Abstract
The majority of deaths from all cancers, including colorectal cancer (CRC), is a result of tumor metastasis to distant organs. To date, an effective and safe system capable of exclusively targeting metastatic cancers that have spread to distant organs or lymph nodes does not exist. Here, we constructed multifunctional RNA nanoparticles, derived from the three-way junction (3WJ) of bacteriophage phi29 motor pRNA, to target metastatic cancer cells in a clinically relevant mouse model of CRC metastasis. The RNA nanoparticles demonstrated metastatic tumor homing without accumulation in normal organ tissues surrounding metastatic tumors. The RNA nanoparticles simultaneously targeted CRC cancer cells in major sites of metastasis, such as liver, lymph nodes and lung. Our results demonstrate the therapeutic potential of these RNA nanoparticles as a delivery system for the treatment of CRC metastasis
Keywords: Colorectal cancer, Metastasis, Nanobiotechnology, Bacteriophage phi29, RNA nanoparticle, RNA therapeutics
Graphical abstract
Colorectal cancer (CRC) is the second leading cause of cancer deaths in the Unites States, with over 130,000 new cases estimated in 2014.1 Metastasis, the primary cause of colon cancer-related deaths, is diagnosed in 15-25% of patients at the time of primary CRC resection and, eventually, up to 60% of patients with CRC develop liver and/or lung metastasis.1, 2 Few patients with metastatic CRC are eligible for surgical intervention, and other treatment modalities are limited.3, 4 New strategies relying on systemic interventions are needed to treat the complex problem of metastatic CRC.
The development of a safe, efficient, specific and nonpathogenic nanodevice to target metastatic cancer cells is badly needed. To address the challenges of targeting metastatic tumors with nanotechnology, it is necessary to combine the rational design of nanocarriers with the fundamental understanding of tumor biology. Targeting nanoparticles to sites of metastasis can be divided into two mechanisms: (1) passive targeting, the process of delivering nanoparticles to the specific organ or organs in which the metastases reside, and (2) active targeting, the precise homing of a nanoparticle to a specific cancer cell receptor. Passive targeting exploits the characteristic features of tumor biology that allow nanocarriers to accumulate in the tumor by the enhanced permeability and retention (EPR) effect. However, recent studies, focused on the delivery of siRNA to liver hepatocytes, showed that precise particle engineering is required to pass through the fenestrae that are present in the liver endothelium.5, 6 In general, intravenously administered nanoparticles, especially for particles larger than 50nm, accumulated in activated Kupffer cells that reside within and near the liver vasculature and did not reach the hepatocytes. The obstacles encountered during nanoparticle delivery into normal liver parenchyma suggest an even greater challenge for targeting metastatic tumors residing within normal liver parenchyma. Indeed, targeting metastatic cancer cells that reside within a population of non-cancerous cells presents a unique clinical challenge.
Active targeting of nanoparticle systems can potentially reduce nanoparticle accumulation in normal organ parenchyma and reduce therapeutic agent side-effects.7 This is achieved by conjugating the surface of nanoparticles using specific cell ligands. Folate receptor α (FRα) is a selective tumor marker, that is overexpressed on the surface of various cancers, including ovary, brain, kidney, breast, and lung cancers.8 Folic acid (FA) has a small size, low cost, high tumor tissue specificity and is non-immunogenic.9 FA-linked nanocarriers have fairly high binding affinity to receptors expressed on tumor cells.10 In this study, we demonstrate that the centerfold domain of the packaging RNA (pRNA) of bacteriophage phi29 DNA packaging motor11, 12 could be engineered to form thermodynamically stable RNA nanoparticles to carry therapeutic modules and FA ligands for specific cell targeting of CRC.
After proving the concept of RNA nanotechnology in 199812, the feasibility of RNA nanotechnology13 for constructing multifunctional therapeutic RNA nanoparticles has been demonstrated by the pRNA based therapeutic system.14-21 Recently, a thermodynamically stable 3-way junction (3WJ) motif was isolated from the central domain of the pRNA.16 The pRNA-3WJ nanoparticles can be assembled with high efficiency from three individual fragments. The resulting RNA nanoparticles remain structurally intact at ultra-low concentrations, resistant to denaturation by 8M urea, and resistant to RNase degradation in the serum after incorporation of 2′-Fluoro modified nucleotides.14-16 The 3WJ motif can be readily functionalized with targeting (RNA aptamers or chemical ligands), therapeutic (siRNA or ribozymes) and detection molecules (near infra-red fluorophores) without affecting the folding of the central scaffold. All incorporated functional modules retain their correct structure and authentic functions within the nanoparticles.14-16 Furthermore, the pRNA nanoparticles are non-toxic, non-immunogenic and display favorable pharmacokinetic profiles in mice.14 Recently the crystal structure of the pRNA-3WJ has been resolved,22 which has greatly facilitated the design of multifunctional RNA nanoparticles with precise control of size, shape, geometry and positioning of functional groups.23-25 Here, we incorporated FA as a targeting ligand and Alexa-647 as a near-infrared imaging module into the pRNA-3WJ scaffold and evaluated the RNA constructs in a murine model of CRC metastasis. We demonstrate that pRNA-3WJ nanoparticles, after systemic injection, strongly bind to CRC liver, lung and lymph node metastases without obvious retention in normal liver, lung or any other organs. Our current study confirms that active targeting is a required feature for high efficient RNA nanoparticle targeting of CRC metastasis and identifies RNA nanoparticles as a potential therapeutic nanocarrier for cancer metastasis targeting.
Results and Discussion
Construction of multi-module RNA nanoparticles
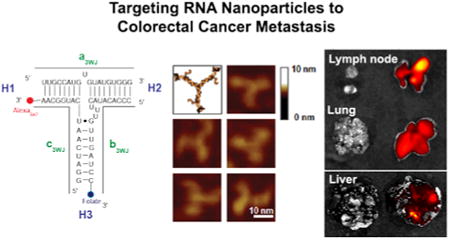
pRNA nanoparticles can carry several therapeutic agents and a recognition ligand for targeted delivery to cancer cells. pRNA nanoparticles were constructed with one of the arms of 3WJ carrying FA to serve as a ligand for binding to the cancer cells, and another 3WJ arm carrying the near infra-red fluorescent dye Alexa647 (Fig 1a). The goal of this study was to design and evaluate a nanoparticle delivery system that could specifically target CRC metastasis in liver, lung and lymph nodes (Fig 1b). Determining the targeting efficiency of an RNA nanoparticle is a complex process and ultimately needs to be assessed in a clinically relevant model in vivo. In this study, we engineered CRC cell lines to express green fluorescent protein (GFP)26 to aid in the precise localization of pRNA binding to metastatic tumors, and we performed in vivo selection of CRC cells that metastasized to the lung. In vivo selection of KM20 and HT29 lung metastasis provided highly metastatic cells with accelerated metastatic growth compared with nonselected cells (6-8 wks vs 14-16 wks) and an even metastatic load between test animals (Fig 1c), thus providing a more realistic biological environment to evaluate RNA nanoparticle targeting in vivo.
