Abstract
Several studies suggest a link between circadian rhythm disturbances and tumorigenesis. However, the association between circadian clock genes and prognosis in breast cancer has not been systematically studied. Therefore, we examined the expression of 17 clock components in tumors from 766 node-negative breast cancer patients that were untreated in both neoadjuvant and adjuvant settings. In addition, their association with metastasis-free survival (MFS) and correlation to clinicopathological parameters were investigated. Aiming to estimate functionality of the clockwork, we studied clock gene expression relationships by correlation analysis. Higher expression of several clock genes (e.g., CLOCK, PER1, PER2, PER3, CRY2, NPAS2 and RORC) was found to be associated with longer MFS in univariate Cox regression analyses (HR<1 and FDR-adjusted P < 0.05). Stratification according to molecular subtype revealed prognostic relevance for PER1, PER3, CRY2 and NFIL3 in the ER+/HER2- subgroup, CLOCK and NPAS2 in the ER-/HER2- subtype, and ARNTL2 in HER2+ breast cancer. In the multivariate Cox model, only PER3 (HR = 0.66; P = 0.016) and RORC (HR = 0.42; P = 0.003) were found to be associated with survival outcome independent of established clinicopathological parameters. Pairwise correlations between functionally-related clock genes (e.g., PER2-PER3 and CRY2-PER3) were stronger in ER+, HER2- and low-grade carcinomas; whereas, weaker correlation coefficients were observed in ER- and HER2+ tumors, high-grade tumors and tumors that progressed to metastatic disease. In conclusion, loss of clock genes is associated with worse prognosis in breast cancer. Coordinated co-expression of clock genes, indicative of a functional circadian clock, is maintained in ER+, HER2-, low grade and non-metastasizing tumors but is compromised in more aggressive carcinomas.
Keywords: breast cancer, circadian clock, clock genes, estrogen receptor, metastasis-free survival, tumor progression
Abbreviations
- ARNTL/2
aryl hydrocarbon receptor nuclear translocator-like/2
- BHLHE40/41
basic helix-loop-helix family, member e
- CLOCK
circadian locomotor output cycles kaput
- CRY1/2
cryptochrome circadian clock 1/2
- DBP
D site of albumin promoter (albumin D-box) binding protein
- DFS
disease-free survival
- ER
estrogen receptor
- HER2
human epidermal growth factor receptor 2
- HR
hazard ratio
- MFS
metastasis-free survival
- NFIL3
nuclear factor, interleukin 3 regulated
- NPAS2
neuronal PAS domain protein 2
- NR1D2
nuclear receptor subfamily 1, group D, member 2
- PER1/2/3
period circadian clock 1/2/3
- RORA/B/C
RAR-related orphan receptor alpha/beta/gamma
- SCN
suprachiasmatic nucleus
Introduction
The circadian clock system coordinates physiological and behavioral processes in mammals throughout the day. It is composed of multiple cellular oscillators distributed throughout the body that are hierarchically organized, the suprachiasmatic nucleus (SCN) being the master pacemaker for subordinated clocks in other brain regions and peripheral tissues.1-3 Three elements are necessary for the proper functioning of biological clocks: a core clockwork capable of autonomous oscillation, thus synchronizing internal and external time, an input pathway that conveys time information from the environment to the clock, and an output pathway that mediates coordination and synchronization of clock-controlled physiological processes.1 At the molecular level, circadian rhythms are generated by a system of interlocking autoregulatory transcriptional/translational feedback loops. The major components of this cellular clock network are the transcriptional activators CLOCK (and its paralog, NPAS2) and BMAL1 (ARNTL), which upon heterodimerization upregulate the expression of the Period (PER1, PER2 and PER3) and Cryptochrome (CRY1, CRY2) genes. The corresponding protein products, in turn, form the negative limb of the core loop. Toward the end of the day PER and CRY proteins translocate to the nucleus to inhibit CLOCK-BMAL1, thus repressing their own transcription. Additional feedback loops have been identified that involve other genes such as NR1D1 (Rev-erbα), RORA, DBP, E4BP4 (NFIL3), and the bHLH members DEC1 and DEC2 (Bhlhb2/BHLHE40, Bhlhb3/BHLHE41).4
The importance and individual contribution of the aforementioned genes in circadian rhythms has been demonstrated by generating mutations in mice. Loss of behavioral and molecular rhythmicity, altered period length, and disrupted entrainment of locomotor activity are examples of phenotypes derived from circadian gene deficiency.4,5
Disruption of circadian rhythms has been linked to mammalian tumorigenesis in numerous reports.6,7 Early studies in the 1960s showed that disturbances of the circadian system in animals, through ablation of the pineal gland or constant light exposure, resulted in increased formation of mammary tumors. In humans, epidemiological studies have provided important evidence that working at night is a risk factor for breast cancer development, although recent systematic reviews are cautious.8 Interestingly, disruption of circadian rhythm does not only seem to increase the risk of tumor development, but also enhances progression of already existing tumors. For instance, clinical studies found that circadian disturbances in cancer patients (e.g., altered 24 h rest/activity patterns or cortisol rhythms) were significantly associated with shorter survival.9 Supporting these findings, destruction of the SCN in mice resulted in accelerated growth of inoculated tumors.10
Evidence that individual genes of the circadian clock play a role in the control of tumorigenesis has been provided as well. Per2 loss-of-function mutations in mice led to higher tumor incidence upon γ-irradiation compared to wildtype mice.11 In addition, the proliferation-associated gene c-myc, as well the c-myc-regulated genes, CyclinD1 and Gadd45 were deregulated in Per2 mutants. This mechanistic link between the circadian clock and proliferation strongly suggests that circadian clock genes suppress cancer development in vivo by controlling expression of genes involved in growth control and cell division.12
The possibility that perturbations in the cellular clockwork might play a role in the initiation of malignant cell transformation has prompted many investigators to examine the expression of circadian genes in tumor tissues. In accordance with a tumor suppressor role of the circadian clock, tumors were found to display decreased levels of several clock genes compared to the adjacent normal tissue.13 Circadian gene abnormalities such as mutations, deregulated expression and even translocation of Period genes have been reported in various cancers including breast, colorectal, endometrial, lung and different types of lymphoma and leukemia.7 In addition, single nucleotide polymorphisms in key circadian genes such as NPAS2 and CRY2 were found to be associated with cancer risk and patient survival.7,14 Together, these findings clearly emphasize the importance of the circadian system in the maintenance of physiological homeostasis. Its perturbation may provoke deregulated proliferation and thus, carcinogenesis.
