MIRMAID (Medical Imaging Research Management and Associated Information Database) provides a highly configurable workflow; has a single interface that can store, manage, and retrieve imaging-based studies; and is capable of handling the requirements of data auditing and project management. It provides a complete system for medical imaging research that has not been previously available.
Abstract
Today, a typical clinical study can involve thousands of participants, with imaging data acquired over several time points across multiple institutions. The additional associated information (metadata) accompanying these data can cause data management to be a study-hindering bottleneck. Consistent data management is crucial for large-scale modern clinical imaging research studies. If the study is to be used for regulatory submissions, such systems must be able to meet regulatory compliance requirements for systems that manage clinical image trials, including protecting patient privacy. Our aim was to develop a system to address these needs by leveraging the capabilities of an open-source content management system (CMS) that has a highly configurable workflow; has a single interface that can store, manage, and retrieve imaging-based studies; and can handle the requirement for data auditing and project management. We developed a Web-accessible CMS for medical images called Medical Imaging Research Management and Associated Information Database (MIRMAID). From its inception, MIRMAID was developed to be highly flexible and to meet the needs of diverse studies. It fulfills the need for a complete system for medical imaging research management.
©RSNA, 2015
Introduction
The development of consistent data management is crucial for large-scale modern clinical imaging research studies (1–3). A typical study can involve thousands of participants, with imaging data acquired over many time points and across multiple institutions. The supplementary information accompanying these data (metadata) can cause data management to be a study-hindering bottleneck. Several researchers have promoted data sharing to accelerate progress, reduce cost, and allow reproducible research (4). Research management systems, such as the one described in this article, are necessary to accurately and effectively carry out these complex studies. In clinical environments, picture archiving and communication systems (PACS) are adequate to support clinical workflows. However, they are not well-suited for research workflows because it is difficult to incorporate third-party applications that facilitate data input and output as well as analysis. Although data are accessible, they are not easily manipulated and analyzed on a large scale. They also support only a simple linear workflow. Furthermore, adding the research workload to the clinical environment is not recommended because doing so might affect system stability and because research protocols require the use of de-identified data.
We are not aware of a commercial or free software solution that provides the complete suite of functions required for a research image management system. This may be because the market size for such a research system is much smaller than that for a clinical image management system (ie, a picture archiving and communication system). Furthermore, research workflows focusing on analyzing data from medical images tend to be heterogeneous because there are a variety of imaging and analysis protocols and study themes (eg, screening versus follow-up), as well as infinite possibilities for generated results. Current solutions, such as XNAT (Extensible Neuroimaging Archive Toolkit) (5), MilxXplore (6), MRIdb (7), NiDB (Neuroinformatics Database) (8), HID (Human Clinical Imaging Database) (9), DFBIdb (10), COINS (Collaborative Informatics and Neuroimaging Suite) (11), LORIS (Longitudinal Online Research and Imaging System) (12), TCIA/NBIA (The Cancer Imaging Archive/National Biomedical Imaging Archive) (13), MIDAS (the Multimedia Digital Archiving System) (14), QI-Bench (15), QIDW (Quantitative Imaging Data Warehouse) (16), and TRIAD (Transfer of Images and Data) (17), have each been developed mainly for a specific research topic and with the assumption of a particular workflow. As a result, adapting them to different applications is challenging. Other approaches, such as portable drives, command shell scripts, and spreadsheets, have numerous shortcomings. These systems make it nearly impossible to accurately follow or document compliance with research workflows and, unfortunately, offer many opportunities for human error.
The U.S. Food and Drug Administration (FDA) has delineated several requirements for research data that are used as part of a submission requesting approval of an agent or device. One is that the system must log all actions taking place. It is critical that researchers have an auditable log of all data manipulations and measurements, by both the operator and the software, because this is one of the main requirements of Title 21 of the Code of Federal Regulations, part 11 (21 CFR part 11) (18,19).