Figure 1. Construction of multi-module pRNA nanoparticles for CRC metastasis targeting.
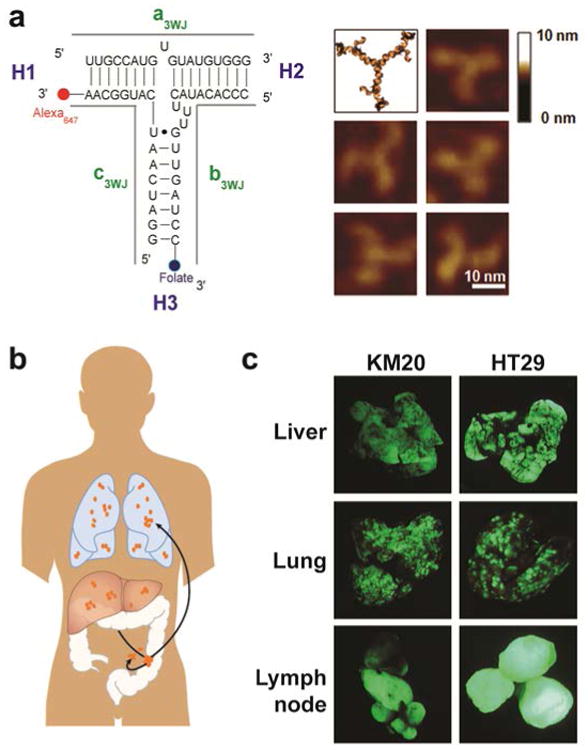
a. 2D sequence of pRNA 3WJ domain composed of three RNA oligomers (a, b, c), folic acid (FA) and near-infrared fluorescent dye Alexa647 (left); 3D model and AFM images of pRNA 3WJ motif harboring monomeric pRNA as functional modules. b. The most common sites of CRC metastasis—liver, lung and lymph nodes. The FA-conjugated pRNA (FA-pRNA) nanoparticle targets FRα positive tumors upon systemic administration. c. CRC liver, lung and lymph node experimental metastasis modes (Green: metastatic tumors)
Demonstration of high FRα expression in CRC metastatic cells in lung and liver
FRα-targeted approaches have not been investigated for the treatment of CRC metastases in vivo, and data on FRα expression in matched primary metastatic tumors is absent. First, we analyzed the status of FRα expression in primary CRCs and liver or lung CRC metastases. Samples were collected from 137 metastatic CRC cases that underwent resection at the University of Kentucky Markey Cancer Center for CRC metastases from January 1, 2003 to January 1, 2013. From this cohort, we identified 10 patients with lung metastasis; 12 patients with primary CRC; 22 cases with liver metastasis. Our results demonstrate FRα expression (score 1 to 6) in 72% of primary CRCs, 91% of CRC liver metastases, and 80% of CRC lung metastasis. High FRα expression (score 3 to 6) was detected in 63% of primary CRCs, 81% of CRC liver metastases, and 60% of CRC lung metastases (Figs 2a and b).
Figure 2. Analysis of FRα expression in CRC liver and lung metastasis.
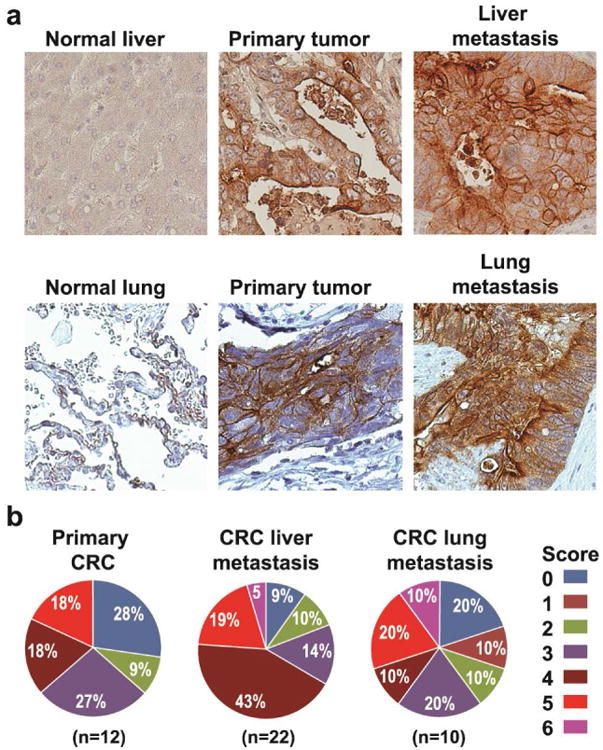
a Examples of immunohistochemical staining for FRα in liver and lung CRC metastases. Positive FRα staining of CRC was cytoplasmic or membranous or both; most positive cases showed both patterns. FRα staining was negative in normal liver and lung tissues. b. Differences in proportion of positive cells and intensity of staining were noted in positively stained cases and formed the basis of our grading system. Comprehensive total score that weighs both factors was calculated by summation of proportion and intensity values. High FRα expression (score 3-6) was detected in 63% of primary CRCs, 81% of CRC liver metastases and 60% of CRC lung metastasis.
Targeting of RNA nanoparticles to CRC cells
FA was incorporated in the pRNA nanoparticles to serve as a cancer cell delivery agent via FR-receptor mediated endocytosis.8, 10 Fluorescent pRNA nanoparticles with FA conjugated into one of the branches of the complex were tested in vitro for cell binding efficiency in the KM20 and HT29 colon cancer cell lines (37°C, 500 nM, 24h). pRNA harboring FA and Alexa647 labels served as the test sample, while the negative control harbored NH2 and Alexa647 labels. Confocal imaging of KM20 and HT29 cells confirmed binding of the pRNA nanoparticles and efficient entry of the FA-conjugated pRNA nanoparticles into the targeted cells (Fig 3a).
