Abstract
The Hippo pathway plays a key role in controlling organ growth in many animal species and its deregulation is associated with different types of cancer. Understanding the regulation of the Hippo pathway and discovering upstream regulators is thus a major quest. Interestingly, while the core of the Hippo pathway contains a highly conserved kinase cascade, different components have been identified as upstream regulators in Drosophila and vertebrates. However, whether the regulation of the Hippo pathway is indeed different between Drosophila and vertebrates or whether these differences are due to our limited analysis of these components in different organisms is not known. Here we show that the mouse Fat4 cadherin, the ortholog of the Hippo pathway regulator Fat in Drosophila, does not apparently regulate the Hippo pathway in the murine liver. In fact, we uncovered an evolutionary shift in many of the known upstream regulators at the base of the arthropod lineage. In this evolutionary transition, Fat and the adaptor protein Expanded gained novel domains that connected them to the Hippo pathway, whereas the cell-adhesion receptor Echinoid evolved as a new protein. Subsequently, the junctional adaptor protein Angiomotin (Amot) was lost and the downstream effector Yap lost its PDZ-binding motif that interacts with cell junction proteins. We conclude that fundamental differences exist in the upstream regulatory mechanisms of Hippo signaling between Drosophila and vertebrates.
Keywords: Hippo signaling, evolution, Fat, Dachs, Angiomotin, Yap
Introduction
Growth is a fundamental aspect of animal development, and the Hippo signal-transduction pathway has emerged as a major pathway that controls organ size.1–4 Loss of Hippo pathway activity in flies and mice results in severely overgrown tissues with an excess number of cells.1–4 A major question about the Hippo pathway concerns the identity of upstream regulators and their function in growth control. Studies in flies and mice identified several upstream regulators of the Hippo pathway. Although it is frequently assumed that upstream regulators have similar functions in flies and mammals, it is not clear, however, which of these inputs are actually conserved.
The core of the Hippo pathway in Drosophila consist of two kinases, Hippo (Hpo) and Warts (Wts), and their adaptor proteins, Salvador (Sav) and Mob as a tumor suppressor (Mats), which regulate the activity of the transcriptional coactivator Yorkie (Yki) and its transcription factor binding partner Scalloped (Sd).1–4 The core of the Hippo pathway is highly conserved in evolution and homologous components are found in representatives of most major animal phyla (Supplementary Figure S1 A).5,6 Specifically, Hpo, Wts, Sav, Mats, Yki and Sd predate the animal radiation, as homologs are present in amoeboid holozoans, where they also form a kinase cascade.5 Vertebrates have two Hpo homologs, Mst1 and Mst2, that work in a redundant manner, as only double mutants show an overgrowth phenotype.7–12 Warts has two vertebrate homologs, Lats1 and Lats2, that function in similar ways by phosphorylating and thereby inhibiting the activity of Yki or the vertebrate homologs Yap and Taz, respectively. In mammals, Yap and Taz have added levels of diversity not observed in flies: Yap and Taz have eight and two alternative splicing isoforms, respectively.13–15 Also, while Yap and Taz are targeted for degradation in mammals, this has not been observed in flies.16,17 Indeed, the phosphodegron responsible for the degradation of Yap and Taz is not conserved in fly Yki,16 although we found this to be conserved in other classes of insects (not shown). Sd has four mammalian homologs, TEAD1–4, that all show high evolutionary conservation.18 Importantly, biochemical experiments and transgenic rescue experiments indicate that the core components function in comparable ways in different phyla, at least in Drosophila and vertebrate systems. The core of the Hippo pathway thus forms a highly conserved signal-transduction module.1–4,19
Some of the upstream regulators of the Hippo pathway, like Merlin (Mer), Kibra, Expanded (Ex), Ras association family members (Rassf), Fat (Ft), Tao, Ajuba, Crumbs (Crb), ZO proteins and α-catenin, are present in both flies and mammals.1–4 However, confusion and controversy exists as to whether the function of these proteins as regulators of the Hippo pathway is also conserved. For example, it is not clear whether the vertebrate homologs of two important regulators in flies, Ft and Ex, regulate Hippo signaling. Knockdown of the ft ortholog, Fat4 (a.k.a. Fatj), in the neural tube of chicks leads to increased cell proliferation of progenitor cells and Yap activation,20 whereas deletion of Fat4 in mice does not lead to overgrowth defects, but instead leads to smaller kidneys and defects in planar cell polarity (PCP)21,22 Thus, while the function of Ft In PCP appears to be conserved between flies and vertebrates,21–25 whether Fat4 directly connects with the Hippo pathway is not clear. Ex has a human ortholog, FRMD6, that may act as a tumor suppressor in vertebrates.26,27 However, whether FRMD6 acts through effects on Hippo signaling is controversial.26,27
To clear up confusion about the conservation and divergence of upstream regulators of the Hippo pathway, we performed a systematic analysis. We used a combination of mouse knockout studies of Fat4 and In vivo structure function analysis of Drosophlia Ft and identified a motif in Drosophlia Ft that is necessary to signal to the Hippo pathway. We then traced the evolutionary origin of this domain and that of other Hippo pathway components and their functional domains to clarify the evolutionary history of the known upstream components. Our analysis revealed an evolutionary shift of several regulators of the Hippo pathway at the base of the arthropod lineage that affected their function In the Hippo pathway. Our analysis indicates that during arthropod evolution Fat Ex and Echinoid (Ed) gained function in the Hippo pathway, whereas Angiomotin was lost and Yap changed its molecular interactions. In addition to these changes in the arthropod lineage, Dachs was lost in chordates. We conclude that fundamental differences exist In the mechanisms of Hippo pathway regulation between flies and mice.
Results
Fat4 does not regulate Hippo signaling in mammalian livers
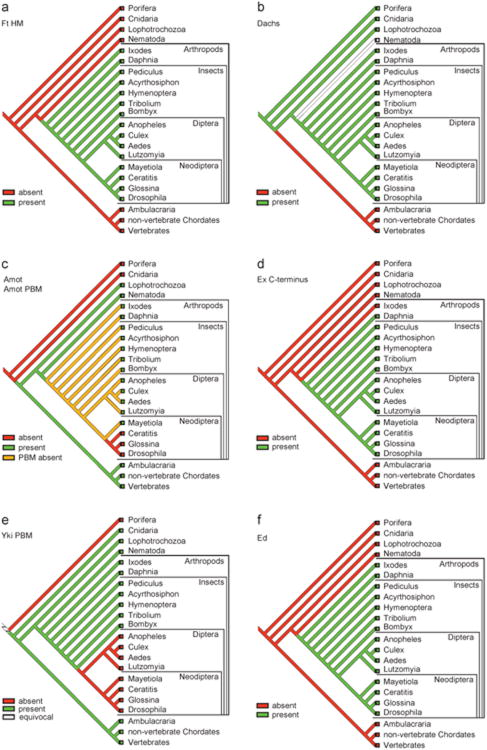
Drosophiia has two Fat-like proteins: Ft, which regulates tissue growth through the Hippo pathway28–32 and PCP independently of the Hippo pathway, and Kugelei, which regulates PCP but not the Hippo pathway.33,34 Mice have four Ft homologs: Fat1–3 are orthologs of Kugelei, and Fat4 is the sole ortholog of Ft.33 Recent studies investigating the function of Fat4 did not report overgrowth phenotypes in Fat4 knockout mice,21,22 indicating that Fat4 might be dispensable for the regulation of Hippo signaling in mice. To more directly examine a role for Fat4 in Hippo signaling, we used Albumin-Cre (Alb-Cre) to delete Fat4 specifically in the mouse liver, an organ that is exquisitely sensitive to Hippo signaling.1,35 Unlike mice defective in Hippo signaling,6–11,36,37 Alb-Cre; Fat4flox/flox mice (N>5) did not show any phenotypic abnormalities at 3.5 or 11 months of age compared with age-matched wild-type littermates (Figures 1a and b), demonstrating that Fat4 is dispensable for normal liver growth.