Despite the considerable number of recent studies reporting abnormalities of circadian genes in tumors, it has not yet been comprehensively investigated how the deregulated expression of clock genes is associated with survival outcome in human breast cancer. Furthermore, an analysis of coordinated clock gene expression as an approach to evaluate functionality of the molecular clockwork in breast tumors has not been performed to date. In the present study, we examined expression of 17 canonical clock genes in a collection of 766 breast cancer patients who were node-negative at the time of diagnosis and did not receive systemic chemotherapy, enabling a unique opportunity to assess the impact of clock gene expression on the natural course of the disease. We demonstrate a strong association between decreased expression of core clock genes and shorter metastasis-free survival. In addition, we report a loss of co-expression of clock genes in more aggressive carcinomas, represented by high histological grade, estrogen receptor negativity, and HER2 positivity.
Results
Circadian clock genes are associated with prognosis in breast cancer
To study a possible association of circadian clock genes with metastasis-free survival (MFS) in breast cancer we analyzed a cohort of 766 node-negative patients consisting of 3 different subcohorts: the Mainz cohort (N = 200), the Rotterdam cohort (N = 280) and the Transbig cohort (N = 286). We considered 17 established regulators of the cellular clock machinery that were previously classified into 3 groups15: E-box regulators (CLOCK, ARNTL (BMAL1), PER1, PER2, PER3, CRY1, CRY2, BHLHE40 (BHLHE2, DEC1), BHLHE41 (BHLHE3, DEC2), NPAS2 and ARNTL2); D-box regulators (DBP and NFIL3); and RORE-box regulators (RORA, RORB, RORC and NR1D2. In addition, TIMELESS - which intersects both with the circadian clock and the cell cycle machinery16 - was also included into the analysis. In the univariate Cox analysis, we observed that high mRNA expression of numerous circadian clock genes was associated with longer MFS (Table 1; Tables S1-3). Most significant values were observed for CLOCK, PER1, PER2, PER3, CRY2 and RORC (all showing hazard ratio HR < 1 and FDR adjusted p-value P < 0.05) (Table 1). In contrast, TIMELESS was associated with shorter MFS (HR = 1.405; P > 0.001) (Table 1). Kaplan-Meier analysis was used to visualize the association between MFS and circadian clock gene expression (Fig. 1; Fig. S1). The time interval until occurrence of metastasis was longer for patients with expression of circadian clock genes that is higher than the median compared to patients with lower expression levels. In contrast, patients with high TIMELESS expression had shorter MFS compared to patients with low TIMELESS (Fig. 1).
Table 1.
Association of RNA expression of circadian genes with metastasis free survival. RNA expression of CLOCK, PER1, PER2, PER3, CRY2, NPAS2 and RORC is significantly associated with better metastasis free survival (MFS) in patients with node-negative breast cancer (N = 766) in the univariate Cox analysis (HR < 1, P < 0.05). In contrast, TIMELESS is associated with worse MFS (HR > 1, P < 0.05, bold face). HR: hazard ratio, FDR: False discovery rate, 95% CI: (95% confidence interval)
| A) E-box regulators: | ||||||
|---|---|---|---|---|---|---|
| Prognostic factor (RNA expression) | Affy probe set | HR (MFS) | P value | FDR | 95%-CI lower upper | |
|
CLOCK |
217563_at |
0.835 |
0.008 |
0.032 |
0.731 |
0.954 |
|
CLOCK |
204980_at |
1.035 |
0.615 |
0.717 |
0.906 |
1.182 |
|
ARNTL |
210971_s_at |
0.908 |
0.159 |
0.262 |
0.793 |
1.039 |
|
ARNTL |
209824_s_at |
0.951 |
0.452 |
0.603 |
0.835 |
1.084 |
|
PER1 |
36829_at |
0.809 |
0.003 |
0.016 |
0.704 |
0.930 |
|
PER1 |
202861_at |
0.810 |
0.004 |
0.016 |
0.703 |
0.933 |
|
PER2 |
208518_s_at |
0.841 |
0.012 |
0.041 |
0.735 |
0.962 |
|
PER2 |
205251_at |
0.874 |
0.040 |
0.112 |
0.769 |
0.994 |
|
PER3 |
221045_s_at |
0.806 |
0.002 |
0.012 |
0.705 |
0.922 |
|
CRY1 |
209674_at |
1.026 |
0.698 |
0.724 |
0.901 |
1.169 |
|
CRY2 |
212695_at |
0.781 |
<0.001 |
0.003 |
0.685 |
0.889 |
|
NPAS2 |
39549_at |
0.952 |
0.486 |
0.619 |
0.830 |
1.093 |
|
NPAS2 |
