Abstract
The limitations of cancer cell lines have led to the development of direct patient derived xenograft (PDX) models. However, the interplay between the implanted human cancer cells and recruited mouse stromal and immune cells alters the tumor microenvironment and limits the value of these models. To overcome these constraints, we have developed a technique to expand human hematopoietic stem and progenitor cells (HSPCs) and use them to reconstitute the radiation-depleted bone marrow of a NOD/SCID/IL2rg−/− (NSG) mouse on which a patient’s tumor is then transplanted (XactMice). The human HSPCs produce immune cells that home into the tumor and help replicate its natural microenvironment. Despite previous passage on nude mice, the expression of epithelial, stromal, and immune genes in XactMice tumors aligns more closely to that of the patient tumor than to those grown in non-humanized mice – an effect partially facilitated by human cytokines expressed by both the HSPC progeny and the tumor cells. The human immune and stromal cells produced in the XactMice can help recapitulate the microenvironment of an implanted xenograft, reverse the initial genetic drift seen after passage on non-humanized mice, and provide a more accurate tumor model to guide patient treatment.
Keywords: Humanized mouse model, head and neck cancer, xenograft model
INTRODUCTION
The absence of native tumor stroma and interaction with the immune system are major limitations of xenograft models for studies of human tumor growth and metastasis. In order to produce a representative environment, xenograft tumor models must account for interactions between tumor, stromal, vascular, and immune cells (1, 2). Cell lines become homogeneous, and epithelial-stromal interactions are no longer present in vitro (3–5), consequently, conventional studies in cell line-derived tumors poorly predict clinical efficacy (6). Patient derived xenografts (PDX) implanted into immunocompromised mice are more representative of patient tumor growth, although genetic drift is observed in microenvironment genes (7–10), perhaps because the tumor stroma consists of recruited mouse cells (11).
Recent studies have taken incremental steps toward overcoming this obstacle. It has been shown that human tumors implanted with their own stromal tissues can temporarily simulate aspects of the host tumor microenvironment in immunocompromised mice (12). Alternatively, human hematopoietic cells can be engrafted in NOD/SCID/IL2rg−/− (NSG) mice to generate many features of the human immune system in these animals (13). In vitro modification of hematopoietic progenitor cells can produce human leukemia and lymphoma models with accurate bone-marrow tumor microenvironments (14, 15), while the introduction of human T cells and experimental monoclonal antibodies can be used to test immunotherapies in NSG xenograft models (16). Finally, the infiltration and activation of myeloid cells in xenografts has been examined in genetically modified NSG mice (17). However, a comprehensive examination of the growth, tumor-stroma interaction, and impact of humanization on gene expression of PDX in humanized mouse models has not been conducted.
We have developed an ex vivo technique to expand human hematopoietic stem and progenitor cells (HSPCs) derived from either cord blood or G-CSF mobilized adult peripheral blood (18). These HSPCs contain a population of rare hematopoietic stem cells (HSCs), capable of reconstituting the hematopoietic system of a mouse into which patient tumors are subsequently transplanted. In these humanized xenochimeric mice, or XactMice (Fig. 1a), the engrafted human HSPCs can express the chemical stimuli necessary to give rise to stromal and immune cells that recreate the original tumor microenvironment observed clinically. The promise of this model is that it can provide a tumor microenvironment more representative of the human host, and it can reverse, at least partially, the genetic drift observed in classical PDX models.
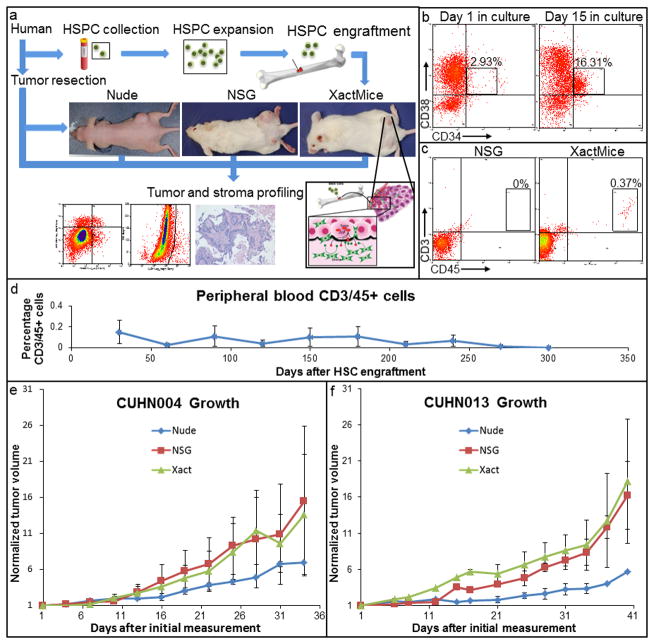
Figure 1. Overview and characterization of XactMice.
(a) Schematic describing the generation of XactMice from human HSPCs, whose progeny migrate into the xenograft and differentiate into stromal cells. The growth and composition of tumors can be compared in nude, NSG, and XactMice. (b) Flow cytometry measuring the expansion of HSPCs in vitro by the percentage of CD34/CD38+ cells. (c) Flow cytometry detecting human hematopoietic CD3/CD45+ cells in peripheral XactMice - but not NSG - blood, indicating that the HSPCs have successfully engrafted and are generating circulating lymphocytes. (d) The average percentage of human CD3/CD45+ cells in the peripheral blood of XactMice, as determined by flow cytometry, over the course of seven months after engraftment. (e, f) There were no significant differences in either CUHN004 or CUHN013 tumor growth rates between nude, NSG, and XactMice. Tumor measurements (W × W × H)/2 were recorded from all mouse strains in three separate experiments. Although tumors seem to grow faster in the NSG and XactMice, no statistical difference was observed in growth between the three strains in these experiments. Average tumor volumes (mm3) with the standard errors were used to create the recorded growth curves.
RESULTS
Expansion of human HSPCs enables the generation of cohorts of XactMice
The generation of cohorts of XactMice with HSPCs from the same donor requires the expansion of HSPCs. The expansion of HSPCs from donated cord blood, or from G-CSF mobilized patient peripheral blood, was achieved using a protocol recently described by Bird et al, which utilizes MYC and Bcl2 proteins fused with the HIV protein transduction peptide Tat (18). Used in combination, Tat-MYC and Tat-Bcl2 are capable of expanding HSPCs long-term; however, these fusion proteins are degraded within 48 hours of exposure to culture medium. Therefore, there is no trace of these activities when the expanded HSPCs are transplanted into mice. Under these conditions, HSPCs proliferate stably in vitro (Fig. 1b) while maintaining the HSC-associated abilities of self-renewal and differentiation into B and T cells in immunocompromised mice. After expansion in culture, we injected the HSPCs into sub-lethally irradiated NSG mice to generate XactMice. We verified the HSPC-mediated bone marrow reconstitution by periodic flow cytometry of mouse peripheral blood for the presence of the hematopoietic cells expressing CD3 and CD45 human antigens (Fig. 1c). We examined the peripheral blood of a cohort of XactMice over six months and found that this human CD3/45+ cell population remains stable (Fig. 1d). Additionally, after almost a year, we could still observe human T and B cells in the bone marrow and spleen (Supplemental Fig. 1a–b), indicating that the progenitor cells continue to proliferate after engraftment.
We have established a xenograft tumor bank, consisting of tumors from head and neck squamous cell carcinoma (HNSCC) patients undergoing surgical resection (19). Freshly resected tumor tissue is implanted heterotopically and then passaged in the flanks of nude mice. Although the passaged heterotopic tumors initially retain the histological features of the original tumor (also shown in (20, 21)), the human stroma is lost after 2–3 passages. We selected two cases from this tumor bank (CUHN004, passage 14; CUHN013, passage 5), and implanted them into cohorts of nude, NSG, and XactMice. We observed no significant differences in the tumor growth rate or gross histology between the different mouse strains, with the NSG and XactMice having identical growth rates (Fig. 1e–f; Supplemental Fig. 1c). To ensure reproducibility, experiments were repeated at least twice and all cohorts in this and subsequent experiments were composed of at least five randomly distributed male and female XactMice.
