Abstract
Objectives
The aim of this study was to characterize the prevalence and patterns of genotypic integrase inhibitor (INI) resistance in relation to HIV-1 clade.
Methods
The cohort comprised 533 INI-naive subjects and 255 raltegravir recipients with viraemia who underwent integrase sequencing in routine care across Europe, including 134/533 (25.1%) and 46/255 (18.0%), respectively, with non-B clades (A, C, D, F, G, CRF01, CRF02, other CRFs, complex).
Results
No major INI resistance-associated mutations (RAMs) occurred in INI-naive subjects. Among raltegravir recipients with viraemia (median 3523 HIV-1 RNA copies/mL), 113/255 (44.3%) had one or more major INI RAMs, most commonly N155H (45/255, 17.6%), Q148H/R/K + G140S/A (35/255, 13.7%) and Y143R/C/H (12/255, 4.7%). In addition, four (1.6%) raltegravir recipients showed novel mutations at recognized resistance sites (E92A, S147I, N155D, N155Q) and novel mutations at other integrase positions that were statistically associated with raltegravir exposure (K159Q/R, I161L/M/T/V, E170A/G). Comparing subtype B with non-B clades, Q148H/R/K occurred in 42/209 (20.1%) versus 2/46 (4.3%) subjects (P = 0.009) and G140S/A occurred in 36/209 (17.2%) versus 1/46 (2.2%) subjects (P = 0.005). Intermediate- to high-level cross-resistance to twice-daily dolutegravir was predicted in 40/255 (15.7%) subjects, more commonly in subtype B versus non-B clades (39/209, 18.7% versus 1/46, 2.2%; P = 0.003). A glycine (G) to serine (S) substitution at integrase position 140 required one nucleotide change in subtype B and two nucleotide changes in all non-B clades.
Conclusions
No major INI resistance mutations occurred in INI-naive subjects. Reduced occurrence of Q148H/R/K + G140S/A was seen in non-B clades versus subtype B, and was explained by the higher genetic barrier to the G140S mutation observed in all non-B clades analysed.
Introduction
Raltegravir was the first integrase inhibitor (INI) to receive approval for the treatment of HIV infection, marking an important therapeutic advance through the provision of a novel class of antiretroviral agents. Raltegravir inhibits the HIV integrase enzyme at the final integration step, the strand transfer reaction, which otherwise results in the covalent linkage of host and viral DNA.1 Raltegravir has shown efficacy in clinical trials of ART-naive and ART-experienced patients.2,3 However, raltegravir possesses a low to moderate genetic barrier to resistance and single amino acid substitutions in integrase are sufficient to compromise activity.4,5 Previously, the largest series of INI resistance data came from the Phase 3 BENCHMARK 1 and 2 trials, which recruited ART-experienced patients infected primarily with HIV-1 subtype B.3,6 Overall, 105/462 (23%) patients receiving raltegravir with an optimized background regimen experienced virological failure at week 48, of whom 68% had evidence of raltegravir resistance.3,6 Three genotypic resistance pathways were identified: N155H (±E92Q), Q148H/R/K (±G140S) and Y143R (±T97A). A survey of genotypic resistance tests performed through routine care in a referral laboratory in the USA has shown similar integrase mutational profiles, and whereas the study population had unknown ART status and carried nearly uniformly HIV-1 subtype B,7 a more recent study has shown consistent findings in a large INI-experienced cohort.8
Two further INIs, elvitegravir and dolutegravir, have recently been approved. Elvitegravir has a similar genetic barrier to and extensive cross-resistance with raltegravir.9–12 In vitro, the activity of dolutegravir is not compromised by mutations occurring at integrase codon 143 or 155 in isolation, although whether mutations can affect susceptibility in combination with multiple others is the subject of ongoing research. Q148 mutations in combination with other mutations (typically at codon L74, E138 or G140) reduce dolutegravir in vitro susceptibility by 10- to 20-fold.13–16 Findings from clinical studies are consistent with the in vitro susceptibility data, and indicate that the Q148 mutation pathway significantly markedly affects dolutegravir activity among INI-experienced patients, even when the drug is used twice daily.17–23
Data on the genotypic pathways of INI resistance occurring with HIV-1 clades other than subtype B are limited to small cohorts. These studies have suggested that there may be important differences in the resistance profiles of subtype C, CRF01_AE and CRF02_AG, including a high prevalence of naturally occurring resistance in INI-naive subjects with CRF02_AG.24–26 The CORONET project (Common Sequence Repository and Research Network for Integrase Inhibitors) is a European network for surveillance and collaborative research on INI resistance. A common repository is produced from genotypic resistance tests performed at multiple collaborating centres, which provide care for ethnically diverse cohorts. The aim of this analysis, the first of the CORONET database, was to determine the integrase sequence profiles of patients infected with diverse HIV-1 clades that were either INI naive or experiencing viraemia while receiving raltegravir-containing ART, and identify known and novel integrase mutations associated with raltegravir selective pressure, with a specific focus on the impact of HIV-1 clade on mutation patterns.