205460_at |
0.942 |
0.380 |
0.531 |
0.824 |
1.077 |
|
NPAS2 |
205459_s_at |
0.852 |
0.017 |
0.053 |
0.747 |
0.972 |
|
ARNTL2 |
220658_s_at |
0.893 |
0.126 |
0.235 |
0.773 |
1.032 |
|
BHLHE40 |
201169_s_at |
0.891 |
0.092 |
0.197 |
0.779 |
1.019 |
|
BHLHE40 |
201170_s_at |
0.883 |
0.054 |
0.126 |
0.778 |
1.002 |
|
BHLHE41 |
221530_s_at |
0.918 |
0.190 |
0.296 |
0.807 |
1.044 |
|
B) D-box regulators: | ||||||
|
DBP |
209783_at |
1.102 |
0.140 |
0.244 |
0.969 |
1.254 |
|
DBP |
209782_s_at |
0.898 |
0.119 |
0.235 |
0.785 |
1.028 |
|
NFIL3 |
203574_at |
0.972 |
0.680 |
0.724 |
0.851 |
1.111 |
|
C) RORE-box regulators: | ||||||
|
RORA |
210426_x_at |
1.038 |
0.583 |
0.710 |
0.908 |
1.187 |
|
RORA |
210479_s_at |
1.027 |
0.697 |
0.724 |
0.898 |
1.174 |
|
RORB |
206443_at |
1.082 |
0.225 |
0.332 |
0.953 |
1.228 |
|
RORC |
206419_at |
0.800 |
<0.001 |
0.007 |
0.702 |
0.910 |
|
NR1D2 |
209750_at |
0.988 |
0.855 |
0.855 |
0.865 |
1.128 |
|
D) TIMELESS: | ||||||
|
TIMELESS |
215455_at |
1.137 |
0.046 |
0.117 |
1.002 |
1.290 |
| TIMELESS | 203046_s_at | 1.406 | <0.001 | <0.001 | 1.243 | 1.590 |
Figure 1.
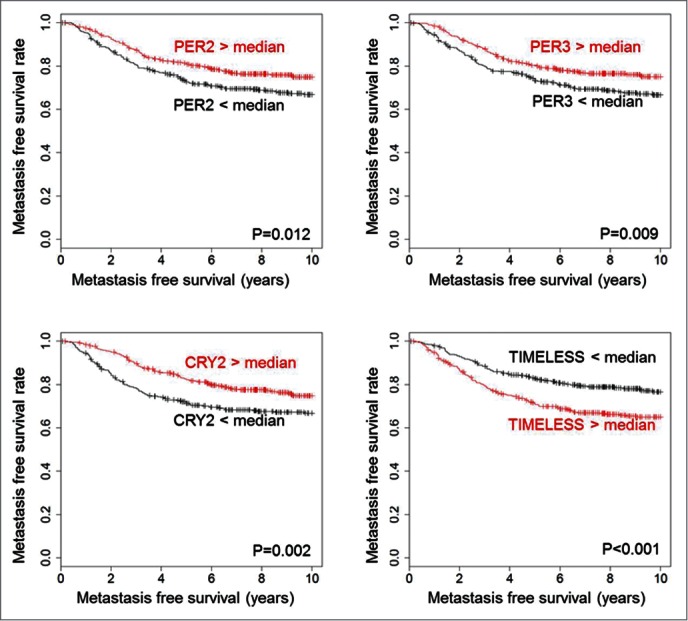
Association of PER2, PER3, CRY2 and TIMELESS with metastasis-free survival time. High expression of PER2 (205251_at), PER3 (221045_s_at) and CRY2 (212695_at) is associated with longer metastasis free survival time, whereas high TIMELESS (203046_s_at) expression is associated with shorter metastasis free survival time. The analysis included 766 patients with node-negative breast cancer who have not been treated by chemotherapy. All genes were dichotomized at the median. The log-rank test was used to assess statistical significance of the Kaplan-Meier plots. Kaplan Meier analysis of further circadian genes is shown in Fig. S1.
In the multivariate Cox analysis only PER3 (P = 0.016; HR = 0.658), RORC (P = 0.003; HR = 0.421), as well as TIMELESS (P = 0.012; HR = 1.607) were associated with MFS, independent of clinical parameters (Table S4).
Data on disease-free survival (DFS) comprising time to loco-regional or metastatic recurrence or secondary carcinoma of the contralateral breast was available for the Mainz cohort only. High expression of PER2, PER3, CRY2, BHLHE40 and RORC was significantly associated with longer DFS in the univariate Cox analysis (HR < 1; FDR adjusted P < 0.05), while high expression of TIMELESS was associated with shorter DFS (HR > 1; FDR adjusted P < 0.05) (Table S5). In conclusion, multiple circadian core clock components – remarkably, those involved in E-box regulation – are associated with better prognosis.
Association of circadian clock genes with prognosis in breast cancer molecular subtypes
Breast tumors are classified into different molecular subtypes, ER+/HER2-, ER-/HER2- and HER2+17, based on estrogen receptor (ER) and HER2 positivity. This classification influences the treatment strategy and serves as an indicator of tumor aggressiveness, with the ER+/HER2- subgroup representing the least aggressive subtype (Fig. S2). In order to elucidate if circadian clock gene expression is of particular relevance in any of these molecular subgroups, we performed individual univariate Cox analyses for ER+/HER2-, ER-/HER2- and HER2+ patients. PER1, PER3, CRY2 and NFIL3 showed a significant association with better prognosis (all HR < 1; P < 0.05), and TIMELESS with worse prognosis (HR = 1.626; P < 0.001) in the ER+/HER2- subtype (Table S6; Fig. S3A and 3B); whereas, NPAS2 and CLOCK were significantly associated with longer MFS in the ER-/HER2- subtype (HR < 1; P < 0.05) (Table S7; Fig. S3C and D), and ARNTL2 in the HER2+ subtype (HR = 0.385 and P < 0.001) (Table S8; Fig. S3E). Altogether, the subtype-specific analysis indicated that the prognostic relevance of circadian gene expression differs between breast cancer subtypes. However, it is important to note that the number of patients differs between subtypes, which can influence subtype-specific significance statements. Of particular interest was the strong association between ARNTL2 expression and better prognosis in HER2-positive tumors, suggestive of a so far not reported protective role for ARNTL2 in this particular subtype.