Figure 1.
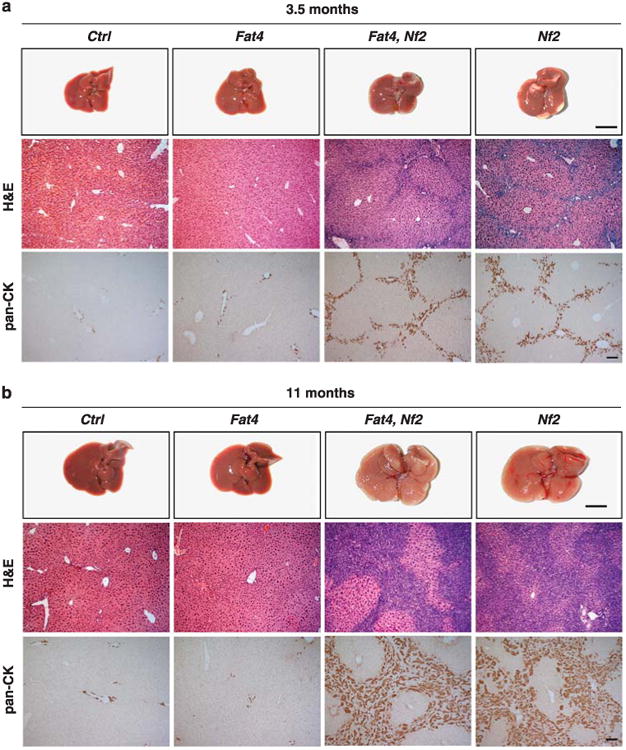
Fat mutant livers do not show a Hippo-like phenotype. Gross Images, hematoxylin and eosin (H&E) and pan-CK staining of livers from wild-type control, Alb-Cre; Fat4flox/flox, Alb-Cre; Fat4flox/flox; Nf2flox/flox and Alb-Cre; Nf2flox/flox at (a) 3.5 months of age or (b) 11 months of age. Gross images show that Nf2 mutants and Nf2 Fat4 double mutants developed comparable hepatomegaly and bile duct hamartomas at the surface of the liver, whereas Fat4 mutant livers were normal. Scale bar=1cm. H&E and pan-CK staining showed comparable bile duct overproliferation in Nf2 mutant and Nf2; Fat4 double mutant livers. Scale bar=100μm.
In Drosophila, Ft functions semiredundantly with Mer to regulate Hippo signaling in imaginal discs, and ft mer double mutants show much more severe tissue overgrowth phenotypes than either single mutant.30,32 To exclude the possibility that our analysis of Fat4 mutant mice might have missed a subtle role for Fat4 in mammalian Hippo signaling, we further examined a double mutant combination of Fat4 with Nf2, the gene encoding Mer. We generated Nf2 knockout livers and Fat4; Nf2 double knockout livers using Alb-Cre (Alb-Cre; Nf2flox/flox and Alb-Cre; Fat4flox/flox; Nf2flox/flox). As reported previously,36 Nf2 mutant livers developed hepatomegaly and widespread bile duct hamartomas, which appeared microscopically as collections of irregularly shaped duct-like structures comprised of cytokeratin-positive biliary epithelial cells (Figures 1a and b). Unlike the strong synergy in the overgrowth phenotype observed between ft and mer mutations in flies,30,32 examination of Fat4; Nf2 double mutant livers revealed similar phenotypes as Nf2 mutant livers in liver size and histology (Figures 1a and b). Thus, even in a sensitized genetic background with compromised Hippo signaling due to loss of Nf2, Fat4 does not contribute significantly to Hippo signaling. While negative data can be difficult to interpret, our genetic analysis, together with the already published data, indicates that the role of Fat4 in PCP is conserved between flies and mammals, and that Fat4 has no or only a minor role in the Hippo pathway in mammals.
The growth control and PCP functions of Fat can be separated
An important question concerns the mechanism by which Ft signals into the Hippo pathway. In a genetic screen for novel Hippo pathway components, we isolated a new allele of ft that we named ‘fat super-size me’ (ftsum) because it specifically affects the function of Ft in growth control. Homozygous ftsum mutant larvae had strongly overgrown eye and wing imaginal discs (Figures 2a and d) and ftsum mutant clones had overgrown eye imaginal discs (Figures 2e and f). Interestingly, ftsum mutant tissues showed no observable PCP phenotypes as assayed by analyzing bristle orientation on the abdomen of mutant pharate adults (Figures 2g-j) and the orientation of ommatidia in mutant clones visualized by md-lacZ expression, which marks R4 photoreceptors (Figures 2k and I). Importantly, ftsum mutant clones upregulated fj-lacZ, a readout for Yki activity, similar to ft422 mutant clones (Figures 2e and f). Ft functions through a mechanism that involves the atypical myosin Dachs (D).29,38–40 In wild-type imaginal discs, D is asymmetrically localized along the proximal-distal axis of each cell, but this asymmetric localization was lost and D levels were increased in ft-null mutant clones (Figure 2n).41–44 Increased D levels correlate with increased growth,40 whereas the asymmetric localization of D is involved in the regulation of PCP.41–44 Interestingly, we found that D was still asymmetrically localized but was increased in ftsum mutant clones, further demonstrating that the PCP function of Ft was not significantly affected by the ftsum mutation (Figure 2n). We sequenced the entire ft open reading frame of ftsum mutants and found a single point mutation giving rise to a substitution of isoleucine 4852 to an asparagine in the Ft intracellular domain (Supplementary Figure S3). Importantly, ftsum has a point mutation in the normal genomic context of the ft gene and therefore the observed phenotypes did not rely on rescue constructs that may cause unrelated overexpression or insertion artifacts. These data indicate that isoleucine 4852 is important in regulating the levels of D but not its subcellular localization. Thus, the tumor suppressor function of Ft can be separated from its PCP function.
Figure 2.
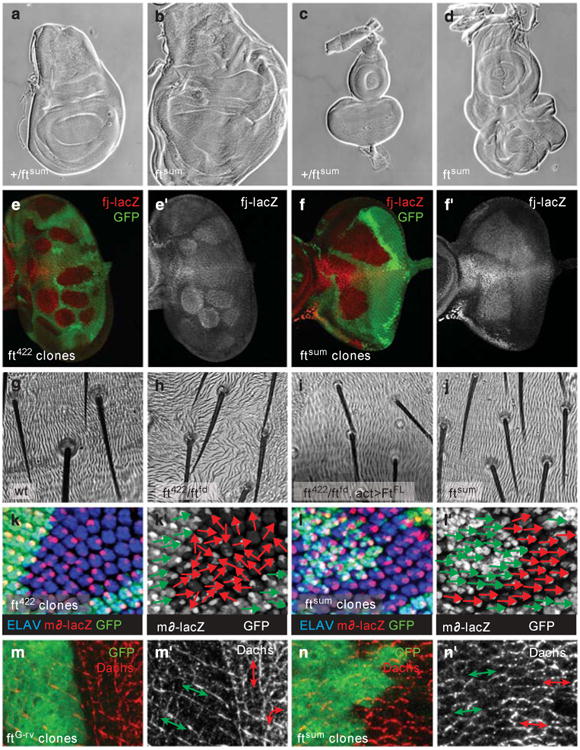
ftsum mutants display overgrowth and activation of Yki-activity readouts, but no planar cell polarity phenotypes. (a and b) Third instar wing discs in (a) heterozygous ftsum or (b) homozygous ftsum mutants, (c and d) Third instar eye discs in (c) heterozygous ftsum mutants or (d) homozygous ftsum mutants, (e) diap1-lacZ expression (red and gray, e′) in eye discs containing ft422 mutant clones marked by the absence of GFP expression, (f) diapl-lacZ expression (red and gray, f′) in eye discs containing ftsum mutant clones marked by the absence of GFP expression, (g-j) Pharate adult abdomen to analyze planar cell polarity phenotypes by hair orientation in (g) wild-type, (h) ft422/ftfd mutant, (i) ft422/ftfd mutant with act-Gal4-driven expression of FtFL and, (j) ftsum mutant animals, (k and I) Third instar wing discs with ft422 or ftsum clones respectively to analyze planar cell polarity by analyzing photoreceptor R4 location marked by m∂-lacZ expression (red) and ELAV (blue) to mark all photoreceptor cells. Mutant clones are marked by the absence of GFP expression (green), (k′–I′) The orientation of rhabdomers is indicated by green arrows for wild-type tissue and red arrows for mutant tissue, (m and n) Dachs stainings in third instar wing discs. D (red and gray, m′ and n′) has a proximodistal sub-cellular localization in wild-type cells marked by GFP expression (green), (m) ftG–rv null mutant clones, marked by the absence of GFP, show mislocalization of D around the cell circumference and increased D levels, (n) ftsum mutant clones, marked by the absence of GFP, show increased D levels but normal localization.