Methods
Study population
Collection of integrase sequences started in 2008 from nine European laboratories and from the UK HIV Drug Resistance Database (http://128.40.115.16/hivrdb/public/default.asp) and the EuResist Database (http://www.euresist.org). Available clinical data included HIV-1 RNA load and ART status at the time of integrase sequencing. The populations undergoing sequencing were INI-naive patients and patients experiencing a viral load >50 copies/mL while receiving raltegravir-containing ART. Sanger sequencing was performed at each participating centre according to local practice and using in-house protocols. All sequences were re-analysed centrally. The current analysis is based upon all submissions made between 2008 and 2011. The Ethics Committee of the Royal Free Hospital in London approved the analysis of fully anonymous data; written informed consent was not required (06/Q0501/125 and MREC/01/2/10). Separate ethics approvals regulate the UK HIV Drug Resistance Database and the EuResist Database.
HIV-1 clades
HIV-1 clades were assigned by submitting the integrase sequences to the Stanford University HIV Drug Resistance Database (http://hivdb.stanford.edu) and Rega HIV-1 subtyping tools (http://dbpartners.stanford.edu:8080/RegaSubtyping/stanford-hiv/typingtool/). Where results were inconclusive, the NCBI genotyping tool (http://www.ncbi.nlm.nih.gov/projects/genotyping) was used and sequences were characterized by phylogenetic analysis with PhyML. A maximum-likelihood tree was produced using a generalized time-reversible (GTR) model of nucleotide substitution with parameters estimated in PAUP* (bootstrap1000) and integrase reference sequences derived from the Los Alamos Database (http://www.hiv.lanl.gov).
Resistance mutations
Integrase sequences were screened for INI resistance-associated mutations (RAMs) as defined by the Stanford University HIV Drug Resistance Database (February 2014) (http://hivdb.stanford.edu) and the International AIDS Society.27 Major RAMs comprised T66A/I/K, E92Q/G/V, F121Y, G140S/A/C, Y143H/R/C/K, S147G, Q148H/R/K and N155H/S/T. Minor RAMs comprised mutations at codons 51, 74, 95, 97, 114, 128, 138, 145, 146, 151, 153, 157, 163, 230 and 263. Cross-resistance to dolutegravir was classified according to the definition used in the VIKING clinical trials,18 modified to include the potential low-level resistance effects conferred by isolated Q148 mutations (in the absence of other INI RAMs) as indicated by the Stanford University HIV Drug Resistance Database. Based on available data from raltegravir-experienced patients,18 resistance was defined for twice-daily use of dolutegravir. Dolutegravir resistance categories were as follows: (i) no resistance, absence of Q148H/R/K; (ii) low-level resistance, Q148H/R/K in the absence of G140S/A/C, L74I and E138A/K/T; (iii) intermediate-level resistance, Q148H/R/K with one of the mutations G140S/A/C, L74I or E138A/K/T; and (iv) high-level resistance, Q148H/R/K with two or three of the mutations G140S/A/C, L74I and E138A/K/T.
Statistical analysis
Fisher's exact test was used to compare amino acid frequencies across all integrase codons in INI-naive versus raltegravir-experienced subjects and to compare prevalence of INI RAMs in subtype B versus non-B HIV-1 clades. Viral load levels at the time of integrase sequencing were compared between different subgroups by the Wilcoxon rank-sum test. All analyses were performed using STATA version 12.1.
Results
Prevalence and patterns of integrase RAMs
Integrase sequences were analysed from 533 INI-naive subjects and 255 raltegravir recipients with viraemia. The most prevalent HIV-1 clades in the two groups were subtype B (75% and 82%, respectively), subtype C (5% and 4%) and CRF02_AG (4% and 4%) (Figure 1). The other non-B clades were diverse and included complex sequences (2% in each group) for which the clade could not be assigned.
Figure 1.
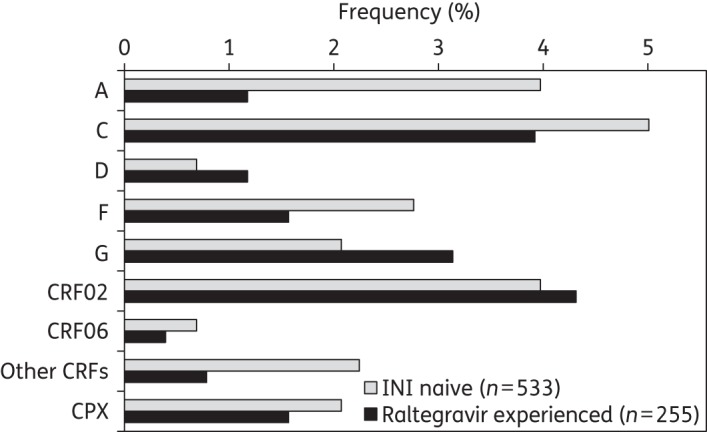
Frequency of non-B HIV-1 clades in INI-naive and raltegravir-experienced patients. Other CRFs comprised CRF01, CRF05 and CRF35.