Circadian clock genes and their association with clinical parameters
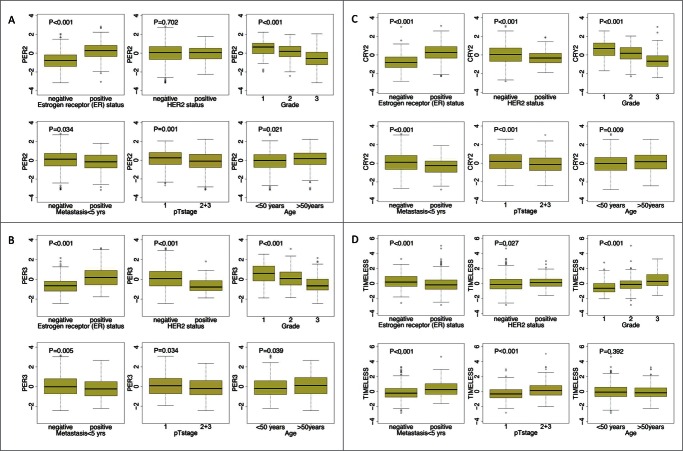
We next investigated the relationship between prognosis-associated circadian clock genes and conventional clinicopathological parameters, including age at diagnosis, pathological tumor stage, histological differentiation grade and ER, as well as human HER2 status. Higher expression of circadian clock genes that are involved in the negative limb of the central clock, such as PER1, PER2, PER3, CRY2, was in general associated with positive ER-status, negative HER2-status, lower grade, lower pT stage, lower metastatic occurrence and age (all P < 0.05) (Fig. 2; Fig. S4); the opposite trend was observed for TIMELESS (Fig. 2). CLOCK and NPAS2 showed significant association with lack of metastasis, and NPAS2 with negative ER-status (Fig. S4).
Figure 2.
Association of RNA levels of PER2, PER3, CRY2 and TIMELESS with clinical parameters. Significant associations (P < 0.05) of PER2 (205251_at), PER3 (221045_s_at) and CRY2 (212695_at) with positive estrogen receptor (ER) status, lower grade, lack of metastasis, lower pT stage (1) and higher age were obtained. In addition, PER3 and CRY2 also associate with negative HER2-status. TIMELESS (203046_s_at), in contrast, showed significant association with negative ER-status, positive HER2-status higher grade, higher pT stage (2+3) and early metastasis. Differences for age, ER status, HER2 status, pTstage and metastasis were tested by the Mann-Whitney test, differences between grading by the Kruskal-Wallis test. The analysis included 766 patients with node negative breast carcinoma. Further analyses are shown in Fig. S4.
Circadian clock genes in relation to known biological motifs in breast cancer
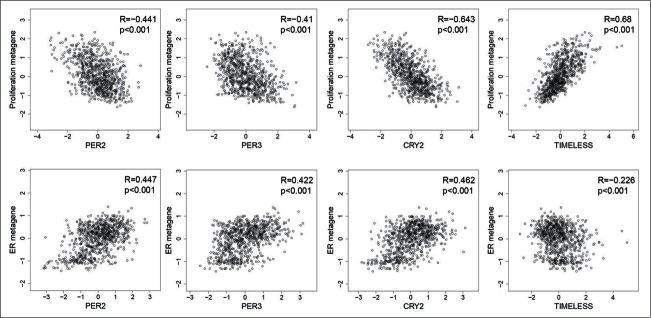
Expression signatures, or metagenes, representative of key biological processes, have been shown to be important prognostic tools,18-20 including (i) the proliferation metagene, consisting of genes involved in cell division (e.g., AURKA, TOP2A), (ii) the B- and T-cell metagenes as markers of immune cell infiltration, and (iii) the estrogen receptor metagene representing estrogen signaling-dependent genes. Additional metagenes, such as a basal-like, a normal-like, or a stromal signature, have also been described.18 We determined the relationship of circadian clock genes to these metagenes. Most clock genes (PER1, PER2, PER3, CRY2, RORC, BHLHE40, BHLHE41) showed a negative correlation (R < −0.3; P < 0.001) with the proliferation metagene (Fig. 3; Table S9), with the strongest observed for CRY2 expression (R = −0.612; P < 0.001), followed by PER2 (R = −0.440; P < 0.001) and PER3 (R = −0.409; P < 0.001) (Fig. 3). In contrast, TIMELESS mRNA showed a strong positive correlation with the proliferation signature (R = 0.674, P < 0.001) (Fig. 3). In addition, several circadian clock genes showed a strong positive correlation with the ER metagene (R > 0.4 P < 0.001 for CRY2, PER2, PER3, BHLHE40), the highest for PER2 (R = 0.487 P < 0.001) (Fig. 3) and BHLHE40 (R = 0.616 P < 0.001) (Table S9). Only weak negative correlations were found with the B- and T-cell metagenes. All correlations found in the combined cohort were also significant in each of the single cohorts (Table S7).