The term content management system (CMS) refers to a computer-based application that, through a centralized database, allows for editing, organizing, deleting, controlling access, searching, providing analytics, reporting, checking content into and out of a repository, and maintaining content.
Our aim was to develop a system for medical image research that has a highly configurable workflow; has a single interface; can store, manage, and retrieve imaging-based studies; and can handle the requirement of data auditing and project management. Databases do an excellent job of storing and retrieving data; a CMS adds capabilities on top of a database, including version management, workflow, flexible tagging, and project management. Because these capabilities appeared useful for research, we decided to build our system using a CMS rather than just a database.
Content in our applications example (discussed later in this article) consists of image data, metadata, biomarker information, notes, and tags. The CMS we built for imaging, called MIRMAID (Medical Imaging Research Management and Associated Information Database), consists of three main components: (a) TACTIC (Southpaw Technology, Toronto, Ontario, Canada), an open-source digital content and asset management system introduced to handle the data objects used in the movie industry (20); (b) tiPY, a Python (Python Software Foundation, Beaverton, Ore) library developed by us for easy system interaction, such as data input and output; and (c) an HTML- and JavaScript-based Web browser user interface, also developed by us, to facilitate interactive image analysis tasks, such as manual generation of regions of interest.
A CMS allows for data provenance, or full documentation, of all steps taken in processing and curating a dataset. MIRMAID adds two important capabilities to traditional CMS frameworks: (a) Graphically defined programmatic workflows that perform a series of steps to the selected content. These steps may include human actions (eg, tracing a tumor) or computational steps (eg, calculation of some metric on the basis of the images and tags). This allows for easy, clear creation and maintenance of study workflow and consistent data analyses. (b) Project management capabilities: This feature enables principal investigators to monitor the progress of a project, curate the data, manage people involved, grant or restrict access, and track work hours.
Requirements of Data Management in Research
Clinical trial management systems should comply with regulatory submission requirements and protect patient privacy. Government regulatory bodies, such as the FDA, have described the requirements for tracking all changes to and measurements of data that might be submitted for FDA approval in 21 CFR part 11 (19). Specifically, all changes to the data must be logged and traceable, ensuring FDA compliance.
Therefore, an audit log is required. In addition, privacy requirements help to protect the privacy rights of patients and research participants. Those requirements are specified in the Health Insurance Portability and Accountability Act (HIPAA) (21). The data objects produced during image acquisition contain real-world identifiers that connect the data objects to the person who was the trial participant. Such identifiers are known under HIPAA as protected health information (PHI). Because the HIPAA privacy regulations impose strict requirements on anyone who possesses PHI, the data objects must be modified to remove PHI and insert pseudonyms that preserve the relationships among the data objects.
A modern clinical trial management system must protect data, while enabling publication and sharing of experimental results, data quality control, project management capabilities, and the ability to mine data from multiple databases. Ideally, all of this should be provided within a single user experience, with advanced dataset search tools. In addition, an open-source implementation of such a CMS enables improvements by other research groups and portability of associated developed algorithms.
MIRMAID
Overview
Figure 1 depicts a flowchart of our CMS. TACTIC (20) was designed to track digital content through every stage of production and serves as the core of MIRMAID. TACTIC was created for the movie industry to manage a “digital assembly line”: the creation of digital content, such as visual effects and computer graphics. Recently, TACTIC was released as an open-source tool under the Eclipse Public License (20,22).
Figure 1.
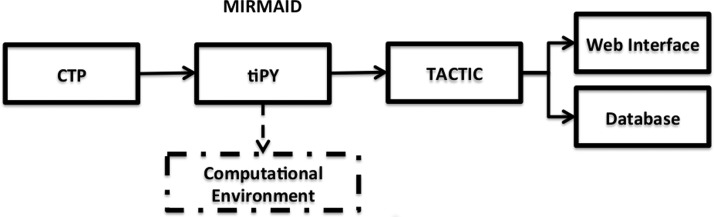
Flowchart of the MIRMAID system. CTP = Clinical Trial Processor.