Among the 533 INI-naive subjects, the median viral load at the time of sequencing was 21 476 copies/mL (IQR 2298–113 308); no patient was found to carry major INI RAMs. Among the 255 raltegravir recipients, the median viral load was 3523 copies/mL (IQR 555–27 793); 113/255 (44.3%) patients showed one or more major INI RAMs. In raltegravir recipients with versus those without major INI RAMs the median viral load was 3420 copies/mL (IQR 1195–20 899) versus 3834 copies/mL (IQR 376–32 342), respectively (P = 0.30). The most commonly observed major INI RAMs were N155H (57, 22.4%), G140S (33, 12.9%), Q148H (28, 11.0%), Q148R (15, 5.9%), E92Q (9, 3.5%) and Y143R (9, 3.5%) (Table 1). The most prevalent mutation profiles were N155H in isolation (45, 17.6%), Q148H/R/K + G140S/A (35, 13.7%) and Y143R/C/H in isolation (12, 4.7%). The three major resistance profiles (codons 143, 148 and 155) did not co-occur in our study. Preferential associations comprised N155H with E92Q, Y143H/R/C with T97A and Q148H/R/K with G140S/A. Conversely, mutations that were mutually exclusive comprised E92Q and T97A with Q148H/R/K, and G140S/A with Y143R/C/H and N155H. An additional four (1.6%) raltegravir recipients, all with subtype B, showed non-classic substitutions at major integrase resistance codons, comprising: (i) E92A, detected with the INI RAMs N155H and T97A (viral load 3170 copies/mL); (ii) S147I (viral load 477 copies/mL); (iii) N155D (viral load 140 copies/mL); and (iv) N155Q (viral load 111 copies/mL).
Table 1.
Major integrase RAMs detected in raltegravir recipients with viraemia
| RAMs, n (%)a |
Pattern, n (%)b | |||||
|---|---|---|---|---|---|---|
| E92Q, 9 (3.5) | G140S/A, 37 (14.5) | Y143R/C/H, 14 (5.5) | S147G, 1 (0.4) | Q148H/R/K, 44 (17.3) | N155H, 57 (22.4) | |
| X | 45 (17.6) | |||||
| X | X | 35 (13.7) | ||||
| X | 12 (4.7) | |||||
| X | X | 8 (3.1) | ||||
| X | 6 (2.4) | |||||
| X | X | 2 (0.8) | ||||
| X | X | 1 (0.4) | ||||
| X | X | 1 (0.4) | ||||
| X | X | X | 1 (0.4) | |||
| X | 1 (0.4) | |||||
| X | 1 (0.4) | |||||
aOverall 113/255 (44.3%) raltegravir recipients had one or more major RAMs. The mutations observed were: E92Q (n = 9); G140S (n = 33) and G140A (n = 4); Y143R (n = 9), Y143C (n = 4) and Y143H (n = 1); Q148H (n = 28), Q148R (n = 15) and Q148K (n = 1); and N155H (n = 57).
bX symbols within each row indicate mutations present in each pattern.
Minor INI RAMs that occurred at a frequency of >2% in raltegravir recipients are shown in Table 2. Of these, L74M, T97A, E138K, V151I and G163R were significantly associated (P < 0.01) with raltegravir exposure.
Table 2.
Minor integrase mutations observed to occur at a frequency of >2% in raltegravir recipients with viraemia
| Mutation | Frequency, n (%) |
P | Positive association with major INI RAMsa | |
|---|---|---|---|---|
| raltegravir naive (n = 533) | raltegravir experienced (n = 255) | |||
| 74I | 35 (6.6) | 13 (5.1) | 0.53 | |
| 74M | 3 (0.6) | 8 (3.1) | 0.007 | |
| 97A | 8 (1.5) | 20 (7.8) | <0.001 | Y143R/C/H |
| 138K | 0 (0.0) | 9 (3.5) | <0.001 | Q148H/R/K |
| 151I | 10 (1.9) | 25 (9.8) | <0.001 | N155H |
| 157Q | 11 (2.1) | 6 (2.4) | 0.80 | |
| 163R | 3 (0.6) | 10 (3.9) | 0.001 | |
| 163E | 29 (5.4) | 7 (2.7) | 0.10 | |
| 230N | 51 (9.6) | 17 (6.7) | 0.22 | |
aPositive association indicated preferential co-occurrence with major INI RAMs.