We also generated XactMice cohorts using expanded HSPCs from peripheral blood of adults treated with granulocyte colony-stimulating factor (G-CSF), which results in an increase in peripheral blood HSPCs and creates a window for their acquisition by a peripheral blood draw. We implanted tumor cells into the flanks of these XactMice, along with nude and NSG controls. Subsequent analysis revealed that the engrafted HSPCs were present in the bone marrow of these XactMice, and their progeny could be seen in the spleen, peripheral blood, and in the tumor (Supplemental Fig. 2, upper two panels). Lastly we generated cohorts of XactMice from the HSPCs of HNSCC patients undergoing induction chemotherapy that received G-CSF to hasten bone marrow recovery (22). Analysis of the bone marrow, spleen, and blood of these mice showed that the HSPCs successfully engrafted. Since none of the patients from which these HSPCs were collected ultimately required surgery, no tissue existed to implant on these XactMice (Supplemental Fig. 2, lower panel). Although the ability to create XactMice from peripheral blood increases the utility of this model, the additional experiments shown below were conducted utilizing cord blood donors to minimize individual variations in the initial cohorts.
Human cells derived from humanized bone marrow are present in the stroma of XactMice tumors
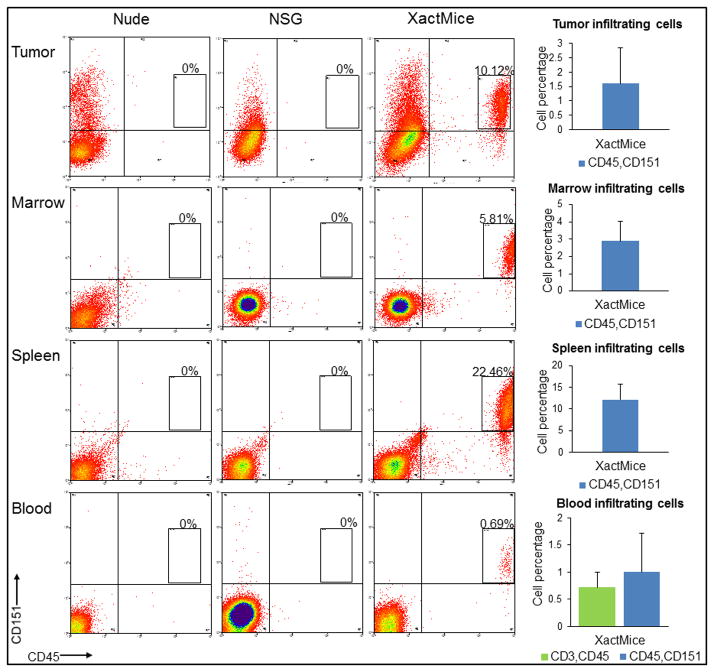
The stroma that forms in a developing tumor has two primary cellular origins: (1) mesenchymal and immune cells originating from the bone marrow, and (2) fibroblasts and other cell types from local tissues (23). The cell surface antigen CD151 has been previously characterized as a mesenchymal cell marker and is found on many different human cell types (24, 25). Furthermore, CD45 is a human hematopoietic cell surface marker found on B and T cells, as well as on hematopoietic progenitors (26). Tumors grown in XactMice contain a unique and quantifiable population of cells displaying both human cell surface antigens CD45 and CD151, which is absent in the nude and NSG mice. Likewise, cells removed from XactMice bone marrow, spleen, and peripheral blood harbor similar double-positive populations, while cells from nude and NSG mice lack these human markers (Fig. 2). We observed that although the human cell population typically accounted for only 2–5% of the total bone marrow cells, this degree of engraftment was sufficient to produce immune cell infiltration into tumors and give rise to αSMA+ cells.
Figure 2. XactMice tumors and tissues harbor human immune cell populations.
Upper panel: Flow cytometry showing that human CD45/151+ cells can be identified and quantified in tumors removed from XactMice, while no corresponding populations can be recovered from nude or NSG mice. Lower panels: A CD45/151+ cell population can also be identified and quantified in the bone marrow, spleen, and peripheral blood of XactMice, while no such cells are observed in nude and NSG controls. Bars represent standard errors.
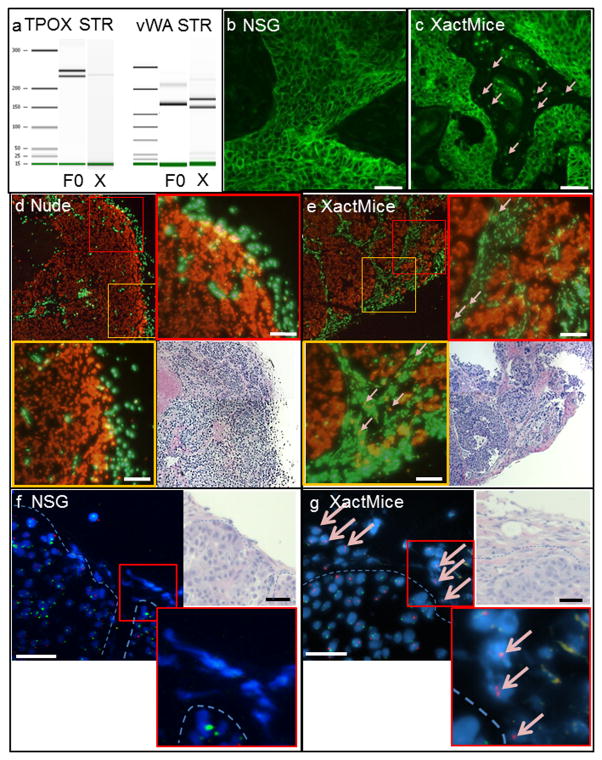
To show that the CD45/CD151+ cells originate within the XactMice humanized bone marrow we performed four different analyses using separate cohorts of XactMice. First, we employed short tandem repeat (STR) analysis, a well-documented forensic examination that compares highly variable DNA loci by PCR to establish the relationship between two or more DNA samples. We purified DNA from CD45/CD151+ cells sorted from CUHN004 tumors grown on XactMice, and DNA from the originator CUHN004 patient sample (F0). We analyzed the DNA at two well-studied loci, TPOX and vWA, to identify STR polymorphisms (Fig. 3a). Our results show that the sorted cells are human and genetically distinct from tumor cells (27). Further, CD151 immunofluorescence analysis revealed the presence of individual human cells scattered throughout the otherwise unstained tumor stroma in the XactMice. These stromal cells were not present in the NSG mice (Fig. 3b,c). Next, we performed fluorescent in situ hybridization (FISH) analysis, using species-specific probes for highly repetitive Cot-1 DNA to clearly identify the species of each cell (28). We found that tumors in nude hosts were composed of human cells (red), encapsulated by and containing small islands of mouse stromal cells (green), as is typical of a xenograft tumor (Fig. 3d)(11). In contrast, tumors in the XactMice consisted of large bundles of human tumor cells (red), surrounded by bands of mouse stroma (green), in which human cells were interspersed (Fig. 3e). Finally, we derived XactMice using HSPCs from female cord blood, and implanted tumors from a male patient. FISH analysis for X and Y chromosomes revealed tightly packed XY epithelial tumor cells surrounded by stromal cells of mouse origin in NSG mice (Fig. 3f) while in XactMice we observed individual XX cells scattered throughout the tumor stroma (Fig. 3g), in a pattern similar to that observed in the Cot-1 FISH analysis. Taken together, these analyses show that the engrafted HSPC progeny migrate from the XactMice bone marrow and are incorporated into implanted tumors.
Figure 3. Documentation of human stroma on XactMice.