The Ft intracellular domain has no annotated protein domains but it does have portions that show varying levels of evolutionary conservation between flies and vertebrates (Supplementary Figure S3). Interestingly, the ftsum mutation resides in a motif that is not conserved between flies and vertebrates, although there are several other motifs that are highly conserved (Supplementary Figure S3). To assess the functional importance of the conserved motifs, we generated a series of deletion constructs in the context of the FtΔECD construct, a Ft construct that lacks the extracellular domain but that is sufficient to rescue growth and PCP phenotypes of ft mutant animals.45 All of our deletion constructs were inserted into the same genomic location by PhiC31-mediated integration46 to ensure equivalent expression levels. We then assayed the activity of these constructs in five different in vivo assays that report Hippo pathway activity and PCP regulation. Specifically, we used actin-Gal4 to overexpress the different Ft constructs in ft mutant animals and assayed the ability to rescue the lethality, PCP of abdominal hairs and the wing size (Figure 3a and Supplementary Figure S4). In addition, we expressed the Ft constructs using hh-Gal4 in ft mutants to test the effect on the size of the posterior compartment and on ex-lacZ expression in wing imaginal discs (Figure 3a and Supplementary Figure S5). We found that overexpression of a deletion construct in which motif 3 is deleted (FtΔ3) was not able to rescue lethality (Figure 3a), wing size (Figure 3a and Supplementary Figure S4), imaginal disc size (Figures 3a-e) and ex-lacZ expression in ft mutants (Figures 3a and 2f-j and Supplementary Figure S5), whereas all other deletion constructs were able to rescue the growth and PCP defects (Figure 3a and Supplementary Figures S4–6, quantified in Supplementary Figure S5). Importantly, deletion of motif 3 did not destabilize the Ft protein as FtΔ3 was expressed at similar levels compared with the other Ft constructs (Figures 3i and j). Therefore, specifically motif 3 is essential for Hippo signaling regulation. We thus named this motif the Hippo pathway-interacting motif (HM). Notably, the HM (residues 4834–4899) overlaps with the domains identified by Matakatsu and Blair47 to be essential for the regulation of Hippo signaling, namely the Hippo N (residues 4775–4836) and Hippo C (residues 4839–4920) domains (Supplementary Figure S3). Notably, the residue that is mutated in ftsum lies in the HM, further demonstrating the importance of the HM in the regulation of the Hippo pathway. Our data also indicate that the other conserved motifs are individually dispensable for growth and PCP regulation. However, an FtΔECD -derived construct that deleted all motifs except the HM failed to rescue lethality, overgrowth and fj-lacZ expression (Figures 3a and k). Therefore, the motifs outside of the HM are necessary in combination with the HM for full Ft activity. Overall, we conclude that the HM is essential for the regulation of growth through the Hippo pathway.
Figure 3.
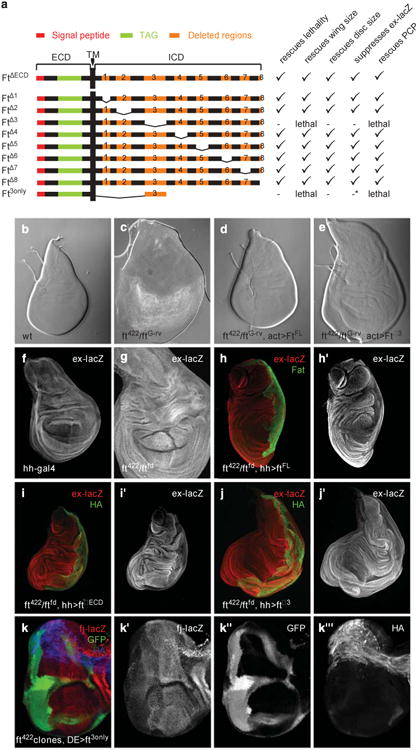
Amino-acid residues 4834-4899 in the intracellular domain of Fat are essential for its tumor suppressor function in Drosophila melanogaster. (a) Graphical representation of the different UAS-Ft constructs (left) and the phenotypes upon expression in a ft mutant background. Rescue of lethality was assayed by act-gal4-driven expression in ft422/ftG–rv trans-heterozygotes. Wing size (Supplementary Figure S4), wing disc size (Supplementary Figure S5) and PCP phenotypes in the abdominal hairs (Supplementary Figure S6) were analyzed in the same genotype. Analysis of ex-lacZ expression was analyzed by hh-Gal4-induced expression in ft422/ftfd trans-heterozygotes. * in the Ft3only construct indicates the use of f]-lacZ. (b-e) Bright-field images of third instar wing imaginal disc of (b) wild-type, (c) ft422/ftG–rv trans-heterozygotes, (d) ft422/ftG–rv discs with act-Gal4-driven expression of full-length Ft and (e) ft422/ftG–rv discs with act-Gal4-driven expression of UAS-FtΔ3. (f-j) The ex-lacZ expression (red and gray, h′, i′, j′) in wing discs of (f) hh-Cal4, (g) ft422/ftG–rv, (h) ft422/ftG–rv, hh-Gal4, UAS-FfFL, (i) ft422/ftG–rv, hh-Gal4, UAS-FtDECD and (j) ft422/ftG–rv, hh-Gal4, UAS-FtD3 third instar larva, (k) The hs-Flp ft422clones, DE-Gal4, UAS-Ft30nly eye discs showing Ff422 clones marked by the absence of GFP (green and gray, k″), showing fj-lacZ (red and gray, k′). The Ft3only construct is marked by an HA tag (blue, gray, k‴).
Co-option of Fat into the Hippo pathway in arthropods
The finding that the HM, which is not conserved in mammals, is essential for the regulation of growth control, whereas single deletion of the other conserved motifs had no noticeable effect, indicates that Fat might have different molecular functions in flies and vertebrates. The molecular mechanisms of Ft signal transduction are poorly understood, but genetic evidence in Drosophila indicates that Ft signals through D29,38–40 to regulate the levels of Wts29 and to affect the localization of Ex.30,32 We thus wanted to trace the evolutionary history of the HM, the other motifs in the Ft intracellular domain and D. We found that the HM is conserved in arthropods, but is not found outside of arthropods (Figure 4a). Notably, the 65 amino acids of the HM of Ft represent a sequence that does not share any significant sequence similarity to sequences of the human and mouse proteomes. In addition, the residue that is mutated in ftsum is conserved in all arthropod species analyzed, pertaining to its functional significance. Thus, Ft acquired a new domain in arthropods that links it to the Hippo pathway. Dachs is present in flies but not in vertebrates. Phylogenetic tracing of D indicates that D may be ancestral in metazoans, but is lost in chordates. Interestingly, D belongs to the myosin X subfamily, which, similar as D in flies,44 regulates spindle orientation in mice.48,49 It is not clear whether D has a function in Hippo signaling outside of arthropods, as some domains are arthropod-specific although it is not known whether these domains act in Hippo signaling. We therefore conclude that the Ft-D axis regulates the Hippo pathway in flies but is absent in vertebrates.