The main features of TACTIC include production data management, scheduling, content management, communication, and reporting (Fig 2). The TACTIC system manages both project data and files, with project data stored in the database and files stored in the file system. TACTIC can store any type of image data format, including file formats commonly used in medical research, such as Analyze 7.5, NRRD (Nearly Raw Raster Data), NIfTI (Neuroimaging Informatics Technology Initiative), and DICOM (Digital Imaging and Communications in Medicine).
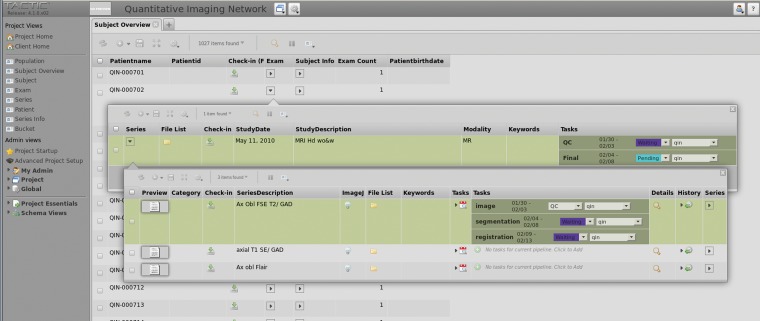
Figure 2.
Snapshot of the TACTIC Web interface. On the left side is the control panel, where arbitrary example tags (eg, “Population,” “Subject Overview”) are shown. The right panel presents the data available under “Subject Overview,” where subjects and relevant information are listed. Two pop-up dialogs demonstrate the exam and the series levels considered by our data handling schema.
TACTIC tracks the digital creation process, which in the case of research means the original acquired image and all of its intermediate processing steps until the final measured version. TACTIC allows tracking of data check-in and check-out by providing a mechanism to identify changes; it also employs a versioning system to record the history of the changes to specific content. Our adaptation of TACTIC for medical image research purposes was straightforward because medical images are digital content.
Architecture
TACTIC is built on existing Web technologies, and as a result is easy to maintain and scale. A Python Web server delivers static content to users. All processes and interactions are managed through the TACTIC transaction system. TACTIC can use any of six popular databases, which include both commercial and open-source options. MIRMAID stores all image data in one location to avoid the problem of keeping federated database systems (multiple databases managed and accessed as if they were one) synchronized. Furthermore, by using one central point, data are queried in only one place and with only one method. Images are stored in an external file system managed by the TACTIC application. MIRMAID can support teams of as many as 10 users on a system with a dual-core processor and 1 GB of RAM (random-access memory); it scales to as many as 150 users when two four-processor systems with 8 GB of RAM are used. We have not explored systems larger than this.
TACTIC’s architecture includes six main components (23).
1. Data model: The data model consists of searchable objects, called sObjects, used by TACTIC to manipulate and check in data.
2. Search model: The objects in each project are searchable types, called sTypes, which are tables in the database in which each column represents image data or tags stored in sObjects. By leveraging this mechanism though the Web interface of TACTIC, the user can perform complex queries of the sTypes. The search mechanism allows for both simple and complex queries that can be performed across multiple sTypes. All the information in an sObject can be used as a query term. Finally, the results of the queries can be extracted into spreadsheet form if needed.
3. Display model: The display model determines the user interface and interaction, which TACTIC calls widgets. Widgets enable customization of user interaction with TACTIC.
4. Command layer: This feature allows changes that affect the data model or the file system to be tracked.
5. TACTIC script editor: The script editor allows the user to write JavaScript or Python-based scripts to be run by the program.
6. TACTIC project: Each project consists of a project configuration file and database. The main components of a project are the project schema, which defines how the objects are related, and the project workflow, which defines the processes and workflows. The TACTIC server can handle creation and management of multiple projects. In addition, TACTIC projects can be easily shared between users and institutions.
Because TACTIC is open source, is written in Python, and has a well-defined and well-documented application programming interface, or API, TACTIC users can easily extend the functions to meet specific requirements not possible with the basic package.