Figure 3.
Correlation of PER2, PER3, CRY2 and TIMELESS with the proliferation and the estrogen receptor (ER) metagenes. PER2 (205251_at), PER3 (221045_s_at) and CRY2 (212695_at) show a negative correlation with the proliferation metagene and a positive correlation with the ER metagene. Conversely, TIMELESS (203046_s_at) shows a positive correlation with the proliferation metagene but negative correlation with the ER metagene. Scatterplots along with the corresponding Spearman´s rank correlation coefficients are shown. The analysis included carcinomas of 766 patients with node-negative breast cancer.
Correlations between the expression of circadian clock genes decrease in more aggressive tumor types
PER1, PER2 and PER3 proteins have been described to form heterodimers among themselves,21 as well as with CRY1 and CRY2,22 and the formation of different dimer combinations is dependent on the availability of the individual components throughout the 24-hour period.23,24 Importantly, dimer formation plays a crucial role in the negative regulatory feedback loop of the circadian clockwork. The CRY and PER genes display highly coordinated temporal expression patterns22,25-29 and loss of proper phase-relationships might affect the efficiency of the negative feedback loop. In agreement with this notion, the stoichiometric relationship among clock proteins has been shown to determine robustness of the circadian rhythm.30,31 Hypothesizing that a strong positive correlation coefficient reflects coordinated co-expression (Fig. S5) and thus proper regulation of the circadian clockwork, we explored pairwise correlations of clock gene expression in tumors stratified according to ER status, HER2 status, histological grade and metastatic recurrence.
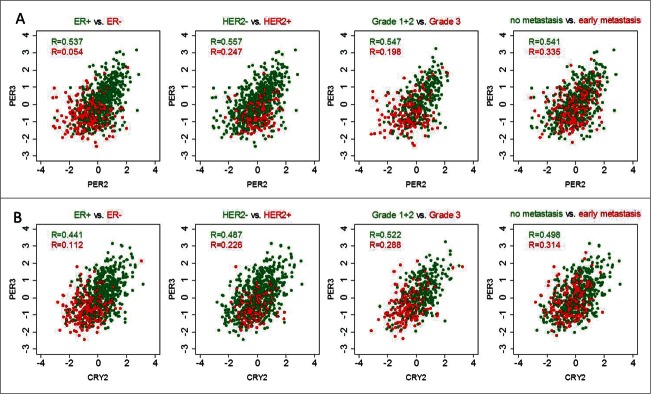
Correlation coefficients between PER2 and PER3, and CRY2 and PER3 were shown to be the highest in the correlation analysis performed in the combined cohort (Fig. S6). Furthermore, after stratification it was shown that the correlation coefficients were higher for ER-positive tumors than for ER-negative tumors (Fig. 4). For example, a relatively high correlation coefficient for PER2-PER3 (R = 0.537) was obtained in ER+ tumors; whereas, in ER- tumors this correlation (R = 0.054) was close to zero (Fig. 4A). A similar scenario was observed for HER2-negative versus HER2-positive as well as grade 1 and 2 vs. grade 3 carcinomas (Fig. 4A). Also, non-metastasizing tumors displayed higher correlation coefficients than metastasizing carcinomas (Fig. 4A). These results obtained in the combined cohort were comparable to those in the Mainz, Transbig and Rotterdam cohort analyzed separately (Fig. S5). Correlation coefficients for CRY2-PER3 showed a similar pattern (Fig. 4B).
Figure 4.
Correlations of circadian clock genes in tumors stratified according to ER status, HER2 status, histological grade and metastasis occurrence. Correlation coefficients between PER2 (205251_at) and PER3 (221045_s_at) and between CRY2 (212695_at) and PER3 (221045_s_at) are higher in estrogen receptor (ER)-positive and HER2-status negative tumors, and also in tumors with low grade and with no metastatic occurrence (green color). Conversely, correlations are lower in ER negative, HER2 status positive, high grade and metastatic tumors (red color). Correlations were analyzed using the Spearman correlation test. The subgroup analysis included the combined cohort of 766 patients with node-negative breast cancer except in grade, since grade information is only available for the Mainz and the Transbig cohorts (combined N = 480). Correlation analysis in the single cohorts is shown in Fig. S7.
Furthermore, we studied correlations in tumors stratified according to molecular subtypes. Positive correlation coefficients above 0.4 were observed for PER2-PER3 (R = 0.578) and PER3-CRY2 (R = 0.434) in ER+/HER2- tumors, the molecular subtype in which these genes were also associated with better outcome. In ER-/HER2- and HER2+ tumors, these correlations were considerably lower (not shown). Taken together, the results show that correlations between the expression of canonical circadian clock genes decrease in more aggressive tumor types.
To further explore this hypothesis we also investigated correlations between PER/CRY genes and ARNTL, representing the negative and positive branches of the E-box regulation circuit, respectively. The expression pattern of ARNTL is known to be antiphasic to that of period genes,32 which we therefore expected to negatively correlate with in a functional clockwork. Our analyses showed a negative correlation - weak but significant- between ARNTL and PER genes (e.g., R = −0.2, P < 0.001 for ARNTL-PER1) in low grade carcinomas, which was not seen in high grade tumors (Table S10).
This finding leads to the question of whether loss of synchronized expression of circadian genes is associated with worse prognosis. To address this statistically, we performed the following analysis: for each patient the residual value – which represents the absolute value of the distance to the regression line in the regression plot - was calculated. Since high residual values are indicative of a higher deviation from a perfect correlation, we hypothesized that the more deviated patients would also display a worse outcome. Indeed, a trend was observed that higher residual values in the regression plot of PER2 and PER3 were associated with worse prognosis (HR = 1.72; P = 0.155). Adjustment of the analysis to either PER2 or PER3 or to clinical factors did result in a significant hazard ratio (Fig. S8).