Figure 3. FA-pRNA nanoparticles binding to CRC cells.
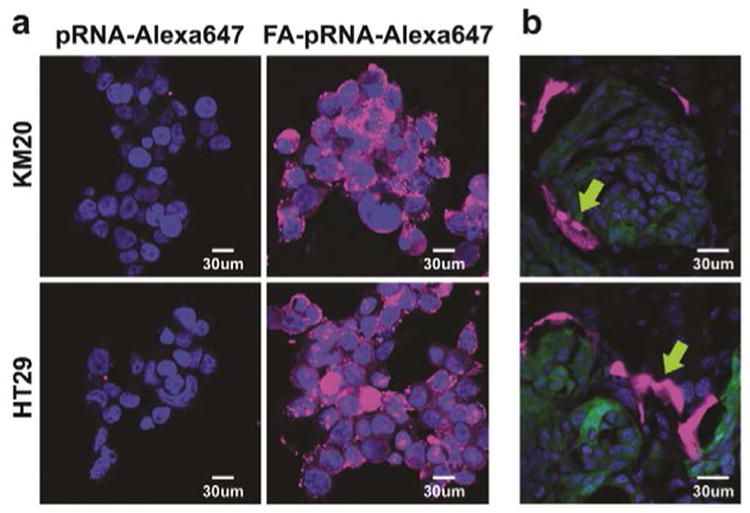
a. Binding and entry of FA-pRNA-Alexa647 nanoparticles into KM20 and HT29 cells in vitro. Magnification 40×. b. Single dose (4μg/g in 100 μl of PBS) of FA-pRNA-Alexa647 labeled nanoparticles was administered intravenously into mice with HT29 liver metastases. Accumulation of fluorescently-labeled nanoparticles was evaluated microscopically 2h after RNA nanoparticle administration. Yellow arrow: pRNA-Alexa647 (top panel), FA-pRNA-Alexa647 (bottom panel); green: GFP-expressing cancer cells; blue: DAPI stain for nuclear dsDNA; magenta: Alexa647. Magnification 40×.
To confirm the benefits of active targeting with pRNA nanoparticles, we administered a single dose of FA-pRNA-Alexa647 (4 μg/g; 100 μl of PBS) nanoparticles into mice with HT29 liver metastases and evaluated mice 2h after injection. Confocal imaging of fixed frozen tissue sections showed accumulation of fluorescently-labeled pRNA nanoparticles in areas adjacent to GFP expressing CRC liver metastases (Fig 3b). Absence of fluorescently-labeled pRNA nanoparticle accumulation in normal liver parenchyma confirmed specificity of FA-pRNA nanoparticles (data not shown). This result also suggested that longer circulation of pRNA nanoparticles might improve CRC liver metastasis penetration. To determine pRNA intravenous (iv) dose frequency, we administered RNA nanoparticles into mice without metastases every 2h and imaged mice at 15 min, 1h, 2h after initial iv dose administration. The rapid entrance into the circulation is demonstrated by the fluorescence signal in supine positioned mice as early as 15 min after FA-pRNA-Alexa647 injection (Supplemental Fig 1). We also observed excretion of cyan-colored urine as early as 30 min after iv injection, suggesting a rapid clearance of the nanoparticles from the circulation. These experiments suggest that repetitive administration of pRNA is required for the maintenance of prolonged pRNA systemic circulation and metastasis targeting.
In vivo targeting of pRNA nanoparticles to CRC liver metastases
To test whether pRNA nanoparticles can target liver metastases, we administered pRNA-Alexa647 and FA-pRNA-Alexa647 (1 μg/g; 300 μl of PBS) every 2h in order to extend pRNA circulation time and improve CRC liver metastasis penetration. Macroscopic organ imaging acquired 6h after the first administration showed maximum fluorescent signal in KM20 and HT29 CRC liver metastases targeted with FA-pRNA nanoparticles, relative to non-targeted pRNA (Fig 4a). Confocal imaging of fixed frozen sections confirmed macroscopic imaging results and demonstrated significantly improved penetration and targeting of CRC liver metastases. No accumulation of FA-conjugated pRNA nanoparticles was detected in normal liver parenchyma (Fig 4b).
Figure 4. FA-pRNA nanoparticles delivery into CRC liver metastases.
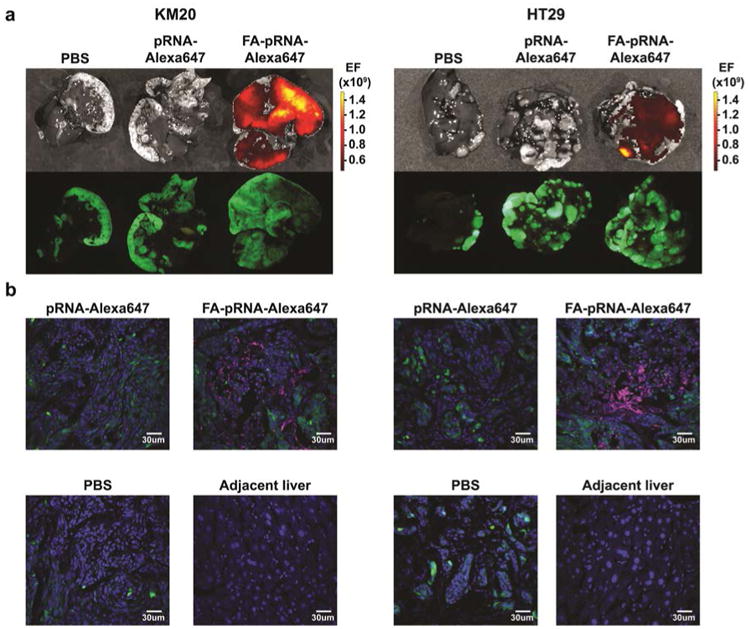
a. pRNA-Alexa647 and FA-pRNA-Alexa647 labeled nanoparticles were administered intravenously (single dose of 1 μg/g in 300μl of PBS) every 2h (three doses total) into mice with KM20 (left panel) and HT29 (right panel) liver metastases and evaluated macroscopically 6h after the first administration. b. Accumulation of fluorescently-labeled nanoparticles was evaluated microscopically in KM20 (left panel) and HT29 (right panel) and in normal liver adjacent to KM20 and HT29 metastases, 6h after the first pRNA dose administration. Control: PBS; pRNA-Alexa647 treated mice. Green: GFP-expressing cancer cells; blue DAPI; magenta: Alexa647. Magnification 40×.