Figure 4.
Phylogenetic distribution of Hippo pathway components. Phylogenetic trees represent different phyla and specific species in arthropods, (a) Fat gains a Hippo signaling interaction motif at the base of arthropods, (b) D is lost in the lineage that gives rise to arthropods. (c) Angiomotin originates at the base of bilateria and is lost in the neodiptera lineage, (d) The Yorkie C-terminal PDZ-binding motif (PBM) originates at the base of eumetazoans and is lost in diptera. (e) Ed is present only in arthropods, (f) Ex gains a C-terminal Hippo signaling interaction domain in the arthropod lineage.
The Crumbs complex exchanges Angiomotin for Expanded at the base of the arthropod lineage
An upstream regulator of Hippo signaling that is present in mammals and flies is the Crb complex composed of the transmembrane domain protein Crb and its binding partners.50 At first sight, it therefore appears that the mechanism of Crb action in the Hippo pathway may be conserved. Interestingly however, different mechanisms have been described by which Crb regulates the Hippo pathway.
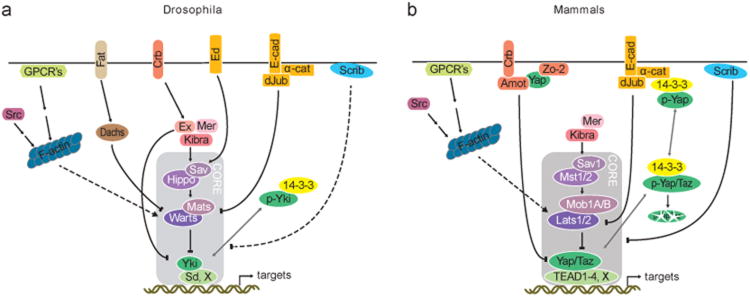
In vertebrates, Crb has three homologs that form a complex with the Angiomotin family proteins, Angiomotin (Amot), and Angiomotin-like 1 and 2 (Amotl1 and Amotl2). Amot proteins are adaptor proteins that associate with the cytoskeleton and localize to tight junctions.51–55 Amot binds Yap through the interaction of its PPxY and LPxY motifs with the WW domains of Yap and Taz56–58 (Supplementary Figures S1 and S2). In addition, Amot interacts with the junction proteins ZO-1 and -2 using its PDZ interaction motif.59,60 Importantly, Amot family proteins are required for the cytoplasmic localization of Yap and Taz.51–53,56 Strikingly, although Amot family proteins are essential for the proper regulation of Yap and Taz in vertebrates, no Amot homolog is present in Drosophila. Was Amot ancestral to both vertebrates and flies but then lost in the lineage giving rise to flies or was Amot gained in the lineage giving rise to vertebrates? To distinguish between these possibilities, we tracked the phylogenetic distribution of Amot genes. We found that Amot-like genes are present in representatives of all major bilaterian phyla including arthropods (Figure 4c). Hence, Amot was most likely present in the last common ancestor of all bilaterian animals and lost during arthropod evolution. Within arthropods, Amot is present in all classes including insects, but in insects it is not present in Schizophora, part of the Neodiptera, which contain fruit flies and houseflies (Figure 4c). Thus, Amot was originally present in all bilaterian animals, but was lost relatively recently in the dipteran lineage that includes Drosophila. Interestingly, although Amot is present in most arthropods, several arthropod Amot proteins lack motifs that are functionally important in vertebrate Amot. First, the C-terminal PDZ-binding motif, which links Amot to the Crb complex, was lost in arthropods. Second, the PPxY motifs, which mediate its interaction with Yap, were lost multiple times independently in arthropods. These data indicate that Amot became less important in arthropods, leading to low selection pressure on important domains and ultimately to the loss of Amot itself. Thus, the Crb complex lost Amot as a link to the Hippo pathway at the base of the arthropod lineage.
The loss of Amot's PDZ-binding motif in arthropods indicates that Amot cannot relay information from the Crb complex into the Hippo pathway. In flies, however, the Crb complex affects Hippo signaling through the FERM domain adaptor protein Ex.61–63 Ex regulates the Hippo pathway by forming a complex with Mer and Kibra to regulate the Hpo and Wts kinases64,65 and by directly binding to Yki.66,67 Interestingly, although Ex and Amot are different proteins with different domains, they resemble each other in their function: both localize subapically, both bind to the Crb complex61–63 and both bind to Yap and Taz/Yki directly.61–63,66–69 We thus investigated the evolution of Ex. The FERM domain of Ex, which is necessary for complex formation with Mer and Kibra, is present, although diversified, in Ex orthologs outside of arthropods (Supplementary Figure S7), indicating that also outside of arthropods. Ex may complex with Mer and Kibra. In contrast to the FERM domain, however, the C-terminal domain of Drosophila Ex, which is necessary and sufficient for its growth control function,70,71 is not conserved in Ex homologs outside of arthropods (Figure 4d and Supplementary Figure S7). Importantly, the three PPxY motifs, which are located in the C-terminal domain of Ex and are necessary for Yki binding, are conserved in arthropods. Thus, the interactions of Ex with Yki and Hpo, respectively, are evolutionary innovations in the arthropod lineage, more specifically in pancrustacea. Therefore, although Ex may form a complex with Mer and Kibra in vertebrates, it is unlikely that vertebrate Ex functions in the same way in the Hippo pathway as in Drosophila, as the C-terminal domain that interacts with the Hippo pathway is arthropod-specific. Thus, the acquisition of the new C-terminal domain in Ex co-occurred with the loss of the PDZ-binding motif of Amot. We speculate that a switch occurred in the regulation of Hippo signaling from Amot to Ex at the base of the arthropod lineage.
Yap loses its PDZ-binding motif during arthropod evolution
Given the changes in Amot and Ex, which both form complexes with Yap and Taz, we wondered whether the loss of Amot and the gain of a novel domain in Ex was accompanied by changes in Yap homologs. In mammals. Yap and Taz are localized to apical junctions by a tripartite complex of Amot, Yap/Taz and the tight junction component ZO-2 (and perhaps ZO-1).59,60 The WW domains of Yap and Taz interact with the PPxY motifs of Amot and the PDZ-binding motif of Yap interacts with the PDZ domain of zo-2.51,59,60,72,73 Single Yap orthologs are found in species of different animal phyla including cnidaria (Supplementary Figure S1A),12 indicating that a single Yap gene was ancestral. Notably, the different domains of Yap, its N-terminal Sd interaction domain, its main Wts phosphorylation site that binds to 14-3-3, its WW domains and its C-terminal PDZ-binding motif that binds to ZO-2 are all highly conserved throughout the phylogenetic tree except in Diptera, where the PDZ-binding motif and the full-length transactivation domain are no longer present (Figure 4e). Thus, we speculate that the loss of the interaction of Amot with the Crb complex at the base of the arthropod lineage opened the possibility for the loss of the PDZ-binding motif of Yap, which also interacts with the Crb complex. Also at the base of the arthropod lineage. Ex gained a novel Yki-interacting domain. Thus, the mechanisms by which the Crb complex regulates the Hippo pathway, including the direct interaction with Yap, changed during arthropod evolution.