Discussion
A tumor suppressor role of the circadian clock has previously been reported in multiple studies.7,33 These findings suggest an important role for the circadian clock in the maintenance of physiological homeostasis and potentially explain how its failure may lead to disease. In this study, we aimed to investigate the importance of circadian clock function in relation to metastasis-free survival in breast cancer. We demonstrate that loss of expression of circadian clock genes is associated with worse prognosis in a large collection of node-negative breast cancer patients. Focusing on well-known key components of the molecular clockwork, high expression of CLOCK, PER1, PER2, PER3, CRY2, NPAS2 and RORC - all of which, with the exception of RORC are involved in E-box regulation - was associated with longer metastasis-free survival. The association of circadian genes with favorable prognosis supports the idea that the maintenance of circadian clock function may protect from tumor progression. Furthermore, stratification of tumors according to their molecular subtype revealed prognostic relevance of different circadian genes in the different subtypes. PER1, PER3 and CRY2 were associated with better prognosis in ER+/HER2- tumors. In contrast, CLOCK and NPAS2 showed significant association with better prognosis in ER-/HER2- tumors and ARNTL2 in the HER2-positive breast cancer subtype. A role for ARNTL2 in this particular breast cancer subtype is a novel and unexpected result that has not been described in prior studies. Interestingly, higher expression of CLOCK and NPAS2, which are paralogs with partially overlapping functions in the positive limb of the molecular clock, may discriminate patients with better outcome in the more aggressive ER-negative breast cancer subtype.
A key feature, shared by the majority of prognosis-associated circadian clock genes, was their strong association with ER and HER2 status as well as tumor grade. They displayed significantly lower expression in ER negative and/or HER2 positive carcinomas. Likewise were lower expression levels observed in high-grade tumors, indicating their decreased expression with tumor de-differentiation. Such strong associations with clinical parameters may explain why most circadian clock genes do not provide independent prognostic information in the multivariate analysis, which limits their usability as prognostic markers in a clinical setting. Noteworthy, exceptions to this general observation were PER3 and RORC, both displaying prognostic power independent of conventional clinicopathological parameters. Concerning PER3, similar associations with disease-free and overall survival in breast cancer have been previously reported.34 However, the relevance of RORC with regard to breast cancer prognosis has not yet been described.
Studying the correlation of clock gene expression with previously established metagenes,18 we observed negative correlations with the proliferation metagene. Remarkably, all core clock genes showed a highly significant negative correlation with relatively high coefficients. These results are in agreement with previously published studies showing how loss of circadian gene expression may lead to loss of control of cellular proliferation.11 The negative correlation of clock genes with the proliferation metagene is also consistent with their association with lower grade and with better prognosis. Correlation of circadian core clock genes with the ER metagene also fits with their association with ER-positive status.
Loss of estrogen receptor (ESR1, coding for ERα) expression in tumors is associated with resistance to endocrine therapy and a more aggressive course of disease. The finding that circadian gene expression (e.g., PER1, PER3 and CRY2) – and likely activity of the circadian clock - is higher in ER-positive and decreased in ER-negative tumors suggests an important connection between the circadian clock and ER-signaling. Importantly, Gery et al.35 could show that PER2 itself is estrogen-inducible and at the same time promotes degradation of the estrogen receptor. One important aspect of this reported ERα-PER2 mutual regulation35 is the control of circadian gene expression by estrogen signaling. If estrogen influences the circadian clock in breast tissue, it may potentially act as a zeitgeber. Interestingly, Bao et al.36 have been able to detect diurnal and ultradian rhythms of salivary estrogen in addition to the well-known fluctuations of estrogen levels during the menstrual cycle. Moreover, the ERα protein itself has been reported to oscillate in a circadian manner in mammary epithelial cells in vitro upon entrainment by serum shock.37 Thus, it is tempting to speculate that loss of ER in tumors may have an impact on circadian clock resetting. Accordingly, the circadian machinery in mammary tumors, when deprived of ER input, would lose its robustness or synchronicity with the rest of the body. This could explain both the observed reduced expression of and correlation between clock genes, including PER2, PER3 and CRY2 in ER-negative tumors. Our findings are consistent with those of Rosetti et al.,37 showing loss of circadian oscillation of clock genes in ER-negative breast cancer cell lines.