In vivo targeting of pRNA nanoparticles to CRC lung metastases and lymph nodes
Mice with KM20 lung and lymph node metastases were treated with pRNA-Alexa647 and FA-pRNA-Alexa647 (1 μg/g; 500 μl of PBS) every 2h. Macroscopic organ imaging acquired 4h after the first dose administration showed maximum fluorescence signal in KM20 CRC lung and lymph node metastasis targeted with FA-conjugated pRNA nanoparticles (Fig 5a). Confocal imaging of fixed frozen sections confirmed macroscopic imaging results and demonstrated penetration and targeting of CRC lung and lymph node metastases (Fig 5b; Supplemental Fig 2). No accumulation of FA-conjugated pRNA nanoparticles was detected in normal lung parenchyma (data not shown). Together, these findings demonstrate, for the first time, successful targeting of CRC liver, lung and lymph node metastases, suggesting RNA nanoparticles as a potential therapeutic nanocarrier in the treatment of CRC metastasis.
Figure 5. FA-pRNA nanoparticle delivery into CRC lung and lymph node metastases.
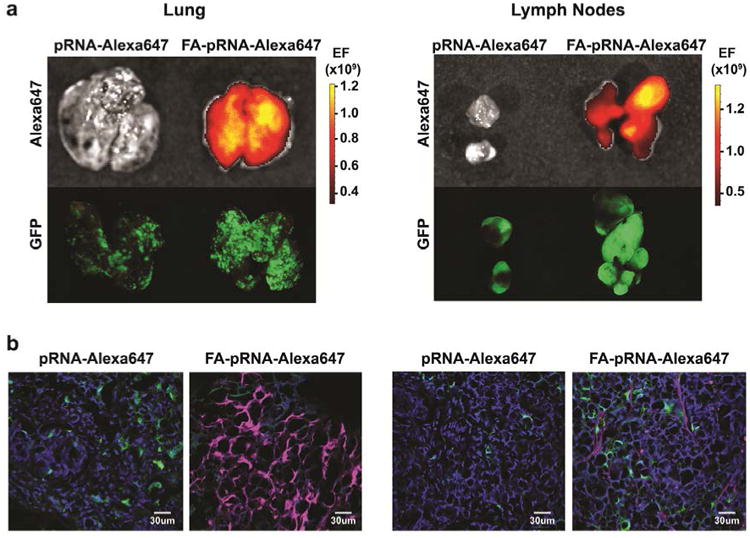
a. Mice with KM20 lung and lymph node metastases were treated with pRNA-Alexa647 and FA-pRNA-Alexa647 (single dose of 1 μg/g in 500 μl of PBS) every 2h (2 doses total). Accumulation of fluorescently-labeled nanoparticles was evaluated macroscopically in mice with KM20 lung and lymph node metastases 4h after the first pRNA dose administration. b. Confocal imaging of KM20 lung and lymph node metastases. Control: pRNA-Alexa647 treated mice. Green: GFP-expressing cancer cells; blue: DAPI; magenta: Alexa647. Magnification 40×
Conclusions
Previously, we demonstrated that bacteriophage phi29 motor pRNA11, 12 contains a 3WJ core that can serve as a cargo carrier for ligand-directed delivery of multiple therapeutic agents in vitro and in subcutaneous xenograft models.10-12, 16 Subcutaneous xenograft models are a valuable tool for the initial evaluation of nanocarriers, but these models do not mimic the clinical situation of metastatic cancer. To circumvent this limitation, we developed several CRC metastasis mouse models and evaluated the targeting capabilities of our rationally-designed RNA constructs harboring FA as a targeting ligand. The choice of FA is based on our analysis of patient samples, where we found FRα to be expressed in approximately 81% of CRC liver metastases and 60% of lung metastases.
Effectively targeting metastases has proven to be extremely challenging due to the difficulty in overcoming biological barriers surrounding the cancer cells; the toxicities of the carrier platform; the lack of tissue specificity; the challenge in formulations (particle heterogeneity, aggregation, and dissociation); unfavorable pharmacological profiles; and, normal tissues accumulation. Many studies suggest that, for successful cancer metastasis targeting, several parameters for the nanocarrier have to be achieved: nanoscale (∼10-50 nm) in size, non-toxic, non-immunogenic, biodegradable, chemically and thermodynamically stable, multivalent, capable of efficient intracellular delivery, and possessing specific targeting ability. To date, there are only a small number of reports in the literature demonstrating active targeting of metastatic cells using nanodelivery platforms, such as lipid, polymer, albumin, inorganic, peptide, antibody-based, and glycol chitosan nanoparticles.27-40 Five recurring themes were observed in the majority of these reports: (1) nanoparticles capable of reaching metastatic cells, but unable to differentiate healthy cells from cancer cells within the affected organ; (2) difficulty in generating reproducible experimental and spontaneous metastasis mouse models; (3) inability to concurrently target metastasis in multiple organs; (4) non-specific distribution in the reticuloendothelial system (lung, liver, spleen) and the kidneys; and (5) varying levels of toxicity based on release of proinflammatory cytokines (IL-6, IL-12, and IFN-γ) and hepatotoxicity markers (aspartate and alanine aminotransferases) in the serum. Our results indicate that FA-pRNA nanoparticles can overcome some of the aforementioned limitations. After systemic injection, the FA-RNA nanoparticles bind to CRC liver, lung and lymph node metastases simultaneously without retention in normal liver, lung or any other healthy tissues. The anionic nature of RNA nanoparticles prevents binding and non-specific entry into negatively charged membranes of normal cells. In addition, homogeneity in particle size also minimizes nonspecific toxicity.14
RNA is an attractive material to build nanoparticles in a variety of shapes, structural modules and motifs.12 Previously, we have designed different shapes of RNA nanoparticles -dimers, trimers, and hexamers - through hand-in-hand interactions.12, 41-44 However, the resulting particles were not thermodynamically stable for potential therapeutic applications. Naturally-selected pRNA-3WJ motifs from pRNA of bacteriophage phi29 DNA packaging motor provided us with a stable RNA building block.16 The 3WJ region extracted from the pRNA can be assembled from three pieces of RNA oligonucleotides to construct trivalent RNA nanoparticles. pRNA-3WJ shows exceptional stability under physiological conditions and in the presence of strong denaturing agents.10, 16 Discovery of the 3WJ motif allowed us to design 14 diverse RNA nanostructures with multiple functionalities as a polyvalent delivery system for nanotechnology and nanomedicinal applications.15 Two differently shaped nanoparticles, 3WJ and 3WJ-derived X-shaped nanoparticles, were tested in vivo in subcutaneous tumor xenograft models. 10, 14-16 In the current study we have demonstrated rapid entrance of 3WJ nanoparticles into tissues (e.g., lung and liver) and homing of the nanoparticles to the metastatic cell clusters within 4 to 6 hours. However, future studies are needed to evaluate the effect of size and shape of RNA nanoparticles on the delivery of therapeutics to liver and lung metastases.