The adherens junction protein Echinoid is arthropod-specific
The loss of Amot during arthropod evolution coincides with major changes in the organization of cell-cell junctions.74 Amot is thus not the only junctional protein that changed. One of the junctional components that is present in flies but not in mammals is Ed (Figure 4f). Ed is loosely related to Hemicentin in mammals, but reciprocal blast searches indicate that they are not orthologs. Interestingly, Ed not only promotes formation of adherens junctions but also plays a role in the regulation of Hippo signaling. Indeed, Ed acts as tumor suppressor upstream of the Hippo pathway and complexes with Ex, Sav, Mer and Yki through its intracellular domain.75 The C-terminal portion of the intracellular domain, which is required for Hippo regulation, is highly conserved in arthropods but not present outside of arthropods. We conclude that Ed originated as a regulator of the Hippo pathway at the base of the arthropod lineage.
Discussion
We addressed the question how similar or dissimilar is the regulation of Hippo signaling in the premier model systems used to study the Hippo pathway, namely Drosophila melanogaster and Mus musculus. Our analyses imply that despite the high conservation of the core components of the Hippo pathway in the animal kingdom, many of the currently known upstream regulatory mechanisms of the Hippo pathway are not conserved between flies and mice. Most notably, Ft and Ex evolved novel protein domains that together with the emergence of Ed formed novel regulatory inputs into the Hippo pathway at the base of the arthropods. Later, Amot and the PDZ-binding motif of Yap were lost during arthropod evolution. In addition to these divergent inputs, the Hippo pathway also has conserved upstream regulators. The currently known conserved upstream regulators that regulate the Hippo pathway through conserved mechanisms are Mer, Kibra, Rassf, Ajuba and Tao. Another group of Hippo pathway regulators are proteins that regulate the formation and function of F-actin, including G-protein-coupled receptors,76 Src,77 cell-cell junction components.78 Although these components are conserved, more needs to be understood on the role and the mechanism of actin in the regulation of the Hippo pathway to fully appreciate whether their mechanisms are evolutionarily conserved. Taken together, fundamental differences exist in the upstream regulatory mechanisms modulating the activity of the Hippo pathway in different species (Figure 5). An interesting evolutionary divergence that we uncovered involves the co-option of Ft into the Hippo pathway at the base of the arthropod lineage. Fat proteins control PCP in vertebrates and flies. The mechanisms by which Fat functions in PCP are not known but the regulation of D localization may be important in flies.40,43,79 However, vertebrates do not have a D homolog, although they do have the closely related myosin X. Interestingly, Fat4 and myosin X mutant mice have similar PCP phenotypes, specifically a misorientation of the mitotic spindle,22,80,81 and D is necessary for correct spindle orientation in flies.43,79 We thus speculate that Fat4 affects PCP by interaction with myosin X, similar to the action of Fat–D in flies. It is unclear whether D functions in Hippo signaling outside of arthropods. Indeed, although D is ancestral to metazoans, arthropod D contains two domains that are specific to arthropod D, which may be important for Hippo signaling. Additionally, we found that Fat acquired a new domain that regulates Hippo signaling. This indicates that outside of arthropods Fat does either not directly regulate the Hippo pathway or regulates the Hippo pathway through mechanisms different than those in flies, for example, by using a different motif in its intracellular domain. Thus, further investigations are required to test whether under conditions where Fat4 affects Yap activity in vertebrates, Fat4 has direct effects on the Hippo pathway or affects Hippo signaling indirectly through its interaction with adherens junctions or through effects on PCP20,82,83
Figure 5.
Simplified representations of Hippo pathway regulation in Drosophila and mammals, (a) Graphical representation of identified Hippo pathway components in Drosophila melanogaster (b) and in mammals. In both graphical representations, solid lines indicate direct interactions and gray lines indicate changes in subcellular localization.
The existence of differences in upstream regulatory inputs between flies and vertebrates is unusual for a developmentally critical signal-transduction pathway, and most other signal-transduction pathways have specific signaling molecules and receptors that are highly conserved, such as the Wnt, Hedgehog, TGFb and Notch pathways, for example, Mauviel et al.84 What then are the implications of this evolutionary switch in the Hippo pathway? How can the Hippo pathway have such dramatic evolutionary plasticity and what does this tell us about the function of the pathway during development?
First, evolutionary changes occurred in several upstream regulators of the Hippo pathway, and at least in flies these inputs often act partially redundant,1–3 which may enable evolutionary flexibility. Second, the evolution of different inputs is the basis of differential regulation of the pathway in different tissues. In flies, for example, the Ft–D axis is essential for the regulation of the pathway in growing imaginal discs, while it is not required for the control of follicle cell proliferation.85,86 On the other hand, Mer is essential in follicle cells, but has only a minor role in imaginal discs.65,87,88 Similarly in vertebrates, α-catenin is essential for the regulation of Yap in keratinocytes where the core Hpo kinases Mst1/2 are not required.89,90 Thus, loss of Hippo pathway components can have strikingly different effects on the activity of the pathway in different tissues. Again, this is unusual for signal-transduction pathways where similar signaling mechanisms are generally used in different tissues, although redundancy and flexibility, of course, exists among components that are present in a genome in multiple copies and as multiple isoforms, which is often the case in vertebrates. Third, care must be taken when generalizing regulatory mechanisms in the Hippo pathway. This is true for comparing mechanisms among different species as well as between different tissues in the same species. The evolutionary changes that we identified may not be the only variations in Hippo signaling.
Given all these variations in the regulatory inputs into the Hippo pathway, is there a common theme? We think there is. First, the currently known major regulatory inputs into the Hippo pathway depend directly or indirectly on cell-cell junctions and cell–cell interactions. This is true for the regulators of the Hippo pathway in Drosophila, where Ft and Crb are transmembrane receptors that localize to subapical junctions and physically interact with ligands on neighboring cells. Also Ajuba, Mer and Kibra localize to cell–cell junctions, although it is not known whether and how their activity is regulated by cell–cell interactions.71,91–97 In addition, Dig, Scrib and Lgl, proteins known to be required for proper apical–basal cell polarity,98 affect the activity of the Hippo pathway.68,99–101 Further, in mammals the Amot complex, a major input into the Hippo pathway, and α-catenin, which may represent a parallel input, localize to cell–cell junctions.56,90 Interestingly, major differences exist in the physical properties and location of cell–cell junctions between flies and vertebrates.102 Flies do not have classical tight junctions apical to adherens junctions that are typical for vertebrate epithelia. Rather, flies have an analogous septate junction that localizes basal to adherens junctions.74 In addition, the adherens junction component Ed, which is essential for adherens junction formation, is arthropod-specific. Our data suggest that the function and thus evolution of Hippo pathway inputs is linked to the evolution of cell–cell junctions, placing cell–cell junctions as a central regulator of organ size control. Altogether, we conclude that major differences exist in the molecular mechanisms that relay information from cell–cell junctions into the Hippo pathway between flies and mammals.
Materials and Methods
Tracing the evolutionary history of genes and protein domains
A full description of the phylogenetic analysis is given in the Supplementary Information; briefly, candidate orthologs or their absence was identified using BLAST/tblastn (translated nucleotide database) and the PhyML algorithm in different configurations. Databases used are listed in the Supplementary Information.
Alignments
All sequences were aligned using the MUSCLE (multiple sequence alignment by log-expectation) alignment algorithm using 800 iterations.
Fly stocks and transgene creation
All crosses were kept at 25 °C. Mutant clones were induced using the FLP/FRT (Flp Recombination Target) system. For generating ft-null mutant clones, ft422, which contains a 5 bp deletion at amino-acid position 1400, were flipped against corresponding ubi-GFP marked FRT chromosomes. Other stocks used were: ex697, ftfd, m@0.5-lacZ and fj-lacZ. All Ft rescue constructs were created using phiC31–mediated integration in the 68A locus, giving rise to UAS-FtDECD, UAS-FtD1 UAS-FtD2, UAS-FtD3, UAS-FtD4, UAS-FtD5, UAS-FtD6, UAS-FfD7 and UAS-FtD8. We also used UAS-FfFL (from S. Blair). To induce overexpression, we used tub-Gal4, act-Gal4 and hh-Gal4. The precise genotypes of all flies used is added as a Supplementary Information file. All our deletion constructs included an N-terminal signal peptide, followed by an HA and a TAP tag in place of the Ft extracellular domain. The constructs were polymerase chain reaction amplified and inserted into the pUAST attB vector by Notl-Kpnl. We used the PhiC31 site-specific integration system to create UAS-transgenic flies on the third chromosome at genomic position 68A.