TACTIC Interaction
Data input and output can be performed manually through the Web user interface or through Python interfaces. The Web-based interface allows for easy data sharing, whereas the Python interface allows alternative input and output. We have implemented a Python library, named tiPY, to allow easy input and output of data.
Image Viewer
Viewing images to assess image quality, verify basic findings, and perform simple measurement or annotation is essential in an imaging-focused research management system. To address this need, an image viewer was added by using TACTIC’s plug-in mechanism. The viewer allows users to view and manipulate images in the browser by selecting an appropriate option in the main MIRMAID interface. The viewer is based on HTML5 and JavaScript using the XTK library (https://github.com/xtk/X) (24).
Workflows
TACTIC offers a graphical mechanism to create workflows. These workflows can include computational tasks, such as image processing routines, data management commands, and assignment of interactive tasks (Fig 3). Workflows can be initiated by user actions or computer-based status changes. Thus, external or internal events can trigger workflows, and workflows can trigger other workflows within TACTIC and send commands to external systems.
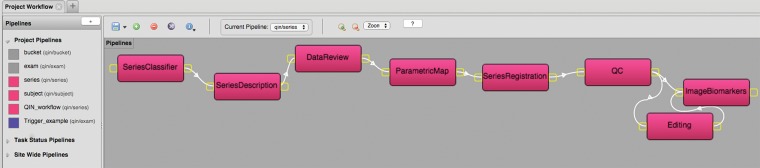
Figure 3.
Snapshot of the pipeline creation tool. The pipeline workflow is used to lay out the steps that a particular series needs to follow as it flows through its processes.
Project Management
To assist in project management, TACTIC offers a tracking system that allows logging of a user effort for a specific task. TACTIC provides the ability to grant different levels of access to data. This allows for secure management of the digital assets of a particular imaging research study. MIRMAID uses these capabilities and also provides several predefined reports that project managers can access to obtain real-time project status (Fig 4). Custom analytics can also be added through the Python interface.
Figure 4.
Snapshot of the TACTIC report Web interface, where common management tasks are provided.
Content Synchronization
Content synchronization is an important feature in multicenter clinical trials and settings with multiple collaborators. TACTIC offers a flexible mechanism to synchronize data among servers hosting the databases and users, ensuring that changes are always up to date and that the correct version of the content is used. Encryption and decryption through a public- and private-key mechanism are used for all data transfers.
Experimental Setup
In addition to TACTIC, MIRMAID incorporates dcm4che (http://www.dcm4che.org/) for DICOM connectivity and the Clinical Trial Processor (CTP) (http://mircwiki.rsna.org/index.php?title=CTP-The_RSNA_Clinical_Trial_Processor) for DICOM de-identification. The dcm4che module is an open-source Java library used as the DICOM receiver (25,26). The receiver can receive the images from a picture archiving and communication system or directly from the particular imaging modality. Subsequently, CTP is used to de-identify the data for compliance with HIPAA (19). The tags that should be removed from the DICOM object are configured through a lookup table. In addition, CTP provides a log of all actions, which meets the logging requirements in 21 CFR part 11 (18,19). During the de-identification process, a table with the correspondence between patient identifier and anonymized identifier is kept and securely maintained. This table is useful for adding information to the patient dataset, such as tags from the pathology reports and survival information. In addition, when data corresponding to follow-up studies of patients who have been anonymized are included, CTP will assign the same pseudonyms. Although CTP is capable of removing PHI, it can appear in many unexpected locations (eg, burned-in pixel values). For this reason, MIRMAID is typically configured to place imported images in a “quarantine” zone until the assigned user reviews the data.
In most cases, the final step of image importation is converting images from DICOM to NIfTI because most image processing packages do not deal well with native DICOM files. The tiPY library includes a routine to perform this conversion.
To ensure data security, MIRMAID regularly backs up all parameter files used by CTP, dcm4che, the virtual machine, or VM, running TACTIC, and the storage area.