To date, all FDA-approved nanoparticles for clinical applications (such as Abraxane®) utilize the EPR mechanism, whereby nanoparticles passively navigate through disorganized and leaky tumor vasculatures.45 There are several challenges when using the EPR effect for nanoparticle delivery. For example, the normal liver and lung parenchyma contain numerous Kupffer cells and macrophages, respectively, that rapidly clear the nanocarriers before they reach the metastatic cells, and metastatic lesions are less-vascularized compared to primary tumors; therefore, tumor targeting via the EPR effect is largely compromised. We hypothesize that only nanocarriers that can pass freely through normal organ parenchyma have a chance for receptor specific targeting of metastatic cancer cells. Our results confirm that active targeting assists in successful localization of nanoparticles to cancer metastasis.
In conclusion, our in vivo studies demonstrate that pRNA-based nanoparticles exhibit both specificity and favorable pharmacokinetic properties that may provide an ideal platform for specific drug delivery to metastatic cells. We have previously demonstrated the successful downregulation of specific gene expression using siRNA-conjugated pRNA nanoparticles in vitro,16, 46 which, together with our current in vivo studies, suggest a therapeutic potential of siRNA, ribozyme or chemotherapeutic drugs conjugated to pRNA nanoparticles for the clinical treatment of CRC metastases.
Methods
Cell lines, transfections
The human CRC cell line KM20 was provided by Dr. Isaiah J. Fidler (MD Anderson Cancer Center, Houston, TX).47 The human CRC cell line HT29 was purchased from American Type Culture Collection (Manassas, VA). Tissue culture media and reagents were obtained from Life Technologies, Inc. (Grand Island, NY). Cell line identities were authenticated at the Johns Hopkins Genetic Resources Core Facility, as previously reported.48 Non-targeting and CY3 labeled non-targeting siRNA were purchased from Thermo Scientific (Pittsburgh PA). Self-assembling nanoparticle system for murine cancer models was purchased from AparnaBio (inVivoTrack, Rockville, MD) and used according to manufacturer's instructions. EGFP-N1 vector was purchased from Clontech (Mountain View, CA). GFP-expressing cells were selected with 500 μg/ml Geneticin (G418), purchased from Life Technologies (Carlsbad, CA) and enriched by three cycles of fluorescence-activated cell sorting (FACS).
Immunohistochemistry (IHC)
FRα IHC assay kit was purchased from Biocare Medical (Concord, CA). Matched CRC lung and liver metastasis samples with corresponding normal tissues were obtained from the Biospecimen and Tissue Procurement Shared Resource Facility of the University of Kentucky Markey Cancer Center. IHC was performed according to the manufacturer's directions (Dako Corp., Carpinteria, CA).49 Briefly, slides were deparaffinized in xylene, rehydrated, incubated for 15 min with fresh 0.3% hydrogen peroxide in methanol, washed with PBS, and heated to 95°C in 10 mM citrate buffer (pH 6.0) for 30 min. Sections were blocked in serum free protein block (Dako) and treated with biotin blocking system (Dako); primary antibody was incubated for 12 h at 4°C, washed with TBST (Tris-Buffered Saline and Tween 20) and incubated with secondary antibody (60 min; RT). Antigen-antibody complexes were detected with the EnVision Kit (Dako Corp., Carpinteria, CA). All sections were counterstained with hematoxylin and observed by light microscopy. For negative controls, primary antibody was omitted from the above protocol. The number of positive cells was visually evaluated in each core by a pathologist (E-Y L) and the staining intensity of FRα was classified using a semi-quantitative system developed by Allred et al.50 This system assesses the proportion of positive cells (none=0; <10%=1; 10% to 50%, =2; >50%=3) and intensity of staining (none=0; weak=1; intermediate=2; and strong=3). Comprehensive total score that weighs both factors is calculated by summation of proportion and intensity values.
Liver metastasis model and in vivo imaging
Male NCr nude (8 wks old) mice were obtained from Taconic (Hudson, NY) and housed in clean, pathogen-free rooms in an environment with controlled temperature (27°C), humidity, and a 12 h light/dark cycle. The mice were fed standard chow and tap water ad libitum and allowed to acclimate for one week. All animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Kentucky and were conducted in accordance with guidelines issued by the National Institutes of Health for the care of laboratory animals. Tumor cells were injected iv or intrasplenically (is) by methods previously described.51 Briefly, mice were anesthetized with isoflurane, a mini-laparotomy was performed to isolate and exteriorize the spleen; viable tumor cells (5×106 cells/100 μl PBS) were injected into the spleen by a 27-gauge needle; splenectomy was performed 1 min after tumor cell injection. The peritoneum and skin were closed with sutures and wound clips. Mice were fed a FA-free diet (Harlan Laboratories; Indianapolis, IN) for a minimum of 2 wks before the FA-pRNA-3WJ injection. For iv injection of cancer cells, mice were anesthetized using isoflurane (2% in oxygen at 0.6 L/min flow rate) and injected with 1×106 cells in 100 μl of PBS. For iv injection of RNA, mice were anesthetized using isoflurane and injected with a 2′-F U/C modified pRNA-3WJ nanoparticles in PBS. Macroscopic optical imaging (Alexa Fluor 647, Ex=640 nm, Em=680 nm) was carried out using an IVIS Spectrum station (Caliper Life Sciences; Hopkinton, MA). Composite images obtained were comprised of black and white digital photos with an overlay of images reflecting fluorescent activity. The density map, measured as photons/second/cm2/steradian (p/s/cm2/sr), was created using the Living Image 3.1 (Caliper Life Sciences; Hopkinton, MA) software and represented as a color gradient centered at the maximal spot.