Novel mutations associated with raltegravir exposure
Comparing sequences from INI-naive and raltegravir-experienced patients, three integrase positions were newly identified statistically as being subjected to selective pressure, involving codons 159 (P = 0.009), 161 (P = 0.004) and 170 (P = 0.009). These mutations occurred in subtype B (K159Q/R, I161L/M/N/T/V and E170A/G) and CRF02_AG (I161L/T). They occurred commonly alongside other mutations, e.g. K159R + I161L occurred with Q148H + G140S, and E170A occurred with N155H and Q148H + G140S (Table S1, available as Supplementary data at JAC Online). In addition, the analysis revealed that raltegravir selective pressure was associated with significantly reduced diversity at integrase positions 24 and 167 (P < 0.001 for both).
Influence of HIV-1 clade on integrase mutational profiles
Comparing subtype B with non-B clades, mutations at codons 148 and 140 occurred in 42 (20.1%) versus 2 (4.3%) (P = 0.009) and 36 (17.2%) versus 1 (2.2%) (P = 0.005), respectively (Table 3). The most prevalent mutations at these positions—G140S and Q148H—were exclusively found in subtype B sequences. G140A occurred in 3 subtype B sequences and 1 subtype G sequence, and Q148R occurred in 13 subtype B sequences, 1 subtype C sequence and 1 subtype G sequence. Conversely, E92Q was significantly less frequent in subtype B (4/209, 1.9%) than in non-B clades (5/46, 10.9%) (P = 0.02), occurring in subtypes C (n = 2), G (n = 1), A (n = 1) and CRF09 (n = 1). In patients with one or more major RAMs, the median viral load at the time of sampling was 3535 copies/mL (IQR 1472–25 704) versus 1820 (IQR 555–11 660) in subjects with B versus non-B clades, respectively (P = 0.33).
Table 3.
Major integrase RAMs in raltegravir recipients at positions that differ in mutation frequency between subtype B and non-B clades
| RAM | Subtype B (n = 209) | Non-B clades (n = 46) |
|---|---|---|
| E92Q, n (%) | 4 (1.9) | 5 (10.9) [A, C, G, CRF09]a |
| G140S, n (%) | 33 (15.8) | 0 (0) |
| G140A, n (%) | 3 (1.4) | 1 (2.2) [G]a |
| Q148H, n (%) | 28 (13.4) | 0 (0) |
| Q148R, n (%) | 13 (6.2) | 2 (4.3) [C, G]a |
| Q148K, n (%) | 1 (0.5) | 0 (0) |
aNon-B clades with the mutation are indicated in square brackets.
Codon usage in integrase sequences of INI-naive patients was examined in order to define its relation to the mutation profiles observed in raltegravir-experienced patients (Table 4). Sequences differed by clade in their propensity to acquire mutations at codon 140. Overall 384/399 (96%) subtype B sequences had GGT or GGC (or a mixture of both) at position 140, requiring a single nucleotide substitution to replace glycine with serine (AGT or AGC). The remaining 4% of subtype B sequences showed GGA, GGM, GGK and GGW. In contrast, all 134 (100%) non-B sequences had GGA or GGG (or a mixture of both), requiring more than one nucleotide substitution to encode serine. There was no consistent difference in codon usage at positions E92 and Q148.
Table 4.
Codon usage in INI-naive subjects at positions showing a different mutation frequency between subtype B and non-B clades after raltegravir treatment
| Position | Subtype B (n = 399) |
Non-B clades (n = 134) |
||
|---|---|---|---|---|
| triplet | n (%) | triplet | n (%) | |
| 92 | GAG | 248 (62.2) | GAG | 7 (5.2) |
| GAA | 117 (29.3) | GAA | 123 (91.8) | |
| GAR | 34 (8.5) | GAR | 3 (2.2) | |
| 140 | GGC | 331 (83.0) | GGA | 84 (62.7) |
| GGT | 35 (8.8) | GGG | 43 (32.1) | |
| GGY | 18 (4.5) | GGR | 7 (5.2) | |
| other | 15 (3.8) | |||
| 148 | CAA | 378 (94.7) | CAA | 106 (79.1) |
| CAG | 13 (3.3) | CAG | 27 (20.1) | |
Cross-resistance to dolutegravir
Of the 255 raltegravir recipients, 211 (83%) had no predicted resistance to dolutegravir (for twice-daily use). Among those with any predicted dolutegravir resistance, 4 (1.6%) had low-level resistance, 36 (14.1%) had intermediate-level resistance and 4 (1.6%) had high-level resistance (Figure 2). Reflecting the lower prevalence of the Q148H/R/K + G140A/S pathway, the prevalence of predicted dolutegravir cross-resistance among subjects with non-B clades was significantly lower than in subjects with subtype B, both overall (2/46, 4.3% versus 42/209, 20.1%; P = 0.009) and when restricting the comparison to subjects with intermediate- to high-level dolutegravir cross-resistance (1/46, 2.2% versus 39/209, 18.7%; P = 0.003).