Human cells only invade the tumors in XactMice. (a) Bioanalyzer gel of two well-defined STR loci, TPOX and vWA (27). Patient DNA from the originator tumor (F0, lanes 2 and 5). XactMice xenograft CD45/CD151+ DNA (X, lanes 3 and 6). (b,c) Human CD151 immunofluorescence. Although the CD151 antibody binds the human tumor cells in both the NSG and XactMice tumors, in NSG tumors (b), the stroma remains unstained, while in XactMice (c) the unstained mouse stroma is punctuated with CD151+ human cells (pink arrows). Magnification is 20×. (d,e) FISH analysis of nude and XactMice tumors. Both xenografts are composed primarily of human (red) tumor cells surrounded by mouse (green) stromal cells. No human cells infiltrate the stroma within the nude xenograft (d). Human cells can be observed throughout the stroma within the XactMice xenograft (e; enlarged and highlighted with pink arrows). Magnification is 10× for the tumor sections and 20× for the enlarged portions. (f,g) FISH analysis images of tumor sections using fluorescently-labeled X (red) and Y (green) probes. In the NSG (f), all tumor cells are male. The mouse stromal cells do not bind to either of the probes. A dashed line has been added to demarcate the approximate tumor-stroma boundary. In XactMice (g), the tumor cells are male, and the stroma is composed largely of mouse cells, but also contains female human cells. Expanded inserts were captured under increased magnification (100×). In all images, the scale bar equals 50 μm.
The stroma of XactMice tumors is infiltrated with human T and B cell populations
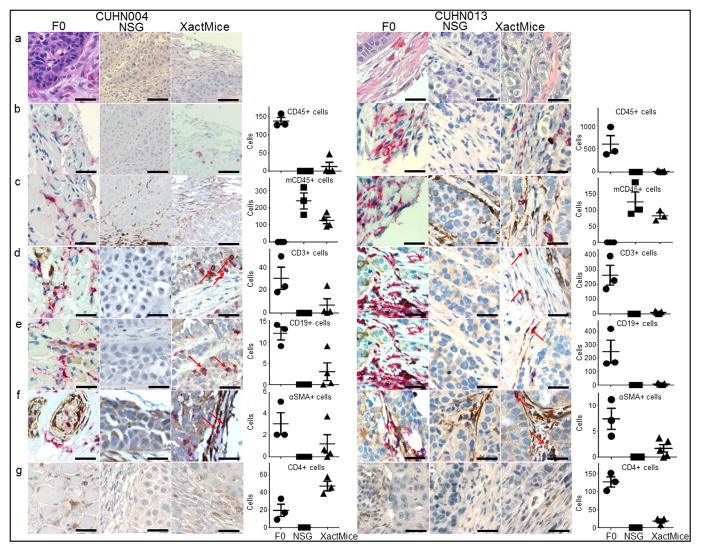
To determine the types of human stromal cells infiltrating XactMice tumors, we stained tumor sections from the CUHN004 and CUHN013 F0 patient tumors and corresponding NSG and XactMice xenografts with either human or human plus mouse pan-leukocyte CD45 antibodies (Fig. 4a–c). While the F0 tumors contained only human CD45+ cells (red) and the NSG tumors contained only mouse CD45+ cells (brown), the XactMice tumors contained both mouse and human CD45+ cells, indicating that HSC-generated cells are invading these tumors. We performed dual staining with human-specific antibodies to CD45 and either CD3 (T cells), CD19 (B cells), or alpha smooth muscle actin (αSMA; fibrocytes) (Fig. 4d–f). The human T cell and B cell populations identified were distributed in similar patterns throughout the F0 and XactMice tumors (generally less abundantly in XactMice), and were absent in NSG or nude tumors from the same generation. The αSMA staining was not human-specific, as can be seen in the NSG tumors, but CD45/αSMA+ double-staining cells were identified in both the F0 and XactMice tumors, and they can be attributed to HSC differentiation. We also stained for human CD4+ cells. Their presence in XactMice tumors indicates that the HSPC-generated T cell progenitors can differentiate into T helper cells (Fig. 4g).
Figure 4. Characterization of human stromal cells in control and XactMice tumors.
A comparison of CUHN004 and CUHN013 patient (F0) tumors with their corresponding NSG and XactMice xenografts. Bar graphs represent the average number of stained cells calculated from three non-overlapping fields visualized in 0.2 mm2 tumor sections taken from four (CUHN004) or five (CUHN013) separate XactMice and compared to three non-overlapping fields from F0 and NSG tumors (b–g). (a) H/E comparisons of the F0, NSG, and XactMice specimens. (b) Tumor IHC using the human pan-leukocyte CD45 antibody (red) indicates that human white blood cells are present in the F0 and XactMice tumors (albeit at difference frequency), but not the NSG tumor. (c) Tumor IHC with both human (red) and mouse (brown) CD45 antibodies indicates that mouse white blood cells are present in the NSG and XactMice tumors. (d) Dual human CD3 (brown) and CD45 (red) IHC indicates that T cells can be found in the F0 and XactMice tumors. (e) Dual human CD19 (brown) and CD45 (red) IHC indicates that B cells can be seen in both the F0 and XactMice tumors. (f) Dual human αSMA (brown) and CD45 (red) IHC. In F0 tumors, cells with either or both antigens are present. In NSGs, some stromal cells stain for the αSMA antigen, indicating that this antibody cross-reacts with mouse αSMA. XactMice tumor cells that stain for the presence of both antigens (indicated by red arrows) must be of human origin and exhibit some fibrocyte characteristics. (g) Human CD4 IHC indicates that T-helper cells can be found in both F0 and XactMice tumors. Magnification is 40× and the scale bar equals 50 μm.
Humanization reverses genetic drift after passage in immune-compromised hosts
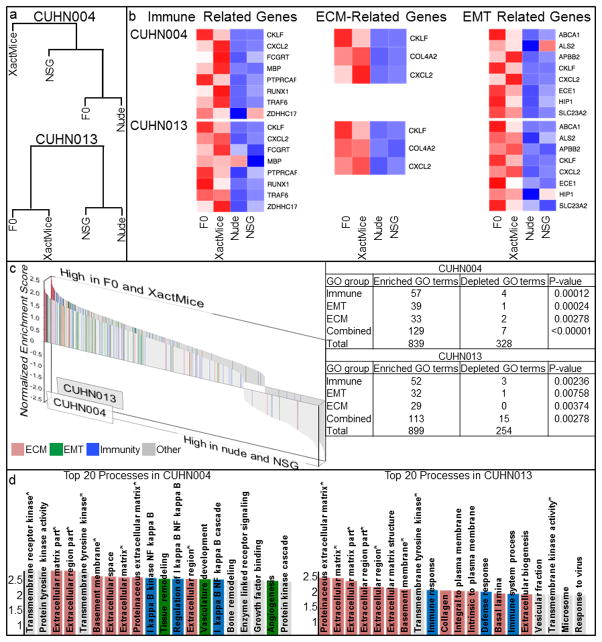
Since the XactMice model is designed to support xenograft tumor growth in a native environment, it is critical to demonstrate that it reduces genetic drift from the originator tumor compared to conventional models. To this end, we micro-dissected CUHN004 and CUHN013 tumor cells from flash-frozen tumors passaged in nude, NSG, and XactMice, as well as from the originator F0 patient tumors resected and flash frozen at the time of surgery. We isolated RNA and performed next generation sequencing to compare gene expression between tumors. A summary of the sequencing data (Table 1) shows that the CUHN013 XactMice tumor transcriptome aligns more completely to the human genome than the nude or NSG-originated tumor transcriptomes. The tumor transcriptome from the CUHN004 XactMice aligns slightly less completely to the human genome than that from the corresponding nude mouse, but its alignment is more complete than that of the NSG mouse. Likewise, while a dendrogram of the CUHN013 shows that the XactMice and the F0 patient tumors are most similar, the dendrogram comparing RNA expression between the CUHN004 tumors indicates that the XactMice transcriptome clusters separately from all others, indicating a fundamental difference in its gene expression (Fig. 5a). It is possible that these differences arise as a consequence of their different prior passages in nude mice.
Table 1. RNA sequencing summary.