Confocal microscopy
For in vitro samples, CRC cells were plated on glass coverslides (Fisher Scientific; 12-545-81) in 12 well plates at 500, 000 cells/well density in 10% FBS RPMI 1640 medium (Life Technologies; 11875-119) overnight. Alexa647 labeled pRNA-3WJ complexes harboring FA and FA-free control were added in FA-free RPMI 1640 medium (Life Technologies; 27016-021) and incubated with the cells at 37°C for 24 h. After washing with PBS 2× at room temperature (RT), the cells were fixed in 4% paraformaldehyde (Polysciences; 18814) for 10 min at RT (Thermo Scientific), washed in RT PBS, incubated with Hoechst 33342 (Life Technologies; H21492; 0.5 μg/ml; PBS) for 5 min, washed in RT PBS and mounted on microscope slides (Fisher Scientific; 12-550-14) in aqueous ultramount permanent mounting medium overnight (Dako; S1964). The cells were then assayed for binding and cell entry by an Olympus FV1000 laser scanning confocal microscope.
For in vivo samples, collected tissues were fixed in 4% paraformaldehyde (Polysciences; 18814) with 10% sucrose (Sigma-Aldrich; St. Louis, MO) for 12h at 4°C and embedded into OCT on dry ice (Tissue-Tek; Andwin Scientific, Schaumburg, IL) for frozen sectioning (10 μm section thickness). Sectioned tissues were dried overnight in the dark, washed in RT PBS, stained with Hoechst 33342 (Life Technologies; H21492; 0.5 μg/ml; PBS) nuclear stain and mounted in aqueous ultramount permanent mounting medium overnight (Dako; S1964).
Construction of pRNA-3WJ RNA nanoparticles
The Alexa647-labeled pRNA-3WJ nanoparticles with and without FA, were synthesized from RNA fragments (Trilink) according to the procedure described previously.14-16, 25, 52 The RNA nanoparticles contained 2′-F U and C nucleotides to render them resistant to RNase degradation in vivo. The RNA complex was then assembled by mixing the individual strands in stoichiometric ratio at RT in PBS buffer and the assembly verified by native PAGE and AFM imaging.
Enzymatic isolation of CRC cells from metastases
Liberase DH Research Grade (05401054001; Roche Applied Science) was resuspended in sterile water to 2.5 mg/ml concentration and stored in single-use 100 μl aliquots at -80°C. Collagenase/Hyaluronidase (07912; StemCell Technologies) was aliquoted into single-use 250 μl aliquots and stored at -80°C. Upon collection, metastatic tumors were placed into complete cell culture media supplemented with 1× Gibco® Antibiotic-Antimycotic (15240-062; Life Technologies) for transportation. Metastatic tumor fragments were minced into 2-mm cubes using scissors and digested in 50 μg/ml Liberase DH (100 μl) and 0.5× Collagenase/Hyaluronidase (250 μl), diluted in 5 mL of McCoy5A serum free media for 4h at 37°C with gentle agitation by magnetic stirring bar. No undigested tissue was observed. Digested cells were washed twice with complete cell culture media and transferred into 10% FBS McCoy5A media supplemented with 1× Gibco® Antibiotic-Antimycotic and 100 μg/ml Primocin (ant-pm-1; InvivoGen). Cells harvested from these cultures were injected iv into another set of nude mice. The sequence of in vivo selection was repeated three times. No change was observed in lymph node metastasis from the first to the last generation of in vivo selected cell lines.
Supplementary Material
Acknowledgments
The authors thank Cynthia Long and Dana Napier for advice on animal tissue processing and Donna Gilbreath and Cathy Anthony for manuscript preparation.
Research was supported by NIH grants U01 CA151648 (P.G.), R01 EB003730 (P.G.), R01 RB 019036 (P.G and B.M.E.), and P30 CA177558 (B.M.E).
Footnotes
Conflict of Interest: The authors declare the following competing financial interest(s): P.G. is a cofounder of Kylin Therapeutics, Inc. and Biomotor and RNA Nanotechnology Development Corp. Ltd.
Supporting Information: Figures of FA-pRNA-Alexa647 nanoparticles systemic circulation evaluation after systemic administration; FA-pRNA nanoparticle delivery into HT29 lung and lymph node metastasis. These materials are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Siegel R, Desantis C, Jemal A. Colorectal Cancer Statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. Cancer Statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Wanebo HJ, Semoglou C, Attiyeh F, Stearns MJ., Jr Surgical Management of Patients with Primary Operable Colorectal Cancer and Synchronous Liver Metastases. Am J Surg. 1978;135:81–85. doi: 10.1016/0002-9610(78)90014-4. [DOI] [PubMed] [Google Scholar]
- 4.Yoon SS, Tanabe KK. Surgical Treatment and Other Regional Treatments for Colorectal Cancer Liver Metastases. Oncologist. 1999;4:197–208. [PubMed] [Google Scholar]
- 5.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R, et al. A Combinatorial Library of Lipid-Like Materials for Delivery of RNAi Therapeutics. Nat Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu S, Naing A, Moulder SL, Culotta KS, Madoff DC, Ng CS, Madden TL, Falchook GS, Hong DS, Kurzrock R. Phase I Trial of Hepatic Arterial Infusion of Nanoparticle Albumin–Bound Paclitaxel: Toxicity, Pharmacokinetics, and Activity. Mol Cancer Ther. 2011;10:1300–1307. doi: 10.1158/1535-7163.MCT-11-0259. [DOI] [PubMed] [Google Scholar]
- 7.Schroeder A, Heller DA, Winslow MM, Dahlman JE, Pratt GW, Langer R, Jacks T, Anderson DG. Treating Metastatic Cancer with Nanotechnology. Nat Rev Cancer. 2012;12:39–50. doi: 10.1038/nrc3180. [DOI] [PubMed] [Google Scholar]
- 8.Sudimack J, Lee RJ. Targeted Drug Delivery via the Folate Receptor. Adv Drug Delivery Rev. 2000;41:147–162. doi: 10.1016/s0169-409x(99)00062-9. [DOI] [PubMed] [Google Scholar]