Drosophila tissue immunohistochemistry
Antibody stainings of imaginal discs were done as described earlier.103 The following antibodies were used: rat anti-Fat (1/2000, M.A. Simon, Stanford, CA, USA), mouse anti-β-Gal (1/2000, Promega, Madison, Wl, USA), rabbit anti-β-Gal (1:600, Invitrogen, Grand Island, NY, USA), rat anti-Ci (1/150, R Holmgren, Evanston, IL, USA), and mouse anti-HA (Invitrogen, 1/200).
Generation of Fat4; Nf2 and Fat4; Nf2 conditional knockout mice
To achieve liver-specific gene deletion, Fat4flox/flox, Nf2flox/flox, 36,104, Fat4flox/flox and Fat4flox/flox; Nf2flox/flox mice were bred to Alb-Cre transgenic mice (no. 003574) from the Jackson Laboratory (Bar Harbor, ME, USA). To avoid potential variation related to gender, all experiments were performed in male mice with paternal inheritance of Alb-Cre. All animal experiments were performed with protocols approved by the Johns Hopkins University IACUC (Baltimore, MD, USA).
Mouse histological analysis and immunohistochemistry
Mouse liver samples were collected, fixed overnight in 10% neutral-buffered formalin solution (Sigma, St Louis, MO, USA), embedded in paraffin and sectioned at 5 μm. Sections were stained with hematoxylineosin for histology analysis. Primary antibodies used for immunohistochemistry were: wild spectrum screening cytokeratin (pan-CK) (Dako, Carpinteria, CA, USA; Z0622, 1:500). The signals were developed with VECTASTAIN Elite ABC kit (Vector Laboratories, Burlingame, CA, USA).
Supplementary Material
Acknowledgments
We are grateful to Drs Alfred Handler, Giuliano Gasperi and Stephen Richards of the Medfly Whole Genome Sequencing Consortium for providing the draft genome sequence of Ceratitis capitata. We thank Lisa McCord for assistance with graphical representation, and also Dr Ryan Udan, Dr Maria Willecke and Chunyao Tao for help in performing the screen, experimental input and valuable discussions. This study was supported in part by grants from Department of Defense (NF093145 to DP) and from the National Institute of Health (GM067997 to GH). QC is a recipient of a Breast Cancer Research Postdoctoral Fellowship from the Department of Defense (BC093902). DP is an investigator of the Howard Hughes Medical Institute.
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
Author Contributions: WB initiated and designed the work, performed evolutionary tracing of the different genes and analyzed the results; WB and CLC performed and analyzed the Drosophila experiments; QC designed, performed and analyzed the mouse experiments; DJP designed and analyzed the mouse experiments; MS provided analysis for the Yap/Yki and Amot data; AK performed evolutionary tracing; GH led and designed the experiments; and WB and GH wrote the manuscript with input from all other authors.
References
- 1.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;(138):9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genevet A, Tapon N. The Hippo pathway and apico-basal cell polarity. Biochem J. 2011;436:213–224. doi: 10.1042/BJ20110217. [DOI] [PubMed] [Google Scholar]
- 5.Sebe-Pedros A, Zheng Y, Ruiz-Trillo la, Pan D. Premetazoan origin of the hippo signaling pathway. Cell Reports. 2012;1:13–20. doi: 10.1016/j.celrep.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hilman D, Gat U. The evolutionary history of YAP and the Hippo/YAP pathway. Mol Biol Evol. 2011;28:2403–2417. doi: 10.1093/molbev/msr065. [DOI] [PubMed] [Google Scholar]
- 7.Lee KP, Lee JH, Kim TS, Kim TH, Park HD, Byun JS, et al. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:8248–8253. doi: 10.1073/pnas.0912203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, et al. Mammalian Mstl and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci USA. 2010;107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, et al. Mstl and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yapl oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oh S, Lee D, Kim T, Kim TS, Oh HJ, Hwang CY, et al. Crucial role for Mst1 and Mst2 kinases in early embryonic development of the mouse. Mol Cell Biol. 2009;29:6309–6320. doi: 10.1128/MCB.00551-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci USA. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaffney CJ, Oka T, Mazack V, Hilman D, Gat U, Muramatsu T, et al. Identification, basic characterization and evolutionary analysis of differentially spliced mRNA isoforms of human YAP1 gene. Gene. 2012;509:215–222. doi: 10.1016/j.gene.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muramatsu T, Imoto I, Matsui T, Kozaki K, Haruki S, Sudol M, et al. YAP is a candidate oncogene for esophageal squamous cell carcinoma. Carcinogenesis. 2011;32:389–398. doi: 10.1093/carcin/bgq254. [DOI] [PubMed] [Google Scholar]
- 15.Webb C, Upadhyay A, Giuntini F, Eggleston I, Furutani-Seiki M, Ishima R, et al. Structural features and ligand binding properties of tandem WW domains from YAP and TAZ, nuclear effectors of the Hippo pathway. Biochemistry. 2011;50:3300–3309. doi: 10.1021/bi2001888. [DOI] [PubMed] [Google Scholar]
- 16.Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu CY, Zha ZY, Zhou X, Zhang H, Huang W, Zhao D, et al. The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCF{beta}-TrCP E3 ligase. J Biol Chem. 2010;285:37159–37169. doi: 10.1074/jbc.M110.152942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burglin TR. The TEA domain: a novel, highly conserved DNA-binding motif. Cell. 1991;66:11–12. doi: 10.1016/0092-8674(91)90132-i. [DOI] [PubMed] [Google Scholar]
- 19.Sudol M, Harvey KF. Modularity in the Hippo signaling pathway. Trends Biochem Sci. 2010;35:627–633. doi: 10.1016/j.tibs.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 20.Van Hateren NJ, Das RM, Hautbergue GM, Borycki AG, Placzek M, Wilson SA. FatJ acts via the Hippo mediator Yap 1 to restrict the size of neural progenitor cell pools. Development. 2011;138:1893–1902. doi: 10.1242/dev.064204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao Y, Mulvaney J, Zakaria S, Yu T, Morgan KM, Allen S, et al. Characterization of a Dchsl mutant mouse reveals requirements for Dchs1-Fat4 signaling during mammalian development. Development. 2011;138:947–957. doi: 10.1242/dev.057166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saburi S, Hester I, Fischer E, Pontoglio M, Eremina V, Gessler M, et al. Loss of Fat4 disrupts PCP signaling and oriented cell division and leads to cystic kidney disease. Nat Genet. 2008;40:1010–1015. doi: 10.1038/ng.179. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence PA, Struhl G, Casal J. Do the protocadherins Fat and Dachsous link up to determine both planar cell polarity and the dimensions of organs? Nat Cell Biol. 2008;10:1379–1382. doi: 10.1038/ncb1208-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reddy BV, Irvine KD. The Fat and Warts signaling pathways: new insights into their regulation, mechanism and conservation. Development. 2008;135:2827–2838. doi: 10.1242/dev.020974. [DOI] [PubMed] [Google Scholar]
- 25.Sopko R, McNeill H. The skinny on Fat: an enormous cadherin that regulates cell adhesion, tissue growth, and planar cell polarity. Curr Opin Cell Biol. 2009;21:717–723. doi: 10.1016/j.ceb.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 26.Visser-Grieve S, Hao Y, Yang X. Human homolog of Drosophila expanded, hEx, functions as a putative tumor suppressor in human cancer cell lines independently of the Hippo pathway. Oncogene. 2011;31:1189–1195. doi: 10.1038/onc.2011.318. [DOI] [PubMed] [Google Scholar]