Application Example
One application of the system is for investigating image-based biomarkers in glioblastoma multiforme to differentiate between progression and pseudoprogression.
When a subject has been identified, it is forwarded to the MIRMAID DICOM receiver. Next, the dataset PHI is de-identified through use of the CTP functionality and a preconfigured CTP configuration file. All the received files are placed in a folder, where they are “ingested” using the tiPY library. A configuration file exists in the receiving pool to assign the proper tags to the data to be ingested, such as institutional review board number, data type, and project name.
The ingesting process will create a new entry inside TACTIC or will update the information if the data already exist. Once the data have been created, Subject, Exam, and Series workflows are triggered. The first step of the workflow is a classifier step, which routes the data for a specific study to the right pipeline. Subsequently, DICOM field tags are extracted and a normalized series description is assigned to each object (eg, “axial,” “T1,” and “postcontrast” might all be assigned to an axial postcontrast T1 image). When this step is finished, a task is assigned to a specific user to review the data and the image quality. Once the user approves the images, the pipeline continues with parametric map calculation for diffusion and perfusion images. Next, all images for the examination are registered to the T1 precontrast image, followed by automated tumor segmentation. At this point, a user performs a quality control assessment. The user will approve the results or route them through a correction pipeline to correct the registered images. When images are approved, image-based biomarkers are calculated and stored in TACTIC.
The final step of the pipeline consists of a query to the clinical data system to add the following tags to each examination: survival status, medication, and pathology report tags. When the examination-level analysis is completed, the subject pipeline starts. This includes analysis of the biomarkers, which uses the survival information included in the database.
Evaluation
In this article, we have introduced MIRMAID, a Web-accessible medical image CMS. From its inception, MIRMAID has been developed to be highly flexible to meet the needs of diverse studies. It is being used for neurology- and nephrology-based imaging research studies at our institution. We anticipate that application of this system to other research areas will be straightforward.
Content management is critical, particularly for team science. MIRMAID accelerates the sharing of data, metadata, and analysis methods and reduces duplicate work. This simplifies the process of algorithm testing and evaluation. MIRMAID offers browsing functions to make it easy to select an interesting subset of, for example, patients, examinations, series, image annotations, image calculations, genes, and medications. The browser will allow selection of not only data but also metadata (eg, tumor annotations) and algorithms. The CMS we are using allows tremendous flexibility in tagging, in turn permitting much richer metadata browsing and cohort selection. In fact, although the system was built to focus on imaging, the design is general and does not need to be image-focused. We have already integrated clinical and genomic information into the system, and analyses focusing on those two types of data are perfectly feasible.
Content management is a critical element of modern science, particularly when team science demands coordination of multiple people and multiple institutions.MIRMAID offers a flexible mechanism for data curation. The data-handling schema can be updated easily to include data from heterogeneous sources, such as imaging and genomic data. The workflow mechanism can be used to automate processes or notify users of pending tasks. By using the tags associated with each dataset, our system routes each process under the correct processing pipeline. Workflows can include not only steps that involve internal methods but also steps to query or store information in external systems. Although the flexibility of a CMS is extremely useful, it can also present a challenge to effective use of the system. In particular, the flexibility of tags is helpful, but one must also manage the tags to ensure that two users do not use the same tag for different purposes. For this reason, all tags used inside MIRMAID must be approved by a curation committee, which also defines allowed values and type. Furthermore, through the pipeline mechanism and tiPY, it is easy to set up control mechanisms that can check for typographical errors or wrong data types in the case of manual data entries.
MIRMAID offers a complete project management suite that can track progress (eg, time spent on specific tasks) and workflow bottlenecks. With the correct setup, a principal investigator can monitor financial status and timelines. The example we presented is characteristic of how the project management tools are used. Because the pipeline is heterogeneous (ie, there are both automated tasks and interactive tasks), the principal investigator can use the project management features to monitor the progress of the study and the timeliness of users in completing interactive tasks.