RNA sequences generated from high-throughput sequencing were aligned to the human genome (NCBI 37.2). Resultant unaligned sequences were then aligned to the mouse genome (NCBI 37.2), and any remaining sequences were classified as unmatched, most likely due to base repetition and/or sequencing errors.
| Tumor | Total Reads | Align to Human | Percent aligned | Align to Mouse | Percent aligned | Unmatched |
|---|---|---|---|---|---|---|
| CUHN004 | ||||||
| F0 | 120,660,814 | 107,088,223 | 88.75 | 3,092,279 | 2.56 | 10,480,312 |
| XactMice | 111,413,778 | 87,512,893 | 78.55 | 9,127,519 | 8.19 | 14,773,366 |
| NSG | 112,796,525 | 79,633,078 | 70.60 | 15,297,511 | 13.56 | 17,865,936 |
| Nude | 112,177,796 | 97,498,001 | 86.91 | 5,957,473 | 5.31 | 8,722,322 |
| CUHN013 | ||||||
| F0 | 137,030,403 | 122,015,117 | 89.04 | 2,423,805 | 1.77 | 12,591,481 |
| XactMice | 129,059,960 | 96,347,935 | 74.65 | 6,748,729 | 5.23 | 25,963,296 |
| NSG | 131,474,894 | 96,192,537 | 73.16 | 12,345,259 | 9.39 | 22,937,096 |
| Nude | 144,123,210 | 101,821,943 | 70.65 | 10,769,634 | 7.47 | 31,531,633 |
Figure 5. Whole transcriptome analyses.
(a) Dendrograms of the four RNA sequencing data sets for the CUHN004 and CUHN013 tumors indicate that the XactMice environment alters gene expression from that observed in the NSG. (b) Heatmaps depicting the relative expression of paired sets of genes in both CUHN004 and CUHN013. The expression of genes known to play roles in these pathways was compared across F0 patient, XactMice, nude, and NSG samples. A red color indicates high RNA expression, while a blue shade signifies a low level of expression. (c) Waterfall graphs showing the relative enrichment of all GO terms associated with the differentially expressed genes identified in the F0 and XactMice tumors. Enrichment scores greater than 1.3 indicate that the GO term is statistically enriched (P-value<0.05) among these genes. After this enrichment analysis, GO terms were color-coded according to their overarching biological process, the most frequently observed of which were 22 immune system (blue), extracellular matrix (ECM; pink), and epithelial mesenchymal transition (EMT; green.) A paired z-test for proportions (inset table) shows that the enrichment of the GO terms representing each of these processes is statistically significant in genes differentially expressed in the F0 patient and XactMice tumors. (d) An enlargement of the top twenty most enriched GO terms in the tumor waterfall graphs from above. Starred (*) GO terms are enriched in both tumors.
To determine the pattern of gene drift upon XactMice implantation, we systematically analyzed the CUHN004 and CUHN013 sequencing data to identify differentially expressed genes, whose expression is similar in the F0 patient and XactMice tumors but different from their expression in the nude and NSG tumors (Supplemental Table 1). Although the variability between the CUHN004 and CUHN013 tumors resulted in few overlapping genes, many of the genes identified were common to several general biological processes, including epithelial differentiation, peptidase inhibition, cell adhesion, and protein processing (Supplemental Table 2). We captured all Gene Ontology (GO) terms associated with the differentially expressed genes and used them to perform a gene set enrichment analysis (GSEA), calculating an enrichment score for each of these GO terms. Many of the most enriched GO terms are associated with the immune system, the extracellular matrix (ECM), or epithelial-mesenchymal transition (EMT). We created heatmaps for both tumors, comparing the expression of several core genes linked to these GO terms (Fig. 5b), and we generated a waterfall graph depicting the relative enrichment of all GO terms, highlighting those relevant to the above-indicated pathways (Fig. 5c). In CUHN004, thirteen of the top twenty most enriched GO terms are associated with the immune system, ECM, or EMT (p-value<0.00001); for CUHN013, fifteen of the top twenty fall within these categories (p-value<0.00001) (Fig. 5d). Conversely, of the GO terms that are enriched in the nude and NSG tumors, few play a role in these processes.
To clarify the relationship between differentially expressed genes and the enriched processes identified by their GO terms, we identified many genes expressed exclusively in the F0 patient and XactMice tumors (Supplemental Table 3). Many of these genes play a role in the immune response or in EMT, or are components of the extracellular matrix, indicative of the HSPC-derived invasive cells playing an active role in stromal growth (Supplemental Table 4). In support of this, several of the differentially expressed genes are related to stromal processes (29, 30).
Humanization modulates lymphangiogenesis and cytokine expression
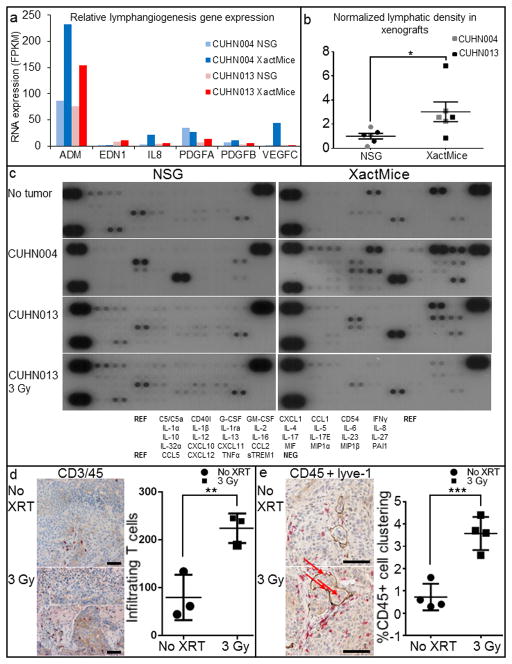
Interestingly, our RNA sequencing data revealed increased expression of several pro-lymphangiogenic factors including VEGF-C and IL8 in CUHN004; PDGF A and B, and endothelin-1 in CUHN013; and adrenomedullin, which was higher in both the CUHN004 and CUHN013 XactMice xenografts compared to those from the NSG controls (Fig. 6a). Given that the lymphatic vasculature plays an essential role in immune surveillance, inflammation, and tumorigenesis (31, 32) and that bone marrow derived cells contribute to lymphangiogenesis (33), we assessed whether the expansion of the lymphatic vasculature occurs in the XactMice. We stained CUHN013 and CUHN004 tumors for lyve-1, a marker of lymphatic vasculature (34), and observed increased intra-tumor lymphatic vessel density (lyve-1+ cells/mm2) in XactMice tumors compared to tumors in NSG hosts (Fig. 6b).
Figure 6. Physiological consequences of differential XactMice gene expression.
(a) A comparison of the FPKM values of several genes involved in lymphangiogenesis. (b) Graphical comparison of lymphatic vasculature in NSG and XactMice tumors. The average number of lyve-1 staining vessels per mm2 in the NSG tumors was used as a baseline against which XactMice tumors were compared. *P-value = 0.0393. (c) Cytokine arrays comparing plasma from NSG and XactMice. Relative cytokine concentrations were compared with ImageJ. (d) Dual CD45+ (red) and CD3+ (brown) IHC of XactMice CUHN013 tumors harvested after 0 and 3 Gy flank irradiation identifies infiltrating T cells (arrows). The associated graph shows that these cells are more abundant after irradiation. **P-value = 0.0115. (e) Dual CD45+/lyve-1 IHC indicates little association between invading blood cells and lymph tissue in non-irradiated tumors, but after 3 Gy, the CD45+ cells cluster around, and are sometimes found within (arrows), the lymph vessels. The graph shows the increased association of human CD45+ cells with lyve-1 staining tissue after irradiation. ***P-value = 0.001. To be included, CD45+ cells must be within 200 um of lyve-1+ cells. Magnification is 10× and the scale bar equals 50 μm.
Analysis of the RNA sequencing data also indicated that many cytokines and chemokines are among the differentially expressed genes. To identify additional cytokines uniquely present in the XactMice, we profiled XactMice and NSG blood plasma on human cytokine arrays (Fig. 6c & Supplemental Table 5). Although there is some antibody cross-reactivity between murine and human cytokines, we can still make several conclusions about the human cytokine signaling present in the XactMice. When no tumors were implanted, three cytokines were markedly elevated in the XactMice plasma: GM-CSF, CD54, and MIF. These cytokines are all known to modulate lymphoid or myeloid cell migration, and their elevated presence supports the role of functioning immune components in the XactMice. Conversely, four cytokines - C5/C5α, CD40l, IL-13 and CXCL12 - were expressed more abundantly in the NSG plasma. This spurious binding prevents us from forming conclusions about possibly alterations in the XactMice plasma. However, even discounting these signals, the cytokine profiles of the NSG plasma from animals bearing either CUHN004 or CUHN013 tumors look quite similar to that observed from NSG mice without tumors, although IL-1rα and MIF are also highly expressed in the CUHN004 plasma (as predicted from RNA sequencing of this tumor.) Conversely, increased concentrations of CD54, IL-1rα, IL-6, IL-8, and MIF are present in the plasma from XactMice bearing either CUHN004 or CUHN013 tumors. Additionally, plasma from the CUHN004 XactMice contains increased levels of GM-CSF, CCL1, IL-13, and PAI1, while that plasma from the CUHN013 XactMice lacks GM-CSF expression, implying a complex interplay between the signals generated by the HSPCs in the bone marrow and the tumors.