Figure 2.
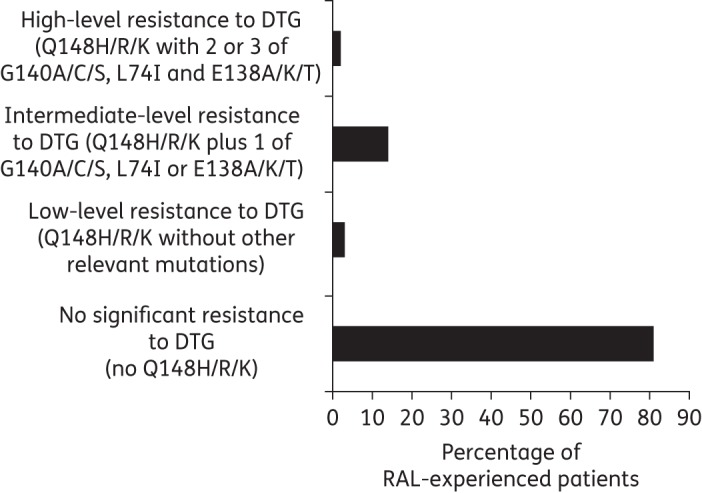
Predicted cross-resistance to dolutegravir (twice-daily dosing) in raltegravir recipients with viraemia. DTG, dolutegravir; RAL, raltegravir.
Discussion
In this large European cohort with diverse HIV-1 clades, there was no evidence of major INI RAMs among INI-naive subjects. In contrast, nearly half of patients experiencing viraemia on raltegravir-containing ART showed one or more major INI RAMs. The prevalence of genotypic INI resistance measured in raltegravir recipients with virological failure was consistent with the resistance rates reported from clinical trials (32%–68%).2,6 Similar rates were reported from an observational cohort, which also found an association between low plasma raltegravir concentrations (taken to indicate poor adherence) and lack of detectable INI RAMs at virological failure.28,29 While we did not have raltegravir concentrations, we did not observe a significant difference in viral load levels between subjects with and without detectable INI RAMs, which might have indirectly suggested a difference in adherence levels.
The three major recognized pathways of genotypic resistance to raltegravir (N155H, Q148H/R/K and Y143R/C/H) were represented in the raltegravir-experienced population. Interestingly, co-occurrence of these major pathways was not seen, possibly representing functional incompatibility of all three pathways in the context of a single virus. Indeed, the N155H and Q148H/R/K pathways have been shown to sequentially emerge in separate genomes within a given individual over time.30 Consistent with available data, there was a strong preferential association of N155H with E92Q, Y143H/R/C with T97A, and G140S/A with Q148H/R/K. Of these three mutually antagonistic genotypic patterns, Q148H/R/K and G140S/A were significantly more prevalent in subtype B than in non-B clades, occurring only in three subjects (with subtype C and subtype G). Strikingly, Q148H and G140S occurred in 13% and 16% of subtype B sequences, respectively, but were absent from all non-B clades analysed. This finding could be wholly explained through differences in codon usage between subtype B and non-B clades in INI-naive patients. In the vast majority of subtype B sequences, one nucleotide substitution was required to replace glycine with serine at integrase codon 140. In all the non-B clades examined (A, C, D, F, G, CRF01, CRF02, CRF06, CRF35_AD and several complex mosaic sequences) two nucleotide substitutions were required, thus raising the genetic barrier to the emergence of G140 mutants. As mutations at codon 140 play a key role in restoring the fitness of Q148 mutants,31 their occurrence can also influence the emergence of Q148H/R/K, thus explaining the reduced prevalence of Q148 mutants observed in non-B subtypes. Small studies previously proposed a higher genetic barrier for G140S and G140C in subtypes C, CRF01 and CRF0224–26,32 and more recently a large study of INI-experienced patients identified more Q148 mutations in subtype B versus non-B subtypes.8 Our findings thus confirm and extend these previous observations, indicating the unique propensity of subtype B to the development of the Q148 + G140 mutation pathway. Taken together, these findings show that theoretical predictions based on assessment of the genetic barrier to resistance translate into the actual acquisition of INI mutations in contemporary clinical practice.