- 27.Angus L, Moleirinho S, Herron L, Sinha A, Zhang X, Niestrata M, et al. Willin/FRMD6 expression activates the Hippo signaling pathway kinases in mammals and antagonizes oncogenic YAP. Oncogene. 2011;31:238–250. doi: 10.1038/onc.2011.224. [DOI] [PubMed] [Google Scholar]
- 28.Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 29.Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- 30.Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 31.Tyler DM, Baker NE. Expanded and fat regulate growth and differentiation in the Drosophila eye through multiple signaling pathways. Dev Biol. 2007;305:187–201. doi: 10.1016/j.ydbio.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, et al. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Castillejo-Lopez C, Arias WM, Baumgartner S. The fat-like gene of Drosophila is the true orthologue of vertebrate fat cadherins and is involved in the formation of tubular organs. J Biol Chem. 2004;279:24034–24043. doi: 10.1074/jbc.M313878200. [DOI] [PubMed] [Google Scholar]
- 34.Viktorinova I, Konig T, Schlichting K, Dahmann C. The cadherin Fat2 is required for planar cell polarity in the Drosophila ovary. Development. 2009;136:4123–4132. doi: 10.1242/dev.039099. [DOI] [PubMed] [Google Scholar]
- 35.Avruch J, Zhou D, Fitamant J, Bardeesy N. Mstl/2 signalling to Yap: gatekeeper for liver size and tumour development. Br J Cancer. 2011;104:24–32. doi: 10.1038/sj.bjc.6606011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benhamouche S, Curto M, Saotome I, Gladden AB, Liu CH, Giovannini M, et al. Nf2/Merlin controls progenitor homeostasis and tumorigenesis in the liver. Genes Dev. 2010;24:1718–1730. doi: 10.1101/gad.1938710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho E, Irvine KD. Action of fat, four-jointed, dachsous and dachs in distal-to-proximal wing signaling. Development. 2004;131:4489–4500. doi: 10.1242/dev.01315. [DOI] [PubMed] [Google Scholar]
- 39.Rogulja D, Rauskolb C, Irvine KD. Morphogen control of wing growth through the Fat signaling pathway. Dev Cell. 2008;15:309–321. doi: 10.1016/j.devcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mao Y, Rauskolb C, Cho E, Hu WL, Hayter H, Minihan G, et al. Dachs: an unconventional myosin that functions downstream of Fat to regulate growth, affinity and gene expression in Drosophila. Development. 2006;133:2539–2551. doi: 10.1242/dev.02427. [DOI] [PubMed] [Google Scholar]
- 41.Brittle A, Thomas C, Strutt D. Planar polarity specification through asymmetric subcellular localization of Fat and Dachsous. Curr Biol. 2012;22:907–914. doi: 10.1016/j.cub.2012.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matakatsu H, Blair SS. The DHHC palmitoyltransferase approximated regulates fat signaling and Dachs localization and activity. Curr Biol. 2008;18:1390–1395. doi: 10.1016/j.cub.2008.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ambegaonkar AA, Pan G, Mani M, Feng Y, Irvine KD. Propagation of Dachsousfat planar cell polarity. Curr Biol. 2012;22:1302–1308. doi: 10.1016/j.cub.2012.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao Y, Tournier AL, Bates PA, Gale JE, Tapon N, Thompson BJ. Planar polarization of the atypical myosin Dachs orients cell divisions in Drosophila. Genes Dev. 2011;25:131–136. doi: 10.1101/gad.610511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matakatsu H, Blair SS. Separating the adhesive and signaling functions of the Fat and Dachsous protocadherins. Development. 2006;133:2315–2324. doi: 10.1242/dev.02401. [DOI] [PubMed] [Google Scholar]
- 46.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matakatsu H, Blair SS. Separating planar cell polarity and Hippo pathway activities of the protocadherins Fat and Dachsous. Development. 2012;139:1498–1508. doi: 10.1242/dev.070367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber KL, Sokac AM, Berg JS, Cheney RE, Bement WM. A microtubule-binding myosin required for nuclear anchoring and spindle assembly. Nature. 2004;431:325–329. doi: 10.1038/nature02834. [DOI] [PubMed] [Google Scholar]
- 49.Zhang H, Berg JS, Li Z, Wang Y, Lang P, Sousa AD, et al. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat Cell Biol. 2004;6:523–531. doi: 10.1038/ncb1136. [DOI] [PubMed] [Google Scholar]
- 50.Bulgakova NA, Knust E. The Crumbs complex: from epithelial-cell polarity to retinal degeneration. J Cell Sci. 2009;122(Part 15):2587–2596. doi: 10.1242/jcs.023648. [DOI] [PubMed] [Google Scholar]
- 51.Oka T, Schmitt AP, Sudol M. Opposing roles of angiomotin-like-1 and zona occludens-2 on pro-apoptotic function of YAP. Oncogene. 2011;31:128–134. doi: 10.1038/onc.2011.216. [DOI] [PubMed] [Google Scholar]
- 52.Chan SW, Lim CI, Chong YF, Pobbati AV, Huang C, Hong W. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J Biol Chem. 2011;286:7018–7026. doi: 10.1074/jbc.C110.212621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang W, Huang J, Chen J. Angiomotin-like proteins associate with and negatively regulate YAPI. J Biol Chem. 2011;286:4364–4370. doi: 10.1074/jbc.C110.205401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ernkvist M, Aase K, Ukomadu C, Wohlschlegel J, Blackman R, Veitonmaki N, et al. P130-angiomotin associates to actin and controls endothelial cell shape. FEBS J. 2006;273:2000–2011. doi: 10.1111/j.1742-4658.2006.05216.x. [DOI] [PubMed] [Google Scholar]
- 56.Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockbum K, Larsen BG, et al. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta-SMAD pathway. Dev Cell. 2010;19:831–844. doi: 10.1016/j.devcel.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 57.Chen HI, Einbond A, Kwak SJ, Linn H, Koepf E, Peterson S, et al. Characterization of the WW domain of human yes-associated protein and its polyproline-con-taining ligands. J Biol Chem. 1997;272:17070–17077. doi: 10.1074/jbc.272.27.17070. [DOI] [PubMed] [Google Scholar]
- 58.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 59.Oka T, Remue E, Meerschaert K, Vanloo B, Boucherie C, Gfeller D, et al. Functional complexes between YAP2 and ZO-2 are PDZ domain-dependent, and regulate YAP2 nuclear localization and signalling. Biochem J. 2010;432:461–472. doi: 10.1042/BJ20100870. [DOI] [PubMed] [Google Scholar]
- 60.Oka T, Sudol M. Nuclear localization and pro-apoptotic signaling of YAP2 require intact PDZ-binding motif. Genes Cells. 2009;14:607–615. doi: 10.1111/j.1365-2443.2009.01292.x. [DOI] [PubMed] [Google Scholar]