XactMice tumors present a dynamic microenvironment
Since patient tumor growth responds to sudden changes in the local environment, we examined the effects of 3Gy ionizing radiation to CUHN013 tumors on NSG and XactMice. An IHC analysis of irradiated XactMice tumors reveals an influx of T cells, as would be expected from a functional immune system (Fig. 6d) (35). Lymph vessels, as visualized by lyve-1 staining, also rapidly expand and cluster with - and occasionally encompass - infiltrating immune cells, providing a potential avenue for their migration to and from the tumor stroma (Fig. 6e). Finally, expression of almost all cytokines falls sharply after irradiation, indicative of the dramatic changes produced by radiation within the tumor microenvironment (Fig 6c). Only, three cytokines (CD40l, GM-CSF, and IL-13) increased very modestly in the XactMice plasma. Since CD40l and IL-13 expression were also found to be elevated in NSG mice without tumors, it is possible that the presence of some of these cytokines after irradiation indicates spurious cross-reactivity.
DISCUSSION
A tumor can be envisioned as the result of the growth and interaction of cancer cells with the surrounding fibroblast scaffolding, infiltrating immune cells, and associated lymphatic vasculature. Without accounting for all of these components, a model system cannot hope to accurately recreate the conditions driving the original tumor’s growth. We propose that the XactMice system represents an important step toward recapitulating a patient’s tumor environment by creating an in vivo system in which a xenograft growth is at least partially regulated by a surrogate human immune system. Human blood cells originating in the engrafted bone marrow circulate throughout the tumor, altering cytokine expression, assisting in stromal deposition, and increasing lymphangiogenesis throughout the tumor microenvironment. Their presence also alters gene expression within the tumor cells, regulating expression of genes critical in maintaining a tumor microenvironment. The observation that XactMice tumor implantation reversed the initial genetic drift observed after tumor passage in immune-compromised hosts indicates that this model enables tumors to revert to their original state.
We have also shown that the immune system engrafted into the XactMice is not only attendant, but capable of interacting with the grafted tumor. Since tumors are constantly engaged with their surrounding and supportive stromal tissues, it is important that the XactMice model replicate this facet of cancer behavior. The immune system in XactMice responds rapidly and appropriately to an environmental insult within the tumor. Changes in immune cell infiltration, cytokine expression, and lymphangiogenesis are fitting physiological responses to radiation and, again, are indicative of the value of the XactMice model.
Also of importance is that this model is self-sustaining: the engrafted HSPCs can be purified from the bone marrow of the XactMice and re-engrafted into another generation of XactMice with HSPCs derived from the original donor source. Bird et al (18) used the bone marrow cells obtained from an initial cohort of xenochimaeric NSG mice for serial transplantation studies. They transplanted 1×106 unfractionated bone marrow cells harvested from human HSPC xenochimaeric mice into a second cohort of irradiated NSG mice. The secondary cohort of xenotransplant recipient mice were euthanized 12 weeks post-transplant, and their bone marrow cells were assessed for the presence of human CD34+ cells by flow cytometry. They observed human CD45+/CD38+/CD34+ cells in the bone marrow of the secondary cohort of xenochimaeric NSG mice, documenting the feasibility of generating additional cohorts of xenochimaeric mice. Therefore implanted patient xenografts could be continually propagated in an environment similar to that of the original tumor. As a first attempt to develop this model, the proof-of-concept studies presented in this report involved mice in which the human hematopoietic system and the tumor were derived from allogeneic sources. This allogeneic system did not significantly impact tumor growth. Future experiments will examine whether an allogeneic response could affect chemotherapy and radiotherapy in humanized mice. The long-term goal of these studies is to enable the generation of human xenochimaeric tumor bearing mice that harbor syngeneic tumor and hematopoietic cells. A syngeneic humanized mouse model could potentially recreate the patient tumor-immune microenvironment and serve as an invaluable platform in the investigation of the next generation of immunomodulatory chemotherapeutics.
Even though various types of humanized mice have been previously reported, they are typically produced by the injection of purified human CD34+ cells into sublethally irradiated mice (36) and have mainly been employed to study the immune system, allograft rejection, and blood cell differentiation (37). Rongvaux recently described the generation of MITRG and MISTRG mice which were engineered to express human cytokines that would support the development and function of monocytes, macrophages, and NK cells derived from the injection of human fetal liver or (17) and activity in these genetically engineered humanized mice, they implanted cell line-derived tumors and observed that human macrophages invaded these tumors in a VEGF-dependent manner. As presented, the Rongvaux model and the humanized mouse model previously described by Wege et al, (38) do not attempt to recreate the microenvironment of a patient’s tumor in a humanized mouse, but it nicely complements our observations concerning human immune cell invasion into cell line-derived tumors.
Cancer research has benefitted significantly from our ability to customize therapy in response to mutated genetic pathways that drive an individual’s tumor growth (39). The host’s relevance in determining disease occurrence and outcome is now also increasingly evident and must be accounted for in developing tomorrow’s therapies (40). Stromal cells and immune cells are emerging as potent drivers of invasion and metastasis and the XactMice model faithfully recreates the native tumor environment absent in current animal models. We predict that this model will be a useful tool in the rapidly expanding field of stroma- and/or immune-directed drug development.
MATERIALS AND METHODS
HSPC isolation and XactMice generation
Donated, de-identified cord blood was obtained from the University of Colorado cord blood bank (http://www.clinimmune.com/cordbloodbank/). G-CSF-mobilized patient blood was collected in accordance with the protocols approved by the Colorado Multiple Institutional Review Board (COMIRB #08-0552 and #06-0720). HSPCs were expanded as previously reported (18). NOD/SCID/IL2rg−/− (NSG; Jackson Laboratories, Bar Harbor, ME) mice were prepared as previously described (18). After eight weeks, mice were bled via the tail vein to assess HSPC engraftment. The peripheral blood was analyzed by flow cytometry, using human CD3, CD45, and/or CD151 (Biolegend, San Diego, CA) antibodies. Tumor tissue was collected and implanted on all mice as described (19).
Mouse irradiation
CUHN013 xenograft-bearing mice were irradiated using an RS-2000 small animal irradiator (Rad Source Technologies, Suwanee, GA), calibrated to deliver 1.15Gy X-ray radiation per minute. Mice were anesthetized via IP injection with a ketamine (60mg/kg)/xylazine (8mg/kg) solution, then positioned beneath a lead/cadmium shield, designed to allow radiation to penetrate only the flank tumors. A 3Gy dose was administered to each tumor, and the mice were allowed to recover for 24 hours before tissue collection.
Flow cytometry/cell sorting
Excised tissues were prepared as previously described (41). Cell suspensions were blocked by the addition of Gamunex (Baxter, Deerfield, IL) for 30 minutes at RT, then washed in flow buffer (PBS + 2%FBS) and incubated at RT for 1 hour with the following antibodies (at a 1:10 final concentration): PerCP/Cy5.5-CD3, APC-CD34, PE-CD45, and/or APC-CD151 (BioLegend). Flow analysis was conducted on a Beckman Coulter Cyan. Cell sorting was performed on the Beckman Coulter Moflo XDP 70. Data analysis was performed using Summit, V5.1 (Beckman Coulter, Brea, CA).