Notably, the Q148 mutation pathway confers the highest level of resistance to available INIs, including a significant level of cross-resistance to dolutegravir.33 In vitro studies show that mutations at codons Y143 and N155 confer EC50 fold changes for dolutegravir consistently <3.14,15 Variable levels of resistance are seen with Q148 mutations depending on the presence of other INI RAMs.14,15 In the Phase 3 VIKING-3 trial, INI-experienced patients showed different virological responses to dolutegravir twice daily plus an optimized background regimen depending on the baseline mutation pattern.18 Virological suppression <50 copies/mL at 24 weeks was 100/126 (79%) in the absence of baseline Q148 mutations (regardless of the presence of Y143 and N155 mutants), 21/36 (58%) with Q148 mutations plus one minor INI RAM and 5/21 (24%) in the presence of Q148 plus at least two minor INI RAMs.18 Our findings are therefore of practical importance in relation to the likelihood of cross-resistance to dolutegravir in raltegravir-experienced patients. Q148H/R/K mutations were present in 17% of raltegravir-experienced patients and ∼16% of the raltegravir-experienced patients were classified as having intermediate- to high-level resistance to dolutegravir. These data are reassuring as they indicate that most raltegravir-experienced patients, including the large majority of subjects with non-B clades, will retain at least partial susceptibility to twice-daily dolutegravir.18 As clinical experience with dolutegravir in INI-experienced patients remains limited, a role for other mutation profiles in conferring dolutegravir cross-resistance cannot be excluded.
In addition to recognized INI RAMs, non-classic amino acid substitutions at the major integrase resistance codons 92, 147 and 155 were detected in four raltegravir recipients with subtype B. The analysis also identified novel mutations as significantly associated with raltegravir selective pressure—K159Q/R, I161L/M/N/T/V and E170AG. Studies employing site-directed mutagenesis are necessary to investigate the phenotypic properties of these novel mutations and confirm a role in INI resistance. It should be noted that our observations do not exclude a role in resistance for rarer variants that would not be captured by a stringent statistical approach.34
The lack of major INI RAMs in INI-naive subjects suggests that transmitted INI resistance was rare in 2008–11. Although transmitted INI RAMs are still uncommonly described even among subjects with primary HIV infection,35–42 the risk may increase with expanded use of INIs, indicating the need for ongoing surveillance. Importantly, the data also indicate that in the absence of INI selective pressure, major INI RAMs do not occur spontaneously in the diverse non-B clades analysed. Others have reported the presence of major INI RAMs in INI-naive patients infected with subtype CRF02_AG in a small cohort in Cameroon.26 We did not find evidence of major INI RAMs in 22 INI-naive patients infected with CRF02_AG in our study.
The study illustrates the ongoing importance of collaborative research between different centres in order to collect and meaningfully interpret data from large numbers of patients in routine care, and we hope that this study will prove useful in designing analyses of large depositories such as the Stanford dataset. However, the study has limitations. Policies and methods for resistance testing are likely to have differed across centres. The full treatment history and the duration of viraemia prior to the resistance test were not available and caution is therefore required in the interpretation of resistance rates. In subtype B, it has been shown that N155 mutants tend to be replaced by the more resistant and fitter Q148 + G140 variants with ongoing drug selective pressure. Thus, the resilience of non-B clades with respect to the emergence of these mutations should be considered relative and time-dependent rather than absolute.
Funding
The study was supported by a research award made by Merck to the Royal Free Charitable Trust. The funder approved the study design and had no involvement in the conduct of the study, data collection and analysis, and preparation of the manuscript. T. D. is a Wellcome Trust Research Training Fellow.
Transparency declarations
None to declare.
Supplementary data
Table S1 is available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
Acknowledgements
Other members of the CORONET Study Group
Vincent Calvez1, Isabelle Malet1, Maria Pino2, José Ramón Santos2 and Bonaventura Clotet2 (1AP-HP, Hôpital Pitié-Salpêtrière, INSERM-Sorbonne Universités, UPMC Univ Paris 06, UMR_S 1136, Paris, France; 2IrsiCaixa, ICREA and UVic-UCC, Barcelona, Spain).