- 9.Lu Y, Low PS. Folate-Mediated Delivery of Macromolecular Anticancer Therapeutic Agents. Adv Drug Delivery Rev. 2002;54:675–693. doi: 10.1016/s0169-409x(02)00042-x. [DOI] [PubMed] [Google Scholar]
- 10.Haque F, Shu D, Shu Y, Shlyakhtenko LS, Rychahou PG, Mark Evers B, Guo P. Ultrastable Synergistic Tetravalent RNA Nanoparticles for Targeting to Cancers. Nano Today. 2012;7:245–257. doi: 10.1016/j.nantod.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo PX, Erickson S, Anderson D. A small viral RNA is required for In Vitro Packaging Of Bacteriophage phi 29 DNA. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 12.Guo P, Zhang C, Chen C, Garver K, Trottier M. Inter-RNA Interaction of Phage phi29 pRNA to Form a Hexameric Complex for Viral DNA Transportation. Mol Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 13.Guo P. The Emerging Field of RNA Nanotechnology. Nat Nanotechnol. 2010;5:833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdelmawla S, Guo S, Zhang L, Pulukuri SM, Patankar P, Conley P, Trebley J, Guo P, Li QX. Pharmacological Characterization of Chemically Synthesized Monomeric phi29 pRNA Nanoparticles for Systemic Delivery. Mol Ther. 2011;19:1312–1322. doi: 10.1038/mt.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shu Y, Haque F, Shu D, Li W, Zhu Z, Kotb M, Lyubchenko Y, Guo P. Fabrication of 14 Different RNA Nanoparticles for Specific Tumor Targeting Without Accumulation in Normal Organs. RNA. 2013;19:767–777. doi: 10.1261/rna.037002.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Thermodynamically Stable RNA Three-Way Junction for Constructing Multifunctional Nanoparticles for Delivery of Therapeutics. Nat Nanotechnol. 2011;6:658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Afonin KA, Kasprzak W, Bindewald E, Puppala PS, Diehl AR, Hall KT, Kim TJ, Zimmermann MT, Jernigan RL, Jaeger L, et al. Computational and Experimental Characterization of RNA Cubic Nanoscaffolds. Methods. 2014;67:256–265. doi: 10.1016/j.ymeth.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasprzak W, Bindewald E, Kim TJ, Jaeger L, Shapiro BA. Use of RNA Structure Flexibility Data in Nanostructure Modeling. Methods. 2011;54:239–250. doi: 10.1016/j.ymeth.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shopsowitz KE, Roh YH, Deng ZJ, Morton SW, Hammond PT. RNAi-Microsponges Form Through Self-Assembly of the Organic and Inorganic Products of Transcription. Small. 2014;10:1623–1633. doi: 10.1002/smll.201302676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JB, Hong J, Bonner DK, Poon Z, Hammond PT. Self-Assembled RNA Interference Microsponges for Efficient siRNA Delivery. Nat Mater. 2012;11:316–322. doi: 10.1038/nmat3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grabow WW, Jaeger L. RNA Self-Assembly and RNA Nanotechnology. Acc Chem Res. 2014;47:1871–1880. doi: 10.1021/ar500076k. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Endrizzi JA, Shu Y, Haque F, Sauter C, Shlyakhtenko LS, Lyubchenko Y, Guo P, Chi YI. Crystal Structure of 3WJ Core Revealing Divalent Ion-Promoted Thermostability and Assembly of the phi29 Hexameric Motor pRNA. RNA. 2013;19:1226–1237. doi: 10.1261/rna.037077.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khisamutdinov EF, Li H, Jasinski DL, Chen J, Fu J, Guo P. Enhancing Immunomodulation on Innate Immunity by Shape Transition Among RNA Triangle, Square and Pentagon Nanovehicles. Nucleic Acids Res. 2014;42:9996–10004. doi: 10.1093/nar/gku516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jasinski DL, Khisamutdinov EF, Lyubchenko YL, Guo P. Physicochemically Tunable Polyfunctionalized RNA Square Architecture with Fluorogenic and Ribozymatic Properties. ACS Nano. 2014;8:7620–7629. doi: 10.1021/nn502160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shu D, Khisamutdinov EF, Zhang L, Guo P. Programmable Folding of Fusion RNA In Vivo and In Vitro Driven by pRNA 3WJ Motif of phi29 DNA Packaging Motor. Nucleic Acids Res. 2014;42:e10. doi: 10.1093/nar/gkt885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elliott VA, Rychahou P, Zaytseva YY, Evers BM. Activation of c-Met and Upregulation of CD44 Expression Are Associated With the Metastatic Phenotype in the Colorectal Cancer Liver Metastasis Model. PLoS ONE. 2014;9:e97432. doi: 10.1371/journal.pone.0097432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C, Zhao M, Liu YR, Luan X, Guan YY, Lu Q, Yu DH, Bai F, Chen HZ, Fang C. Suppression of Colorectal Cancer Subcutaneous Xenograft and Experimental Lung Metastasis Using Nanoparticle-Mediated Drug Delivery to Tumor Neovasculature. Biomaterials. 2014;35:1215–1226. doi: 10.1016/j.biomaterials.2013.08.091. [DOI] [PubMed] [Google Scholar]
- 28.Murphy EA, Majeti BK, Barnes LA, Makale M, Weis SM, Lutu-Fuga K, Wrasidlo W, Cheresh DA. Nanoparticle-Mediated Drug Delivery to Tumor Vasculature Suppresses Metastasis. Proc Natl Acad Sci U S A. 2008;105:9343–9348. doi: 10.1073/pnas.0803728105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Werner ME, Karve S, Sukumar R, Cummings ND, Copp JA, Chen RC, Zhang T, Wang AZ. Folate-Targeted Nanoparticle Delivery of Chemo- and Radiotherapeutics for the Treatment of Ovarian Cancer Peritoneal Metastasis. Biomaterials. 2011;32:8548–8554. doi: 10.1016/j.biomaterials.2011.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Na JH, Koo H, Lee S, Min KH, Park K, Yoo H, Lee SH, Park JH, Kwon IC, Jeong SY, et al. Real-Time and Non-Invasive Optical Imaging of Tumor-Targeting Glycol Chitosan Nanoparticles in Various Tumor Models. Biomaterials. 2011;32:5252–5261. doi: 10.1016/j.biomaterials.2011.03.076. [DOI] [PubMed] [Google Scholar]