STR PCR analysis
Genomic DNA from tumor tissue was isolated using a QIAamp DNA Mini Kit (Qiagen, Germantown, MD). STR amplification was performed using Taq polymerase (Promega, Madison, WI) on a GenAmp 9700 machine (Applied Biosystems, Grand Island, NY) with the following primers (5′-3′): TPOX-For ACTGGCACAGAACAGGCACTTAGG, TPOX-Rev GGAGGAACTGGGAACCCCACAGGTTA, vWA-For GGACAGATGATAAATACATAGGATGGATGG, and vWA-Rev GCCCTAGTGGATGATAAGAATAATCAGTATGTG. The resultant DNA was visualized using an Agilent Bioanalyzer 2100 with a DNA1000 chip (Agilent, Santa Clara, CA)
Immunofluorescence
Slides were de-paraffinized in xylene twice for five minutes and rehydrated in graded concentrations of ethanol. They were washed 3× in PBS and blocked with 15% goat serum in mouse IgG before the addition of CD151 (Novus Biologicals, Littleton, CO) at a 1:50 concentration. Alexa Fluor© goat-α-rabbit 488 (Invitrogen, Grand Island, NY) was added for secondary staining, and the slides were incubated for 1 hour at RT, washed 3× in PBS, and covered with Fluoromount-G (SouthernBiotech, Birmingham, AL).
Histology/Immunohistochemistry
For single staining with the human CD45 antibody (Dako) or mouse lyve-1 (Abcam, San Fransisco, CA), the samples were incubated with antibody, then rinsed and a secondary reagent, AP-conjugated MACH2 Mouse Polymer (BioCare Medical, Concord, CA), RT for CD45 staining or EnVision + Dual Link- HRP Polymer (Dako) for lyve-1 staining was applied before development with Vulcan Fast Red Chromogen (BioCare Medical) or DAB+ (Dako). For dual staining of mouse and human CD45, anti-mouse CD45 antibody (BD Pharmingen, San Jose, CA) was applied, followed by a secondary rabbit anti-rat biotinylated antibody (Dako) and Streptavidin-HPR conjugated solution (Dako). The CD3, lyve-1 (Abcam), and CD19 (MyBiosource, San Diego, CA) antibodies were applied at the appropriate dilution, followed by HRP-conjugated EnVision + Dual Link System (Dako). In all cases, primary staining was developed using DAB+ chromogen solution (Dako).
FISH analysis
The sections were subjected to a dual-color FISH assay using Vysis CEPX SpectrumOrange/Y SpectrumGreen probe (Abbott Molecular, Abbott Park, IL) following standard laboratory protocol. After post-hybridization washes, the slides were dehydrated in ethanol, and the chromatin was counterstained with DAPI (0.3μg/ml in Vectashield mounting medium, Vector Laboratories, Burlingame, CA).
Cytokine arrays
Human cytokine arrays (Panel A, R&D Systems, Minneapolis, MN) were processed according to the manufacturer’s instructions, using 150uL of the indicated NSG or XactMice plasma (from blood collected in 50U/mL heparin and centrifuged 10 min at 1800rcf to remove platelets). Digital copies of the developed film were quantified using ImageJ software.
Laser-capture microscopy
Eight micron frozen sections of tissue were cut using a cryostat, dehydrated through graded solutions of alcohols and xylene, and air-dried for 5 minutes before being put into an Arcturus XT Laser Capture Microdissection Instrument (Applied Biosystems).
RNA extraction
Tissue was placed in 300 μL of QIAzol and homogenized using the MP Biomedicals Fast Prep 24. RNA was extracted from LCM-captured cells and tissue using the RNeasy Mini RNA purification protocol (Qiagen). Total RNA was quantified on a Nanodrop 1000 (Thermo Scientific, Waltham, MA).
RNA sequencing
Libraries were constructed using 1μg total RNA, following the Illumina TruSeq RNA Sample Preparation v2 Guide. The products were amplified by PCR to create a cDNA library, which was then validated on the Agilent 2100 Bioanalyzer using DNA-1000 chip. Cluster generation was performed on the Illumina cBot, using a Single Read Flow Cell with a Single Read cBot reagent plate (TruSeq SR Cluster Kit). Sequencing of the clustered flow cell was performed on the Illumina HiSeq 2000, using TruSeq SBS v3 reagents (Illumina, San Diego, CA).
Bioinformatics
The RNA reads were mapped against the human genome using Tophat (version 2.0.5) (42), using the NCBI reference annotation as a guide. Cufflinks (version 2.0.2) (43) was used to assemble the transcripts. Differentially expressed genes between F0/XactMice and the NSG/Nude were identified using CuffDiff and a fold-change threshold of at least two. Top pathways/gene ontologies were identified using DAVID (44, 45). Gene set enrichment analysis (GSEA, v. 2.07) was conducted on the collapsed data using the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) pathway definitions obtained from the Molecular Signatures Database, v.3.1. Pathways with a nominal p-value ≤ 0.05 were deemed significant. All analyses except GSEA were conducted in R/Bioconductor, v. 3.0.1.
Supplementary Material
(a) Flow cytometry shows the presence of a small population of human CD45/CD3+ cells in the bone marrow, spleen, and blood of the CUHN082 patient XactMice, collected almost a year subsequent to HSPC engraftment. (b) Dual human CD3 (brown) and CD45 (red) IHC indicates that T cells can be found (arrows) in the bone marrow and spleens of these XactMice. Dual human CD19 (brown) and CD45 (red) IHC indicates that B cells can also be seen (arrows) in the bone marrow and spleens of these XactMice. Magnification is 20× and scale bar equals 50 um. (c) H/E comparisons of CUHN004 and CUHN013 tumors from patients (F0), and after implantation in nude, NSG, and XactMice. Magnification is 10× and scale bar equals 50 um.
The progeny of HSPCs isolated from adult peripheral blood are present within the XactMice. Upper panel: Flow cytometry showing that no human CD45/151+ cells are present in the bone marrow, spleen, blood, or tumor of a nude mouse (no tumors grew on the NSG mice) into whose rear flank 50,000 tumor cells were injected. Middle panel: Human CD45/151+ cells can be found within the bone marrow, spleen, peripheral blood and tumor of the XactMice. Lower panel: Human CD45/151+ cells are present within the bone marrow, spleen, and peripheral blood of XactMice generated from the peripheral blood of a cancer patient given G-CSF while undergoing chemotherapy.
These genes in the CUHN004 and CUHN013 tumors were either calculated by Cuff-diff to be differentially expressed in the XactMice and F0 samples, or they were subjected to an expression fold-change analysis between the XactMice-F0 and the NSG-nude groups and found to have an absolute fold change value 2. Genes in bold appear in both tumors.
The differentially expressed genes in the XactMice and F0 samples for each tumor are statistically enriched with members of several different biological processes. Their enrichment score is calculated by the NIH-DAVID algorithm and derived from the negative log of the P-value of their presence together within the queried gene list. Any enrichment score greater than 1.3 correlates with a P-value of less than 0.05.
These genes were identified from RNA sequencing data from their low expression in the nude and NSG tumors and dramatically increased expression in F0 and XactMice tumors. To be considered activated, a gene’s expression in the XactMice tumor must be greater than four times its expression in the nude or NSG tumors. Additionally, its average expression in F0 and XactMice tumors must be greater than 20 times its average expression in the nude and NSG tumors. The activated genes highlighted in pink are implicated in ECM function. Those in green have a known role in EMT, while those in blue play a role in the immune response. Genes activated in both tumors are in bold.
The activated genes identified in the XactMice tumors are statistically enriched with members of immune response, inflammation, and cell adhesion pathways. The enrichment score is calculated by the NIH-DAVID algorithm and derived from the negative log of the P-value of their presence together within the queried gene list. Any enrichment score greater than 1.3 correlates with a P-value of less than 0.05.
Cytokines whose expression differs between NSG mice and XactMice were quantified. Those in red are expressed less abundantly in XactMice, while those in blue are more abundantly expressed in XactMice. Cytokines in black show little difference in expression between mouse groups.