Contributor Information
Collaborators: on behalf of the CORONET Study Group, Vincent Calvez, Isabelle Malet, Maria Pino, José Ramón Santos, and Bonaventura Clotet
References
- 1.Hazuda DJ. Resistance to inhibitors of the human immunodeficiency virus type 1 integration. Braz J Infect Dis 2010; 14: 513–8. [PubMed] [Google Scholar]
- 2.Lennox JL, DeJesus E, Lazzarin A et al. Safety and efficacy of raltegravir-based versus efavirenz-based combination therapy in treatment-naive patients with HIV-1 infection: a multicentre, double-blind randomised controlled trial. Lancet 2009; 374: 796–806. [DOI] [PubMed] [Google Scholar]
- 3.Steigbigel RT, Cooper DA, Kumar PN et al. Raltegravir with optimized background therapy for resistant HIV-1 infection. N Engl J Med 2008; 359: 339–54. [DOI] [PubMed] [Google Scholar]
- 4.Marsden MD, Avancena P, Kitchen CM et al. Single mutations in HIV integrase confer high-level resistance to raltegravir in primary human macrophages. Antimicrob Agents Chemother 2011; 55: 3696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanco JL, Varghese V, Rhee SY et al. HIV-1 integrase inhibitor resistance and its clinical implications. J Infect Dis 2011; 203: 1204–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper DA, Steigbigel RT, Gatell JM et al. Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N Engl J Med 2008; 359: 355–65. [DOI] [PubMed] [Google Scholar]
- 7.Hurt CB, Sebastian J, Hicks CB et al. Resistance to HIV integrase strand transfer inhibitors among clinical specimens in the United States, 2009–2012. Clin Infect Dis 2014; 58: 423–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fourati S, Charpentier C, Amiel C et al. Cross-resistance to elvitegravir and dolutegravir in 502 patients failing on raltegravir: a French national study of raltegravir-experienced HIV-1-infected patients. J Antimicrob Chemother 2015; 70: 1507–12. [DOI] [PubMed] [Google Scholar]
- 9.Molina JM, Lamarca A, Andrade-Villanueva J et al. Efficacy and safety of once daily elvitegravir versus twice daily raltegravir in treatment-experienced patients with HIV-1 receiving a ritonavir-boosted protease inhibitor: randomised, double-blind, phase 3, non-inferiority study. Lancet Infect Dis 2012; 12: 27–35. [DOI] [PubMed] [Google Scholar]
- 10.Lampiris HW. Elvitegravir: a once-daily, boosted, HIV-1 integrase inhibitor. Expert Rev Anti Infect Ther 2012; 10: 13–20. [DOI] [PubMed] [Google Scholar]
- 11.Zolopa AR, Berger DS, Lampiris H et al. Activity of elvitegravir, a once-daily integrase inhibitor, against resistant HIV Type 1: results of a phase 2, randomized, controlled, dose-ranging clinical trial. J Infect Dis 2010; 201: 814–22. [DOI] [PubMed] [Google Scholar]
- 12.Garrido C, Villacian J, Zahonero N et al. Broad phenotypic cross-resistance to elvitegravir in HIV-infected patients failing on raltegravir-containing regimens. Antimicrob Agents Chemother 2012; 56: 2873–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hightower KE, Wang R, Deanda F et al. Dolutegravir (S/GSK1349572) exhibits significantly slower dissociation than raltegravir and elvitegravir from wild-type and integrase inhibitor-resistant HIV-1 integrase-DNA complexes. Antimicrob Agents Chemother 2011; 55: 4552–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Underwood MR, Johns BA, Sato A et al. The activity of the integrase inhibitor dolutegravir against HIV-1 variants isolated from raltegravir-treated adults. J Acquir Immune Defic Syndr 2012; 61: 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canducci F, Ceresola ER, Boeri E et al. Cross-resistance profile of the novel integrase inhibitor dolutegravir (S/GSK1349572) using clonal viral variants selected in patients failing raltegravir. J Infect Dis 2011; 204: 1811–5. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi M, Yoshinaga T, Seki T et al. In vitro antiretroviral properties of S/GSK1349572, a next-generation HIV integrase inhibitor. Antimicrob Agents Chemother 2011; 55: 813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cahn P, Pozniak AL, Mingrone H et al. Dolutegravir versus raltegravir in antiretroviral-experienced, integrase-inhibitor-naive adults with HIV: week 48 results from the randomised, double-blind, non-inferiority SAILING study. Lancet 2013; 382: 700–8. [DOI] [PubMed] [Google Scholar]
- 18.Castagna A, Maggiolo F, Penco G et al. Dolutegravir in antiretroviral-experienced patients with raltegravir- and/or elvitegravir-resistant HIV-1: 24-week results of the phase III VIKING-3 study. J Infect Dis 2014; 210: 354–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Lunzen J, Maggiolo F, Arribas JR et al. Once daily dolutegravir (S/GSK1349572) in combination therapy in antiretroviral-naive adults with HIV: planned interim 48 week results from SPRING-1, a dose-ranging, randomised, phase 2b trial. Lancet Infect Dis 2012; 12: 111–8. [DOI] [PubMed] [Google Scholar]
- 20.Katlama C, Murphy R. Dolutegravir for the treatment of HIV. Expert Opin Investig Drugs 2012; 21: 523–30. [DOI] [PubMed] [Google Scholar]