- 31.Peiris PM, Toy R, Doolittle E, Pansky J, Abramowski A, Tam M, Vicente P, Tran E, Hayden E, Camann A, et al. Imaging Metastasis Using an Integrin-Targeting Chain-Shaped Nanoparticle. ACS Nano. 2012;6:8783–8795. doi: 10.1021/nn303833p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kusumoto K, Akita H, Ishitsuka T, Matsumoto Y, Nomoto T, Furukawa R, El-Sayed A, Hatakeyama H, Kajimoto K, Yamada Y, et al. Lipid Envelope-Type Nanoparticle Incorporating a Multifunctional Peptide for Systemic siRNA Delivery to the Pulmonary Endothelium. ACS Nano. 2013;7:7534–7541. doi: 10.1021/nn401317t. [DOI] [PubMed] [Google Scholar]
- 33.Luo G, Yu X, Jin C, Yang F, Fu D, Long J, Xu J, Zhan C, Lu W. Lyp-1-Conjugated Nanoparticles for Targeting Drug Delivery to Lymphatic Metastatic Tumors. Int J Pharm. 2010;385:150–156. doi: 10.1016/j.ijpharm.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 34.Li SD, Chono S, Huang L. Efficient Gene Silencing in Metastatic Tumor by siRNA Formulated in Surface-Modified Nanoparticles. J Control Release. 2008;126:77–84. doi: 10.1016/j.jconrel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang F, Huang W, Li Y, Liu S, Jin M, Wang Y, Jia L, Gao Z. Anti-Tumor Effects in Mice Induced by Survivin-Targeted siRNA Delivered Through Polysaccharide Nanoparticles. Biomaterials. 2013;34:5689–5699. doi: 10.1016/j.biomaterials.2013.03.047. [DOI] [PubMed] [Google Scholar]
- 36.Ji S, Xu J, Zhang B, Yao W, Xu W, Wu W, Xu Y, Wang H, Ni Q, Hou H, et al. RGD-Conjugated Albumin Nanoparticles as a Novel Delivery Vehicle in Pancreatic Cancer Therapy. Cancer Biol Ther. 2012;13:206–215. doi: 10.4161/cbt.13.4.18692. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Xu Z, Guo S, Zhang L, Sharma A, Robertson GP, Huang L. Intravenous Delivery of siRNA Targeting CD47 Effectively Inhibits Melanoma Tumor Growth and Lung Metastasis. Mol Ther. 2013;21:1919–1929. doi: 10.1038/mt.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim SH, Choi SA, Lee JY, Wang KC, Phi JH, Lee DH, Song SH, Song JH, Jin X, Kim H, et al. Therapeutic Targeting of Subdural Medulloblastomas Using Human Neural Stem Cells Expressing Carboxylesterase. Cancer Gene Ther. 2011;18:817–824. doi: 10.1038/cgt.2011.52. [DOI] [PubMed] [Google Scholar]
- 39.Murphy EA, Majeti BK, Mukthavaram R, Acevedo LM, Barnes LA, Cheresh DA. Targeted Nanogels: A Versatile Platform for Drug Delivery to Tumors. Mol Cancer Ther. 2011;10:972–982. doi: 10.1158/1535-7163.MCT-10-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Zhu X, Zhang X, Liu B, Huang L. Nanoparticles Modified With Tumor-Targeting scFv Deliver siRNA and miRNA for Cancer Therapy. Mol Ther. 2010;18:1650–1656. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo S, Tschammer N, Mohammed S, Guo P. Specific Delivery of Therapeutic RNAs to cancer Cells via the Dimerization Mechanism of phi29 Motor pRNA. Hum Gene Ther. 2005;16:1097–1109. doi: 10.1089/hum.2005.16.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shu D, Huang LP, Hoeprich S, Guo P. Construction of phi29 DNA-Packaging RNA Monomers, Dimers, and Trimers with Variable Sizes and Shapes as Potential Parts for Nanodevices. J Nanosci Nanotechnol. 2003;3:295–302. doi: 10.1166/jnn.2003.160. [DOI] [PubMed] [Google Scholar]
- 43.Shu Y, Cinier M, Shu D, Guo P. Assembly of Multifunctional phi29 pRNA Nanoparticles for Specific Delivery of siRNA and Other Therapeutics to Targeted Cells. Methods. 2011;54:204–214. doi: 10.1016/j.ymeth.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khaled A, Guo S, Li F, Guo P. Controllable Self-Assembly of Nanoparticles for Specific Delivery of Multiple Therapeutic Molecules to Cancer Cells Using RNA Nanotechnology. Nano letters. 2005;5:1797–1808. doi: 10.1021/nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsumura Y, Maeda H. A New Concept for Macromolecular Therapeutics in Cancer Chemotherapy: Mechanism of Tumoritropic Accumulation of Proteins and the Antitumor Agent Smancs. Cancer Res. 1986;46:6387–6392. [PubMed] [Google Scholar]
- 46.McNamara JO, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell Type-Specific Delivery of siRNAs with Aptamer-siRNA Chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 47.Morikawa K, Walker SM, Nakajima M, Pathak S, Jessup JM, Fidler IJ. Influence of Organ Environment on the Growth, Selection, and Metastasis of Human Colon Carcinoma Cells in Nude Mice. Cancer Res. 1988;48:6863–6871. [PubMed] [Google Scholar]
- 48.Gulhati P, Bowen KA, Liu J, Stevens PD, Rychahou PG, Chen M, Lee EY, Weiss HL, O'Connor KL, Gao T, et al. mTORC1 and mTORC2 regulate EMT, Motility, and Metastasis of Colorectal Cancer via Rhoa and Rac1 Signaling Pathways. Cancer Res. 2011;71:3246–3256. doi: 10.1158/0008-5472.CAN-10-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rychahou PG, Jackson LN, Silva SR, Rajaraman S, Evers BM. Targeted Molecular Therapy of the PI3K Pathway: Therapeutic Significance of PI3K Subunit Targeting in Colorectal Carcinoma. Ann Surg. 2006;243:833–842. doi: 10.1097/01.sla.0000220040.66012.a9. discussion 843-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and Predictive Factors in Breast Cancer by Immunohistochemical Analysis. Mod Pathol. 1998;11:155–168. [PubMed] [Google Scholar]
- 51.Bruns CJ, Harbison MT, Kuniyasu H, Eue I, Fidler IJ. In Vivo Selection and Characterization of Metastatic Variants from Human Pancreatic Adenocarcinoma by Using Orthotopic Implantation in Nude Mice. Neoplasia. 1999;1:50–62. doi: 10.1038/sj.neo.7900005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shu Y, Shu D, Haque F, Guo P. Fabrication of pRNA Nanoparticles to Deliver Therapeutic RNAs and Bioactive Compounds into Tumor Cells. Nat Protocols. 2013;8:1635–1659. doi: 10.1038/nprot.2013.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


