The human genome harbors more than a million copies of Alu, a member of retrotransposons called short interspersed nuclear elements (SINEs).1 Alu is about 300 bp in length and originated from 7SL RNA, an RNA polymerase III (Pol III) transcript involved in protein transport. Accordingly, Alu is also transcribed by Pol III. However, most Alu copies are not occupied by the Pol III subunits,2 and the Alu RNA level is much lower than other Pol III transcripts, such as 7SL RNA, tRNAs, and 5S rRNA. Mutations in Alu copies, genomic position effects, and epigenetic silencing are thought to account for the limited Alu transcription, but the whole picture remains obscure.3 Interestingly, Alu RNA directly binds to Pol II and inhibits transcription of many protein-coding genes.4 Moreover, under the heat-shock condition, Alu transcription is upregulated while many Pol II genes are repressed, suggesting a role of Alu RNA in the heat-shock-induced global decrease in the Pol II activity.4
Transition between quiescence and proliferation of a cell also coincides with global changes in transcription: proliferating cells require abundant transcription, whereas quiescent cells are in a more repressed state. CGG triplet repeat-binding protein 1 (CGGBP1) is a DNA binding protein involved in various cellular functions, including proliferation and heat-shock response.5,6 Through binding to the CGG-rich sequence, CGGBP1 activates the transcription of cell-cycle regulators during cell growth, and also activates heat-shock response genes upon heat shock. It was unknown, however, whether CGGBP1 is involved in regulation of the global transcriptional activity upon growth stimulation and heat shock.
In this edition of Cell Cycle, Agarwal et al. have investigated transcriptomes and CGGBP1-binding profiles in serum-starved and serum-stimulated normal human fibroblast cells with and without CGGBP1 depletion.7 They showed that the expression levels of a number of genes are changed upon serum stimulation or CGGBP1 depletion. Their ChIP-seq analysis revealed that, whereas CGGBP1 binds to various genomic regions with some preference to LINE-1 and satellite repeats in quiescent cells, CGGBP1 accumulates at several thousands copies of Alu when proliferation is stimulated. It has been known that certain Alu copies provide binding sites for transcription factors and affect the expression of neighboring genes3. However, the proliferation-induced CGGBP1 binding sites are not located in proximity to the genes of which expression is affected upon growth stimulation, suggesting transcriptional regulation in trans in someway. What is the mediating trans-factor? The most likely candidate the authors found is the Alu RNA. They showed that the binding of CGGBP1 to an Alu-specific sequence between the Pol III promoter motifs interferes with the recruitment of Pol III components, and therefore inhibits the Alu transcription in cis. The Alu downregulation diminishes the inhibitory effect of Alu RNA on Pol II genes, and therefore increases the global transcriptional activity. Inversely, depletion of CGGBP1 increases the Alu RNA and decreases global transcription by Pol II. The authors also showed that CGGBP1 becomes phospoholylataed upon EGF stimulation, and that its phospholylation allows efficient nuclear localization, indicating a phospholylation switch to mediate the extracellular signal.
In summary, CGGBP1 regulates the Pol III transcription of SINE-derived non-coding RNA that regulates the global Pol II transcription (Fig. 1); therefore, CGGBP1 is a pivotal protein that links the Pol II and Pol III transcriptomes in response to growth stimulation. The effect of CGGBP1 is labile to heat shock due to the change in its subnuclear localization,5 which could account for the upregulation of Alu RNA and downregulation of many Pol II genes under heat shock. From an evolutional point of view, it is of particular interest whether the CGGBP1-dependent Pol II regulation in trans through a Pol III transcript(s) is conserved in other species, because most mammalian genomes carry various Pol III-transcribed SINEs, some of which are known to become transcribed upon heat shock and/or to inhibit the Pol II activity.3
Figure 1.
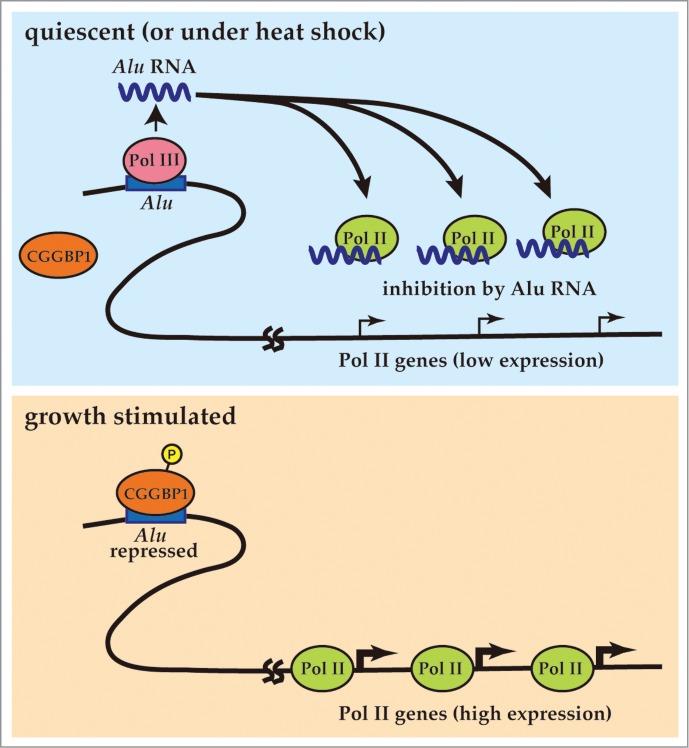
CGGBP1-mediated regulation of global transcription by Pol II. (top) In quiescent cells (or under heat shock), Alu is transcribed by Pol III. The Alu RNA binds to RNA Pol II to inhibit the transcriptional activity. (bottom) Upon growth stimulation by extracellular signals, CGGBP1 becomes phospholylated and binds to the Alu sequences to interfere the Pol III recruitment. Accordingly, the Alu RNA level is decreased and the Pol II genes become more activated.
References
- 1. Batzer MA, et al. Nat Rev Genet 2002; 3:370-379; PMID:11988762; http://dx.doi.org/ 10.1038/nrg798 [DOI] [PubMed] [Google Scholar]
- 2. Oler AJ, et al. Nat Struct Mol Biol 2010; 17:620-628; PMID:20418882; http://dx.doi.org/ 10.1038/nsmb.1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ichiyanagi K. Genes Genet Syst 2013; 88:19-29; PMID:23676707 [DOI] [PubMed] [Google Scholar]
- 4. Mariner PD, et al. . Mol Cell 2008; 29:499-509; PMID:18313387; http://dx.doi.org/ 10.1016/j.molcel.2007.12.013 [DOI] [PubMed] [Google Scholar]
- 5. Singh U, et al. PLoS One 2009; 4:e5050; PMID:19337383; http://dx.doi.org/ 10.1371/journal.pone.0005050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Singh U, et al. BMC Mol Biol 2011; 12:28; PMID:21733196; http://dx.doi.org/ 10.1186/1471-2199-12-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Agarwal P, et al. . Cell Cycle 2014; in this issue [Google Scholar]