Security is a critical requirement of a modern research management system, and TACTIC provides multiple security access levels for projects, including none, low, medium, and high. The TACTIC administrator can manage the level of access through its security tool. Lightweight Directory Access Protocol, or LDAP, is also supported for user authentication. Additional advantages of MIRMAID include shareable templates, the flexible schema design, and extensive task management and project management features.
Web-accessible data management tools, such as MilxXplore (6), LORIS (12), TCIA (13), and HID (9), are aimed at storing large diverse image datasets. XNAT (5) and COINS (11) are examples of well-established systems tested on large datasets under many different configurations. Although their database schemas are constructed to support the storage of new data types, project management features are not provided, and the overall systems were each developed for neurologic studies in particular. As a result, their adaptation to other imaging study pipelines is difficult.
MIRMAID and XNAT (5) have many similarities but also some important differences. MIRMAID and XNAT both have the ability to make data available in a sustained and secure manner. They also offer ways of searching, querying, and updating data. In XNAT, data types and objects are maintained internally in a verbose and fairly complex XML (Extensible Markup Language)-based structure. MIRMAID uses database technologies to host all types and objects, so that adding custom data types is fairly easy (28). However, once an archive is set up with XNAT, there is much less flexibility in updating the general schema. Versioning of files is another advantage of MIRMAID. Versioning can help researchers test different algorithms and keep results under different versions of the output images. The workflow system of TACTIC allows users to define workflows graphically or as Python scripts. In XNAT, a Python module, PyXNAT (29), that allows scripts to be executed has recently been described. However, no graphical workflow system is available.
QI-Bench (15) also has similarities to MIRMAID. It is a platform where researchers can use curated data to develop and test imaging biomarkers and analytic methods. However, it lacks many of MIRMAID’s features, including graphical workflows, project management, and versioning.
Several components of MIRMAID are available as open-access tools through GitHub, a Web-based software repository (https://github.com/Southpaw-TACTIC/TACTIC, https://github.com/dblezek/tactical-vagrant). Documentation and application examples are a work in progress and will also be available through GitHub.
Conclusion
MIRMAID, a Web-accessible medical image asset manager, offers a solution for management of medical imaging–based research studies. MIRMAID consists of three main components: (a) TACTIC, an open-source digital content and asset management system; (b) tiPY, a Python library allowing for easy system interaction, such as input and output of data and analysis; and (c) an HTML- and JavaScript-based widget set that facilitates image analysis tasks, such as manual generation of regions of interest. MIRMAID can manage imaging-based studies, with sophisticated security, graphical and flexible workflows, and powerful project management. This provides a complete system for medical imaging research management not otherwise available.
Funding: P.D.K., D.J.B., and W.J.R. were supported by the National Cancer Institute (NCI), National Institutes of Health (NIH) [grant number CA-160045]. T.L.K. was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), NIH [grant number P30 DK090728]. B.J.E. was supported by the NCI, NIH [grant number CA-16045].
All authors have disclosed no relevant relationships.