An additional interesting aspect derived from the crosstalk reported by Gery et al.35 is the regulation of the ERα itself by the circadian clock. Could disturbances of circadian rhythms be a cause for aberrant or impaired estrogen receptor expression/signaling in breast cancer? One recent study has suggested that long-term night shift work is associated with an increased risk of ER-negative breast cancer.38 Thus, further investigations are required to understand in detail the crosstalk between the circadian clock and estrogen signaling. Regardless of the underlying mechanism, the idea that a functional circadian clock would temporarily restrict the ability of cells to respond to estrogen via controlling ERα stability may have important implications in the management of endocrine therapy. Well-differentiated ER-positive tumors that still contain a robust circadian clock may therefore display circadian variation in ERα levels and activity. Although to our knowledge no chronotherapeutic administration of estrogen antagonists (e.g., tamoxifen) has been reported (in contrast to chrono-chemotherapy in other cancers14), our observation that ER-positive tumors seem to have a functional molecular clockwork suggests that they may be suitable for chronotherapy, as also pointed out in other studies.37
In contrast to the aforementioned core clock components, high expression of TIMELESS correlated very strongly with the proliferation metagene and was associated with poor prognosis. This is not surprising because TIMELESS is a known positive regulator of DNA replication39 and was previously associated with unfavorable outcome in other cancers.40 A remarkable finding was that the association of TIMELESS with MFS is independent of the established clinical parameters age, grading, pTstage, ER and HER2 status. Moreover, we observed higher expression of TIMELESS in more aggressive carcinomas, such as grade 3, ER-negative, HER2-positive and metastasizing tumors. Vice versa, expression of core clock components, such as CRY2, PER2 and PER3, was decreased in these more aggressive tumor types. This shift in balance may have important implications for the integrity of the molecular clock. Recently, TIMELESS has been reported to physically interact with the core clock component CRY1.16 It was also shown that PER2 competes with TIMELESS for binding to CRY1. According to this, higher expression of TIMELESS and lower expression of PER2 in tumors would result in a reduction of PER2-CRY dimers in favor of TIMELESS-CRY1 dimers. This may antagonize the ability of PER-CRY heterodimers to regulate expression of key genes involved in proliferation, such as c-Myc, and cyclins D1, B1 and A2.6,41 This mechanism may also explain the co-existence of high TIMELESS, low core clock components and high proliferation in more aggressive breast carcinomas.
For a more in depth analysis of circadian gene relationships, we focused on the PER and CRY genes, whose molecular interaction patterns are well understood. PER and CRY proteins form dimers that act as negative feedback regulators by repressing their own transcription as well as the transcription of other E-box-regulated, clock-controlled genes and have moreover been shown to display a similar oscillation pattern.22,25-29 Under conditions of coordinated oscillation, a strong correlation between the expression levels of these genes can be anticipated. In agreement with this concept, strong correlations between PER2-PER3 and CRY2-PER3 were indeed observed. While less aggressive tumors displayed strong correlations, lower correlation coefficients were generally observed in more aggressive carcinomas. Hypothesizing that low correlations, due to desynchronization of the circadian clock, would compromise heterodimer formation and hence the function of the molecular clockwork, deregulation of originally synchronized genes may be a feature of tumor dedifferentiation. This “breakdown” of correlation between clock genes was observed with loss of ER expression, HER2 amplification and increasing histological grade, well-known features of poorly differentiated and more aggressively growing tumors. Moreover, tumors with short time to metastasis showed lower correlation coefficients than non-metastasizing tumors. The absence of a negative correlation coefficient between ARNTL and PER genes in high grade tumors also supports the idea that the negative regulatory feedback loop of the circadian clockwork is less robust in more dedifferentiated breast cancer.
The results discussed so far illustrate that the circadian clock machinery is compromised in poorly differentiated and more aggressive carcinomas not only due to reduced expression levels of circadian genes but also as a result of desynchronization between them. In an attempt to differentiate between these 2 possibilities, we analyzed the absolute residuals of the correlation plots of PER2 and PER3. These residuals represent a measure to which degree expression of PER2 and PER3 deviate from a perfect correlation. While PER2 and PER3 were both significantly associated with better prognosis, the high residuals showed a trend toward worse prognosis. This trend, however, did not amount to statistical significance. Therefore, this type of statistical analysis could not answer the question whether compromised correlation between core clock genes leads to a more aggressive tumor growth.
In conclusion, loss of expression of core clock genes, mainly those involved in E-box regulation is associated with worse prognosis in breast cancer. Correlations between clock genes could still be observed in low grade, ER+ and HER2- and non-metastasizing tumors but were lost or strongly decreased in high grade, ER-, HER2+ and metastasizing carcinomas.
Materials and Methods
Patient cohorts and gene expression array analysis
This study is based on Affymetrix HG-U133A gene expression array data from a cohort consisting of 200 node-negative breast cancer patients, operated for a primary breast tumor between 1988 and 1998 at the Department of Obstetrics and Gynecology, Johannes Gutenberg University, Mainz, and untreated in the adjuvant setting. Sample collection, clinical data acquisition and gene expression array analysis have been described previously for the Mainz cohort.18 The use of human tissue samples and clinical patient information was approved by the Medical Association of Rhineland-Palatinate ethical view board. The data are available from the Gene Expression Omnibus database (GEO, accession number GSE11121). In addition, Affymetrix HG-U133A array data from 2 additional cohorts of node-negative breast cancer patients who did not receive adjuvant chemotherapy, encompassing 286 (Rotterdam) and 280 (Transbig) patients, respectively, was retrieved from GEO (accession numbers GSE2034, GSE6532 and GSE7390). Normalization of the raw data was done using RMA (robust multi-array average). RMA was applied separately for each cohort for single-cohort analyses and to all 766 samples jointly in one step for combined-cohort analyses. Hormone receptor status was derived from the gene array data. Cut-points were 8.2 for the estrogen receptor and 11.2 for HER2. Patient characteristics for the complete Mainz, Rotterdam and Transbig cohorts (total N = 766) have been reported previously18 and are presented in Supplemental Table 11A-C.
Standardization of data
In order to eliminate possible batch effects derived from the combination of data derived from different microarray gene expression experiments, we standardized the expression values cohort-wise before the statistical analysis. First, for each probe set we calculated the mean and the standard deviation across the samples belonging to the same cohort. Then we standardized expressions values by subtraction of the corresponding mean and division by the corresponding standard deviation.