Acknowledgments
Support
This work was primarily supported by Department of Defense Award W81XWN-10-1-0798 (principal investigator A.J.; co-investigator Y.R.), the Charles C. Gates Center for Regenerative Medicine and Stem Cell Biology, and National Institutes of Health (NIH) Cancer Center Support Grant P30 CA046934 (A.J., Y.R.). Additionally supported by NIH R21 DE019712 (A.J.), NIH R01 CA149456 (A.J.), the Janet Mordecai Foundation (A.J.), the Peter and Rhonda Grant Foundation (A.J.), NIH R01 CA117802-06 (Y.R.), NIH P30 AR057212-02 (Y.R.), NIH P01 AI022295-24 (Y.R.), NIH T32 CA174648 (principal investigator XJ.W.; co-investigator P.N.L), and a Translational Research Award from the Leukemia and Lymphoma Society (Y.R.).
We thank the patients and normal subjects for their time and generous tissue and blood donations.
Footnotes
COMPETING FINANCIAL INTERESTS
A.J., Y.R., XJ.W. and D.R.R. are co-inventors of technology presented in this report. The other authors declare no competing financial interests.
References
- 1.Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27(45):5904–12. doi: 10.1038/onc.2008.271. Epub 2008/10/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wels J, Kaplan RN, Rafii S, Lyden D. Migratory neighbors and distant invaders: tumor-associated niche cells. Genes & development. 2008;22(5):559–74. doi: 10.1101/gad.1636908. Epub 2008/03/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engelholm SA, Vindelov LL, Spang-Thomsen M, Brunner N, Tommerup N, Nielsen MH, et al. Genetic instability of cell lines derived from a single human small cell carcinoma of the lung. European journal of cancer & clinical oncology. 1985;21(7):815–24. doi: 10.1016/0277-5379(85)90221-4. Epub 1985/07/01. [DOI] [PubMed] [Google Scholar]
- 4.Hausser HJ, Brenner RE. Phenotypic instability of Saos-2 cells in long-term culture. Biochemical and biophysical research communications. 2005;333(1):216–22. doi: 10.1016/j.bbrc.2005.05.097. Epub 2005/06/09. [DOI] [PubMed] [Google Scholar]
- 5.Dadras SS, Paul T, Bertoncini J, Brown LF, Muzikansky A, Jackson DG, et al. Tumor lymphangiogenesis: a novel prognostic indicator for cutaneous melanoma metastasis and survival. The American journal of pathology. 2003;162(6):1951–60. doi: 10.1016/S0002-9440(10)64328-3. Epub 2003/05/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suggitt M, Bibby MC. 50 years of preclinical anticancer drug screening: empirical to target-driven approaches. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11(3):971–81. Epub 2005/02/15. [PubMed] [Google Scholar]
- 7.Wennerberg J, Trope C, Biorklund A. Heterotransplantation of human head and neck tumours into nude mice. Acta oto-laryngologica. 1983;95(1–2):183–90. doi: 10.3109/00016488309130933. Epub 1983/01/01. [DOI] [PubMed] [Google Scholar]
- 8.DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nature medicine. 2011;17(11):1514–20. doi: 10.1038/nm.2454. Epub 2011/10/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, et al. Patient-derived tumour xenografts as models for oncology drug development. Nature reviews Clinical oncology. 2012;9(6):338–50. doi: 10.1038/nrclinonc.2012.61. Epub 2012/04/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrido-Laguna I, Uson M, Rajeshkumar NV, Tan AC, de Oliveira E, Karikari C, et al. Tumor engraftment in nude mice and enrichment in stroma-related gene pathways predict poor survival and resistance to gemcitabine in patients with pancreatic cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17(17):5793–800. doi: 10.1158/1078-0432.CCR-11-0341. Epub 2011/07/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iorns E, Clarke J, Ward T, Dean S, Lippman M. Simultaneous analysis of tumor and stromal gene expression profiles from xenograft models. Breast cancer research and treatment. 2012;131(1):321–4. doi: 10.1007/s10549-011-1784-8. Epub 2011/09/29. [DOI] [PubMed] [Google Scholar]
- 12.Bankert RB, Balu-Iyer SV, Odunsi K, Shultz LD, Kelleher RJ, Jr, Barnas JL, et al. Humanized mouse model of ovarian cancer recapitulates patient solid tumor progression, ascites formation, and metastasis. PLoS One. 2011;6(9):e24420. doi: 10.1371/journal.pone.0024420. Epub 2011/09/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Q, Facciponte J, Jin M, Shen Q, Lin Q. Humanized NOD-SCID IL2rg−/− mice as a preclinical model for cancer research and its potential use for individualized cancer therapies. Cancer letters. 2014;344(1):13–9. doi: 10.1016/j.canlet.2013.10.015. Epub 2014/02/12. [DOI] [PubMed] [Google Scholar]
- 14.Leskov I, Pallasch CP, Drake A, Iliopoulou BP, Souza A, Shen CH, et al. Rapid generation of human B-cell lymphomas via combined expression of Myc and Bcl2 and their use as a preclinical model for biological therapies. Oncogene. 2013;32(8):1066–72. doi: 10.1038/onc.2012.117. Epub 2012/04/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pallasch CP, Leskov I, Braun CJ, Vorholt D, Drake A, Soto-Feliciano YM, et al. Sensitizing protective tumor microenvironments to antibody-mediated therapy. Cell. 2014;156(3):590–602. doi: 10.1016/j.cell.2013.12.041. Epub 2014/02/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abate-Daga D, Lagisetty KH, Tran E, Zheng Z, Gattinoni L, Yu Z, et al. A Novel Chimeric Antigen Receptor Against Prostate Stem Cell Antigen Mediates Tumor Destruction in a Humanized Mouse Model of Pancreatic Cancer. Human gene therapy. 2014 doi: 10.1089/hum.2013.209. Epub 2014/04/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rongvaux A, Willinger T, Martinek J, Strowig T, Gearty SV, Teichmann LL, et al. Development and function of human innate immune cells in a humanized mouse model. Nature biotechnology. 2014 doi: 10.1038/nbt.2858. Epub 2014/03/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bird GA, Polsky A, Estes P, Hanlon T, Hamilton H, Morton JJ, et al. Expansion of Human and Murine Hematopoietic Stem and Progenitor Cells Ex Vivo without Genetic Modification Using MYC and Bcl-2 Fusion Proteins. PloS one. 2014;9(8):e105525. doi: 10.1371/journal.pone.0105525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keysar SB, Astling DP, Anderson RT, Vogler BW, Bowles DW, Morton JJ, et al. A patient tumor transplant model of squamous cell cancer identifies PI3K inhibitors as candidate therapeutics in defined molecular bins. Molecular oncology. 2013;7(4):776–90. doi: 10.1016/j.molonc.2013.03.004. Epub 2013/04/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimple RJ, Harari PM, Torres AD, Yang RZ, Soriano BJ, Yu M, et al. Development and characterization of HPV-positive and HPV-negative head and neck squamous cell carcinoma tumorgrafts. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(4):855–64. doi: 10.1158/1078-0432.CCR-12-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lui VW, Hedberg ML, Li H, Vangara BS, Pendleton K, Zeng Y, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer discovery. 2013;3(7):761–9. doi: 10.1158/2159-8290.CD-13-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozer H, Armitage JO, Bennett CL, Crawford J, Demetri GD, Pizzo PA, et al. 2000 update of recommendations for the use of hematopoietic colony-stimulating factors: evidence-based, clinical practice guidelines. American Society of Clinical Oncology Growth Factors Expert Panel. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2000;18(20):3558–85. doi: 10.1200/JCO.2000.18.20.3558. Epub 2000/10/14. [DOI] [PubMed] [Google Scholar]
- 23.Kidd S, Spaeth E, Watson K, Burks J, Lu H, Klopp A, et al. Origins of the tumor microenvironment: quantitative assessment of adipose-derived and bone marrow-derived stroma. PLoS One. 2012;7(2):e30563. doi: 10.1371/journal.pone.0030563. Epub 2012/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee HJ, Choi BH, Min BH, Park SR. Changes in surface markers of human mesenchymal stem cells during the chondrogenic differentiation and dedifferentiation processes in vitro. Arthritis and rheumatism. 2009;60(8):2325–32. doi: 10.1002/art.24786. Epub 2009/08/01. [DOI] [PubMed] [Google Scholar]
- 25.Geary SM, Cowin AJ, Copeland B, Baleato RM, Miyazaki K, Ashman LK. The role of the tetraspanin CD151 in primary keratinocyte and fibroblast functions: implications for wound healing. Experimental cell research. 2008;314(11–12):2165–75. doi: 10.1016/j.yexcr.2008.04.011. Epub 2008/06/07. [DOI] [PubMed] [Google Scholar]