- 21.Walmsley SL, Antela A, Clumeck N et al. Dolutegravir plus abacavir-lamivudine for the treatment of HIV-1 infection. N Engl J Med 2013; 369: 1807–18. [DOI] [PubMed] [Google Scholar]
- 22.Raffi F, Jaeger H, Quiros-Roldan E et al. Once-daily dolutegravir versus twice-daily raltegravir in antiretroviral-naive adults with HIV-1 infection (SPRING-2 study): 96 week results from a randomised, double-blind, non-inferiority trial. Lancet Infect Dis 2013; 13: 927–35. [DOI] [PubMed] [Google Scholar]
- 23.Clotet B, Feinberg J, van Lunzen J et al. Once-daily dolutegravir versus darunavir plus ritonavir in antiretroviral-naive adults with HIV-1 infection (FLAMINGO): 48 week results from the randomised open-label phase 3b study. Lancet 2014; 383: 2222–31. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen HL, Ruxrungtham K, Delaugerre C. Genetic barrier to the development of resistance to integrase inhibitors in HIV-1 subtypes CRF01_AE and B. Intervirology 2012; 55: 287–95. [DOI] [PubMed] [Google Scholar]
- 25.Maiga AI, Malet I, Soulie C et al. Genetic barriers for integrase inhibitor drug resistance in HIV type-1 B and CRF02_AG subtypes. Antivir Ther 2009; 14: 123–9. [PubMed] [Google Scholar]
- 26.Turriziani O, Montagna C, Falasca F et al. Short communication: analysis of the integrase gene from HIV type 1-positive patients living in a rural area of West Cameroon. AIDS Res Hum Retroviruses 2012; 28: 1729–33. [DOI] [PubMed] [Google Scholar]
- 27.Wensing AM, Calvez V, Gunthard HF et al. 2014 Update of the drug resistance mutations in HIV-1. Top Antivir Med 2014; 22: 642–50. [PMC free article] [PubMed] [Google Scholar]
- 28.Garrido C, de Mendoza C, Alvarez E et al. Plasma raltegravir exposure influences the antiviral activity and selection of resistance mutations. AIDS Res Hum Retroviruses 2012; 28: 156–64. [DOI] [PubMed] [Google Scholar]
- 29.Rusconi S, Vitiello P, Adorni F et al. Factors associated with virological success with raltegravir-containing regimens and prevalence of raltegravir-resistance-associated mutations at failure in the ARCA database. Clin Microbiol Infect 2013; 19: 936–42. [DOI] [PubMed] [Google Scholar]
- 30.Fransen S, Gupta S, Frantzell A et al. Substitutions at amino acid positions 143, 148, and 155 of HIV-1 integrase define distinct genetic barriers to raltegravir resistance in vivo. J Virol 2012; 86: 7249–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delelis O, Malet I, Na L et al. The G140S mutation in HIV integrases from raltegravir-resistant patients rescues catalytic defect due to the resistance Q148H mutation. Nucleic Acids Res 2009; 37: 1193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brenner BG, Lowe M, Moisi D et al. Subtype diversity associated with the development of HIV-1 resistance to integrase inhibitors. J Med Virol 2011; 83: 751–9. [DOI] [PubMed] [Google Scholar]
- 33.Geretti AM, Armenia D, Ceccherini-Silberstein F. Emerging patterns and implications of HIV-1 integrase inhibitor resistance. Curr Opin Infect Dis 2012; 25: 677–86. [DOI] [PubMed] [Google Scholar]
- 34.Malet I, Gimferrer Arriaga L, Artese A et al. New raltegravir resistance pathways induce broad cross-resistance to all currently used integrase inhibitors. J Antimicrob Chemother 2014; 69: 2118–22. [DOI] [PubMed] [Google Scholar]
- 35.Garrido C, Geretti AM, Zahonero N et al. Integrase variability and susceptibility to HIV integrase inhibitors: impact of subtypes, antiretroviral experience and duration of HIV infection. J Antimicrob Chemother 2010; 65: 320–6. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Miller MD, Danovich RM et al. Analysis of low-frequency mutations associated with drug resistance to raltegravir before antiretroviral treatment. Antimicrob Agents Chemother 2011; 55: 1114–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cossarini F, Boeri E, Canducci F et al. Integrase and fusion inhibitors transmitted drug resistance in naive patients with recent diagnosis of HIV-1 infection. J Acquir Immune Defic Syndr 2011; 56: e51–4. [DOI] [PubMed] [Google Scholar]
- 38.Hurt CB. Transmitted resistance to HIV integrase strand-transfer inhibitors: right on schedule. Antivir Ther 2011; 16: 137–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyd SD, Maldarelli F, Sereti I et al. Transmitted raltegravir resistance in an HIV-1 CRF_AG-infected patient. Antivir Ther 2011; 16: 257–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young B, Fransen S, Greenberg KS et al. Transmission of integrase strand-transfer inhibitor multidrug-resistant HIV-1: case report and response to raltegravir-containing antiretroviral therapy. Antivir Ther 2011; 16: 253–6. [DOI] [PubMed] [Google Scholar]
- 41.Saladini F, Meini G, Bianco C et al. Prevalence of HIV-1 integrase mutations related to resistance to dolutegravir in raltegravir naive and pretreated patients. Clin Microbiol Infect 2012; 18: E428–30. [DOI] [PubMed] [Google Scholar]
- 42.Stekler JD, McKernan J, Milne R et al. Lack of resistance to integrase inhibitors among antiretroviral-naive subjects with primary HIV-1 infection, 2007–2013. Antivir Ther 2015; 20: 77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