Abbreviations:
- CMS
- content management system
- DICOM
- Digital Imaging and Communications in Medicine
- FDA
- Food and Drug Administration
- HIPAA
- Health Insurance Portability and Accountability Act
- MIRMAID
- Medical Imaging Research Management and Associated Information Database
- PHI
- protected health information
References
- 1.Doran SJ, d’Arcy J, Collins DJ, et al. Informatics in radiology: development of a research PACS for analysis of functional imaging data in clinical research and clinical trials. RadioGraphics 2012;32(7):2135–2150. [DOI] [PubMed] [Google Scholar]
- 2.Erickson BJ, Meenan C, Langer S. Standards for business analytics and departmental workflow. J Digit Imaging 2013;26(1):53–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langer SG. Challenges for data storage in medical imaging research. J Digit Imaging 2011;24(2):203–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poline JB, Breeze JL, Ghosh S, et al. Data sharing in neuroimaging research. Front Neuroinform 2012;6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics 2007;5(1):11–34. [DOI] [PubMed] [Google Scholar]
- 6.Bourgeat P, Dore V, Villemagne VL, Rowe CC, Salvado O, Fripp J. MilxXplore: a web-based system to explore large imaging datasets. J Am Med Inform Assoc 2013;20(6):1046–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woodbridge M, Fagiolo G, O’Regan DP. MRIdb: medical image management for biobank research. J Digit Imaging 2013;26(5):886–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Book GA, Anderson BM, Stevens MC, Glahn DC, Assaf M, Pearlson GD. Neuroinformatics Database (NiDB): a modular, portable database for the storage, analysis, and sharing of neuroimaging data. Neuroinformatics 2013;11(4):495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozyurt IB, Keator DB, Wei D, et al. Federated web-accessible clinical data management within an extensible neuroimaging database. Neuroinformatics 2010;8(4):231–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adamson CL, Wood AG. DFBIdb: a software package for neuroimaging data management. Neuroinformatics 2010;8(4):273–284. [DOI] [PubMed] [Google Scholar]
- 11.Scott A, Courtney W, Wood D, et al. COINS: an innovative informatics and neuroimaging tool suite built for large heterogeneous datasets. Front Neuroinform 2011;5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das S, Zijdenbos AP, Harlap J, Vins D, Evans AC. LORIS: a web-based data management system for multi-center studies. Front Neuroinform 2011;5:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 2013;26(6):1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MIDAS at Kitware Inc . MIDAS Web site. http://www.insight-journal.org/midas. Accessed July 8, 2015.
- 15.QI-Bench Web site . http://www.qi-bench.org. Accessed July 8, 2015.
- 16.Quantitative Imaging Data Warehouse (QIDW) Radiological Society of North America . https://www.rsna.org/QIDW. Accessed July 8, 2015.
- 17.TRIAD: Transfer of Images and Data . American College of Radiology. https://triad.acr.org. Accessed July 8, 2015.
- 18.Rhodes C, Moore S, Clark K, et al. Regulatory compliance requirements for an open source electronic image trial management system. Conf Proc IEEE Eng Med Biol Soc 2010;2010:3475–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Electronic records; electronic signatures, 62 Federal Register 13464 (1997) (codified at 21 CFR part 11).
- 20.TACTIC Digital Asset and Workflow Software . Southpaw Technology Web site. http://www.southpawtech.com/tactic. Accessed July 8, 2015.
- 21.Summary of the HIPAA Privacy Rule . U.S. Department of Health and Human Services Web site. http://www.hhs.gov/ocr/privacy/hipaa/understanding/summary/privacysummary.pdf. Accessed July 8, 2015.
- 22.Eclipse Public License, version 1.0 . Eclipse Foundation. http://www.eclipse.org/legal/epl-v10.html. Accessed July 8, 2015.
- 23.TACTIC Anatomy Lesson . Southpaw Technology. TACTIC Web site. http://community.southpawtech.com/tactic-setup_setup_tactic-anatomy-lesson.html. Accessed July 23, 2015.
- 24.The X Toolkit: WebGLTM for Scientific Visualization . Developers XTK. README.md https://github.com/xtk/X. Accessed July 8, 2015.
- 25.Warnock MJ, Toland C, Evans D, Wallace B, Nagy P. Benefits of using the DCM4CHE DICOM archive. J Digit Imaging 2007;20(suppl 1):125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langer S, Charboneau N, French T. DCMTB: a virtual appliance DICOM toolbox. J Digit Imaging 2010;23(6):681–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erickson BJ, Fajnwaks P, Langer SG, Perry J. Multisite image data collection and management using the RSNA image sharing network. Transl Oncol 2014;7(1):36–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herrick R, McKay M, Olsen T, et al. Data dictionary services in XNAT and the Human Connectome Project. Front Neuroinform 2014;8:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwartz Y, Barbot A, Thyreau B, et al. PyXNAT: XNAT in Python. Front Neuroinform 2012;6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]