- 61.Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, et al. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci USA. 2010;107:15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y, et al. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc Natl Acad Sci USA. 2010;107:10532–10537. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson BS, Huang J, Hong Y, Moberg KH. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Develop Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, et al. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 66.Oh H, Reddy BV, Irvine KD. Phosphorylation-independent repression of Yorkie in Fat-Hippo signaling. Dev Biol. 2009;335:188–197. doi: 10.1016/j.ydbio.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Badouel C, Gardano L, Amin N, Garg A, Rosenfeld R, Le Bihan T, et al. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev Cell. 2009;16:411–420. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 68.Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 69.Parsons LM, Grzeschik NA, Allott ML, Richardson HE. Lgl/aPKC and Crb regulate the Salvador/Warts/Hippo pathway. Fly (Austin) 2010;4:288–293. doi: 10.4161/fly.4.4.13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pellock BJ, Buff E, White K, Hariharan IK. The Drosophila tumor suppressors Expanded and Merlin differentially regulate cell cycle exit, apoptosis, and Wingless signaling. Dev Biol. 2007;304:102–115. doi: 10.1016/j.ydbio.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boedigheimer MJ, Nguyen KP, Bryant PJ. Expanded functions in the apical cell domain to regulate the growth rate of imaginal discs. Dev Genet. 1997;20:103–110. doi: 10.1002/(SICI)1520-6408(1997)20:2<103::AID-DVG3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 72.Jaramillo BE, Ponce A, Moreno J, Betanzos A, Huerta M, Lopez-Bayghen E, et al. Characterization of the tight junction protein ZO-2 localized at the nucleus of epithelial cells. Exp Cell Res. 2004;297:247–258. doi: 10.1016/j.yexcr.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 73.Remue E, Meerschaert K, Oka T, Boucherie C, Vandekerckhove J, Sudol M, et al. TAZ interacts with zonula occludens-1 and -2 proteins in a PDZ-1 dependent manner. FEBS Lett. 2010;584:4175–4180. doi: 10.1016/j.febslet.2010.09.020. [DOI] [PubMed] [Google Scholar]
- 74.Leys SP, Riesgo A. Epithelia, an evolutionary novelty of metazoans. J Exp Zool Part B. 2012;318:438–447. doi: 10.1002/jez.b.21442. [DOI] [PubMed] [Google Scholar]
- 75.Yue T, Tian A, Jiang J. The cell adhesion molecule echinoid functions as a tumor suppressor and upstream regulator of the Hippo signaling pathway. Develop Cell. 2012;22:255–267. doi: 10.1016/j.devcel.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Enomoto M, Igaki T. Src controls tumorigenesis via JNK-dependent regulation of the Hippo pathway in Drosophila. EMBO Rep. 2013;14:65–72. doi: 10.1038/embor.2012.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boggiano JC, Fehon RG. Growth control by committee: intercellular junctions, cell polarity, and the cytoskeleton regulate Hippo signaling. Dev Cell. 2012;22:695–702. doi: 10.1016/j.devcel.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bosveld F, Bonnet I, Guirao B, Tlili S, Wang Z, Petitalot A, et al. Mechanical control of morphogenesis by Fat/Dachsous/Four-jointed planar cell polarity pathway. Science. 2012;336:724–727. doi: 10.1126/science.1221071. [DOI] [PubMed] [Google Scholar]
- 80.Liu KC, Jacobs DT, Dunn BD, Fanning AS, Cheney RE. Myosin-X functions in polarized epithelial cells. Mol Biol Cell. 2012;23:1675–1687. doi: 10.1091/mbc.E11-04-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Toyoshima F, Nishida E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EBI-and myosin X-dependent manner. EMBO J. 2007;26:1487–1498. doi: 10.1038/sj.emboj.7601599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim NG, Koh E, Chen X, Gumbiner BM. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc Natl Acad Sci USA. 2011;108:11930–11935. doi: 10.1073/pnas.1103345108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ishiuchi T, Misaki K, Yonemura S, Takeichi M, Tanoue T. Mammalian Fat and Dachsous cadherins regulate apical membrane organization in the embryonic cerebral cortex. J Cell Biol. 2009;185:959–967. doi: 10.1083/jcb.200811030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mauviel A, Nallet-Staub F, Varelas X. Integrating developmental signals: a Hippo in the (path) way. Oncogene. 2011;31:1743–1756. doi: 10.1038/onc.2011.363. [DOI] [PubMed] [Google Scholar]
- 85.Polesello C, Tapon N. Salvador-warts-hippo signaling promotes Drosophila posterior follicle cell maturation downstream of notch. Curr Biol. 2007;17:1864–1870. doi: 10.1016/j.cub.2007.09.049. [DOI] [PubMed] [Google Scholar]
- 86.Bu X, Avraham HK, Li X, Lim B, Jiang S, Fu Y, et al. Mayven induces c-Jun expression and cyclin D1 activation in breast cancer cells. Oncogene. 2005;24:2398–2409. doi: 10.1038/sj.onc.1208466. [DOI] [PubMed] [Google Scholar]
- 87.MacDougall N, Lad Y, Wilkie GS, Francis-Lang H, Sullivan W, Davis I. Merlin, the Drosophila homologue of neurofibromatosis-2, is specifically required in posterior follicle cells for axis formation in the oocyte. Development. 2001;128:665–673. doi: 10.1242/dev.128.5.665. [DOI] [PubMed] [Google Scholar]
- 88.Milton CC, Zhang X, Albanese NO, Harvey KF. Differential requirement of Salvador-Warts-Hippo pathway members for organ size control in Drosophila melanogaster. Development. 2010;137:735–743. doi: 10.1242/dev.042309. [DOI] [PubMed] [Google Scholar]
- 89.Silvis MR, Kreger BT, Lien WH, Klezovitch O, Rudakova GM, Camargo FD, et al. Alpha-catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Science Signal. 2011:4ra33. doi: 10.1126/scisignal.2001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, et al. Yapl acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev Cell. 2010;18:309–316. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 92.Boedigheimer M, Laughon A. Expanded: a gene involved in the control of cell proliferation in imaginal discs. Development. 1993;118:1291–1301. doi: 10.1242/dev.118.4.1291. [DOI] [PubMed] [Google Scholar]
- 93.Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McCartney BM, Kulikauskas RM, LaJeunesse DR, Fehon RG. The Neurofibromatosis-2 homologue, Merlin, and the tumor suppressor expanded function together in Drosophila to regulate cell proliferation and differentiation. Development. 2000;127:1315–1324. doi: 10.1242/dev.127.6.1315. [DOI] [PubMed] [Google Scholar]
- 95.Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Langer EM, Feng Y, Zhaoyuan H, Rauscher FJ, III, Kroll KL, Longmore GD, et al. Proteins are snail/slug compressors required for neural crest development in Xenopus. Dev Cell. 2008;14:424–436. doi: 10.1016/j.devcel.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marie H, Pratt SJ, Betson M, Epple H, Kittler JT, Meek L, et al. The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with alpha-catenin. J Biol Chem. 2003;278:1220–1228. doi: 10.1074/jbc.M205391200. [DOI] [PubMed] [Google Scholar]
- 98.Humbert PO, Grzeschik NA, Brumby AM, Galea R, Elsum I, Richardson HE. Control of tumourigenesis by the Scribble/Dlg/Lgl polarity module. Oncogene. 2008;27:6888–6907. doi: 10.1038/onc.2008.341. [DOI] [PubMed] [Google Scholar]
- 99.Grzeschik NA, Parsons LM, Richardson HE. Lgl, the SWH pathway and tumorigenesis: it's a matter of context & competition! Cell Cycle. 2010;9:3202–3212. doi: 10.4161/cc.9.16.12633. [DOI] [PubMed] [Google Scholar]
- 100.Ziosi M, Baena-Lopez LA, Grifoni D, Froldi F, Pession A, Garoia F, et al. dMyc functions downstream of Yorkie to promote the supercompetitive behavior of hippo pathway mutant cells. PLoS Genet. 2010;6:e1001140. doi: 10.1371/journal.pgen.1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Menendez J, Perez-Garijo A, Calleja M, Morata G. A tumor-suppressing mechanism in Drosophila involving cell competition and the Hippo pathway. Proc Natl Acad Sci USA. 2010;107:14551–14656. doi: 10.1073/pnas.1009376107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Muller HA, Bossinger O. Molecular networks controlling epithelial cell polarity in development. Mech Dev. 2003;120:1231–1256. doi: 10.1016/j.mod.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 103.Kango-Singh M, Nolo R, Tao C, Verstreken P, Hiesinger PR, Bellen HJ, et al. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- 104.Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.