scholar]
- 48.Nury H., Dahout-Gonzalez C., Trézéguet V., Lauquin G.J.M., Brandolin G., Pebay-Peyroula E. Relations between structure and function of the mitochondrial ADP/ATP carrier. Annu. Rev. Biochem. 2006;75:713–741. doi: 10.1146/annurev.biochem.75.103004.142747. [DOI] [PubMed] [Google Scholar]
- 49.Fedorenko A., Lishko P.V., Kirichok Y. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell. 2012;151:400–413. doi: 10.1016/j.cell.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brand M.D., Affourtit C., Esteves T.C., Green K., Lambert A.J., Miwa S., Pakay J.L., Parker N. Mitochondrial superoxide: production, biological effects, and activation of uncoupling proteins. Free Radic. Biol. Med. 2004;37:755–767. doi: 10.1016/j.freeradbiomed.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 51.Jabůrek M., Ježek J., Zelenka J., Ježek P. Antioxidant activity by a synergy of redox-sensitive mitochondrial phospholipase A2 and uncoupling protein-2 in lung and spleen. Int. J. Biochem. Cell Biol. 2013;45:816–825. doi: 10.1016/j.biocel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 52.Rousset S., Alves-Guerra M.-C., Mozo J., Miroux B., Cassard-Doulcier A.-M., Bouillaud F., Ricquier D. The biology of mitochondrial uncoupling proteins. Diabetes. 2004;53:130S–135S. doi: 10.2337/diabetes.53.2007.S130. [DOI] [PubMed] [Google Scholar]
- 53.Witting L.A., Harvey C.C., Century B., Horwitt M.K. Dietary alterations of fatty acids of erythrocytes and mitochondria of brain and liver. J. Lipid Res. 1961;2:412–418. [Google Scholar]
- 54.Echtay K.S., Winkler E., Bienengraeber M., Klingenberg M. Site-directed mutagenesis identifies residues in uncoupling protein (UCP1) involved in three different functions. Biochemistry. 2000;39:3311–3317. doi: 10.1021/bi992448m. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y., Tajkhorshid E. Electrostatic funneling of substrate in mitochondrial inner membrane carriers. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9598–9603. doi: 10.1073/pnas.0801786105. [DOI] [PMC free article] [PubMed] [Google Scholar]