- 26.Thomas ML. The leukocyte common antigen family. Annual review of immunology. 1989;7:339–69. doi: 10.1146/annurev.iy.07.040189.002011. Epub 1989/01/01. [DOI] [PubMed] [Google Scholar]
- 27.Ruitberg CM, Reeder DJ, Butler JM. STRBase: a short tandem repeat DNA database for the human identity testing community. Nucleic acids research. 2001;29(1):320–2. doi: 10.1093/nar/29.1.320. Epub 2000/01/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Redon R, Fitzgerald T, Carter NP. Comparative genomic hybridization: DNA labeling, hybridization and detection. Methods in molecular biology. 2009;529:267–78. doi: 10.1007/978-1-59745-538-1_17. Epub 2009/04/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fullar A, Kovalszky I, Bitsche M, Romani A, Schartinger VH, Sprinzl GM, et al. Tumor cell and carcinoma-associated fibroblast interaction regulates matrix metalloproteinases and their inhibitors in oral squamous cell carcinoma. Experimental cell research. 2012;318(13):1517–27. doi: 10.1016/j.yexcr.2012.03.023. Epub 2012/04/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuyada A, Chow A, Wu J, Somlo G, Chu P, Loera S, et al. CCL2 mediates cross-talk between cancer cells and stromal fibroblasts that regulates breast cancer stem cells. Cancer research. 2012;72(11):2768–79. doi: 10.1158/0008-5472.CAN-11-3567. Epub 2012/04/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christiansen A, Detmar M. Lymphangiogenesis and cancer. Genes & cancer. 2011;2(12):1146–58. doi: 10.1177/1947601911423028. Epub 2012/08/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomes FG, Nedel F, Alves AM, Nor JE, Tarquinio SB. Tumor angiogenesis and lymphangiogenesis: tumor/endothelial crosstalk and cellular/microenvironmental signaling mechanisms. Life sciences. 2013;92(2):101–7. doi: 10.1016/j.lfs.2012.10.008. Epub 2012/11/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tawada M, Hayashi S, Osada S, Nakashima S, Yoshida K. Human gastric cancer organizes neighboring lymphatic vessels via recruitment of bone marrow-derived lymphatic endothelial progenitor cells. Journal of gastroenterology. 2012;47(9):1057–60. doi: 10.1007/s00535-012-0638-4. Epub 2012/07/26. [DOI] [PubMed] [Google Scholar]
- 34.Jackson DG. The lymphatics revisited: new perspectives from the hyaluronan receptor LYVE-1. Trends in cardiovascular medicine. 2003;13(1):1–7. doi: 10.1016/s1050-1738(02)00189-5. Epub 2003/01/30. [DOI] [PubMed] [Google Scholar]
- 35.McBride WH, Chiang CS, Olson JL, Wang CC, Hong JH, Pajonk F, et al. A sense of danger from radiation. Radiation research. 2004;162(1):1–19. doi: 10.1667/rr3196. Epub 2004/06/30. [DOI] [PubMed] [Google Scholar]
- 36.Lan P, Tonomura N, Shimizu A, Wang S, Yang YG. Reconstitution of a functional human immune system in immunodeficient mice through combined human fetal thymus/liver and CD34+ cell transplantation. Blood. 2006;108(2):487–92. doi: 10.1182/blood-2005-11-4388. Epub 2006/01/18. [DOI] [PubMed] [Google Scholar]
- 37.Rong Z, Wang M, Hu Z, Stradner M, Zhu S, Kong H, et al. An Effective Approach to Prevent Immune Rejection of Human ESC-Derived Allografts. Cell stem cell. 2014;14(1):121–30. doi: 10.1016/j.stem.2013.11.014. Epub 2014/01/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wege AK, Ernst W, Eckl J, Frankenberger B, Vollmann-Zwerenz A, Mannel DN, et al. Humanized tumor mice--a new model to study and manipulate the immune response in advanced cancer therapy. International journal of cancer Journal international du cancer. 2011;129(9):2194–206. doi: 10.1002/ijc.26159. Epub 2011/05/06. [DOI] [PubMed] [Google Scholar]
- 39.Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nature biotechnology. 2012;30(7):679–92. doi: 10.1038/nbt.2284. Epub 2012/07/12. [DOI] [PubMed] [Google Scholar]
- 40.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. Epub 2012/09/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keysar SB, Le PN, Anderson RT, Morton JJ, Bowles DW, Paylor JJ, et al. Hedgehog signaling alters reliance on EGF receptor signaling and mediates anti-EGFR therapeutic resistance in head and neck cancer. Cancer research. 2013;73(11):3381–92. doi: 10.1158/0008-5472.CAN-12-4047. Epub 2013/04/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. Epub 2009/03/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28(5):511–5. doi: 10.1038/nbt.1621. Epub 2010/05/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. Epub 2009/01/10. [DOI] [PubMed] [Google Scholar]
- 45.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. Epub 2008/11/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) Flow cytometry shows the presence of a small population of human CD45/CD3+ cells in the bone marrow, spleen, and blood of the CUHN082 patient XactMice, collected almost a year subsequent to HSPC engraftment. (b) Dual human CD3 (brown) and CD45 (red) IHC indicates that T cells can be found (arrows) in the bone marrow and spleens of these XactMice. Dual human CD19 (brown) and CD45 (red) IHC indicates that B cells can also be seen (arrows) in the bone marrow and spleens of these XactMice. Magnification is 20× and scale bar equals 50 um. (c) H/E comparisons of CUHN004 and CUHN013 tumors from patients (F0), and after implantation in nude, NSG, and XactMice. Magnification is 10× and scale bar equals 50 um.
The progeny of HSPCs isolated from adult peripheral blood are present within the XactMice. Upper panel: Flow cytometry showing that no human CD45/151+ cells are present in the bone marrow, spleen, blood, or tumor of a nude mouse (no tumors grew on the NSG mice) into whose rear flank 50,000 tumor cells were injected. Middle panel: Human CD45/151+ cells can be found within the bone marrow, spleen, peripheral blood and tumor of the XactMice. Lower panel: Human CD45/151+ cells are present within the bone marrow, spleen, and peripheral blood of XactMice generated from the peripheral blood of a cancer patient given G-CSF while undergoing chemotherapy.
These genes in the CUHN004 and CUHN013 tumors were either calculated by Cuff-diff to be differentially expressed in the XactMice and F0 samples, or they were subjected to an expression fold-change analysis between the XactMice-F0 and the NSG-nude groups and found to have an absolute fold change value 2. Genes in bold appear in both tumors.
The differentially expressed genes in the XactMice and F0 samples for each tumor are statistically enriched with members of several different biological processes. Their enrichment score is calculated by the NIH-DAVID algorithm and derived from the negative log of the P-value of their presence together within the queried gene list. Any enrichment score greater than 1.3 correlates with a P-value of less than 0.05.
These genes were identified from RNA sequencing data from their low expression in the nude and NSG tumors and dramatically increased expression in F0 and XactMice tumors. To be considered activated, a gene’s expression in the XactMice tumor must be greater than four times its expression in the nude or NSG tumors. Additionally, its average expression in F0 and XactMice tumors must be greater than 20 times its average expression in the nude and NSG tumors. The activated genes highlighted in pink are implicated in ECM function. Those in green have a known role in EMT, while those in blue play a role in the immune response. Genes activated in both tumors are in bold.
The activated genes identified in the XactMice tumors are statistically enriched with members of immune response, inflammation, and cell adhesion pathways. The enrichment score is calculated by the NIH-DAVID algorithm and derived from the negative log of the P-value of their presence together within the queried gene list. Any enrichment score greater than 1.3 correlates with a P-value of less than 0.05.
Cytokines whose expression differs between NSG mice and XactMice were quantified. Those in red are expressed less abundantly in XactMice, while those in blue are more abundantly expressed in XactMice. Cytokines in black show little difference in expression between mouse groups.