Statistical analysis
A univariate Cox model was applied to evaluate the association between mRNA expression levels of selected circadian clock genes and metastasis-free survival (MFS). To further visualize survival differences, mRNA expression levels were dichotomized at median expression and survival rates were plotted according to the Kaplan-Meier method and compared with the log-rank test. Metastasis-free interval (MFI) was defined as the time between surgery and diagnosis of distant metastasis. Disease-free survival (DFS) was defined as time to metastasis, local recurrence or secondary carcinoma in the contralateral breast. DFS was included as the endpoint in the analysis of the Mainz cohort only. To assess the prognostic power of circadian clock gene expression independent of established clinical parameters, multivariate Cox regression was performed with inclusion of age at diagnosis (<50 y vs. ≥50 y), tumor size (≤2 cm vs. >2 cm), tumor grade (I-II vs. III), hormone or estrogen receptor status (positive vs. negative), and HER2 status (positive vs. negative). The multivariate Cox analysis considered data from the Mainz and the Transbig cohorts (N = 480) since clinical parameters were not available for the Rotterdam cohort. The Spearman correlation coefficient was calculated to assess the relation between mRNA expression levels and the expression of metagenes - constructed as described elsewhere18 - that represent proliferation, estrogen receptor (ER)-responsiveness and T- and B-cell infiltration. For assessing significant expression differences between 2 clinical or molecular groups the Mann-Whitney test was applied. For comparing multiple groups the Kruskal-Wallis rank sum test was used. The Spearman correlation coefficient was calculated to investigate pairwise correlations of circadian genes in the different patient subgroups.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This project was funded by the BMBF (NGFN project Oncoprofile, no. 01GR0816).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Ann Rev physiol 2010; 72:517-49; PMID:20148687; http://dx.doi.org/ 10.1146/annurev-physiol-021909-135821 [DOI] [PubMed] [Google Scholar]
- 2. Barclay JL, Tsang AH, Oster H. Interaction of central and peripheral clocks in physiological regulation. Prog Brain Res 2012; 199:163-81; PMID:22877665; http://dx.doi.org/ 10.1016/B978-0-444-59427-3.00030-7 [DOI] [PubMed] [Google Scholar]
- 3. Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Ann Rev Neurosci 2012; 35:445-62; PMID:22483041; http://dx.doi.org/ 10.1146/annurev-neuro-060909-153128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lowrey PL, Takahashi JS. Genetics of circadian rhythms in Mammalian model organisms. Adv Genet 2011; 74:175-230; PMID:21924978; http://dx.doi.org/ 10.1016/B978-0-12-387690-4.00006-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Human Mol Genet 2006; 15 Spec No 2:R271-7; http://dx.doi.org/ 10.1093/hmg/ddl207 [DOI] [PubMed] [Google Scholar]
- 6. Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer 2003; 3:350-61; PMID:12724733; http://dx.doi.org/ 10.1038/nrc1072 [DOI] [PubMed] [Google Scholar]
- 7. Gery S, Koeffler HP. Circadian rhythms and cancer. Cell Cycle (Georgetown, Tex) 2010; 9:1097-103; PMID:20237421; http://dx.doi.org/ 10.4161/cc.9.6.11046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kamdar BB, Tergas AI, Mateen FJ, Bhayani NH, Oh J. Night-shift work and risk of breast cancer: a systematic review and meta-analysis. Breast Cancer Res Treat 2013; 138:291-301; PMID:23400581; http://dx.doi.org/ 10.1007/s10549-013-2433-1 [DOI] [PubMed] [Google Scholar]
- 9. Mormont MC, Levi F. Circadian-system alterations during cancer processes: a review. Int J Cancer J Int Du Cancer 1997; 70:241-7; PMID:9009166; http://dx.doi.org/ 10.1002/(SICI)1097-0215(19970117)70:2<241::AID-IJC16>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 10. Filipski E, King VM, Li X, Granda TG, Mormont MC, Liu X, Claustrat B, Hastings MH, Levi F. Host circadian clock as a control point in tumor progression. J Natl Cancer Ins 2002; 94:690-7; PMID:11983758; http://dx.doi.org/ 10.1093/jnci/94.9.690 [DOI] [PubMed] [Google Scholar]
- 11. Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 2002; 111:41-50; PMID:12372299; http://dx.doi.org/ 10.1016/S0092-8674(02)00961-3 [DOI] [PubMed] [Google Scholar]
- 12. Canaple L, Kakizawa T, Laudet V. The days and nights of cancer cells. Cancer Res 2003; 63:7545-52; PMID:14633665 [PubMed] [Google Scholar]
- 13. Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ, Chang JG. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis 2005; 26:1241-6; PMID:15790588; http://dx.doi.org/ 10.1093/carcin/bgi075 [DOI] [PubMed] [Google Scholar]
- 14. Innominato PF, Levi FA, Bjarnason GA. Chronotherapy and the molecular clock: Clinical implications in oncology. Adv Drug Del Rev 2010; 62:979-1001; PMID:20600409; http://dx.doi.org/ 10.1016/j.addr.2010.06.002 [DOI] [PubMed] [Google Scholar]
- 15. Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet 2005; 37:187-92; PMID:15665827; http://dx.doi.org/ 10.1038/ng1504 [DOI] [PubMed] [Google Scholar]
- 16. Engelen E, Janssens RC, Yagita K, Smits VA, van der Horst GT, Tamanini F. Mammalian TIMELESS is involved in period determination and DNA damage-dependent phase advancing of the circadian clock. PloS One 2013; 8:e56623; PMID:23418588; http://dx.doi.org/ 10.1371/journal.pone.0056623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Desmedt C, Haibe-Kains B, Wirapati P, Buyse M, Larsimont D, Bontempi G, Delorenzi M, Piccart M, Sotiriou C. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res: An Off J Am Assoc Cancer Res 2008; 14:5158-65; PMID:18698033; http://dx.doi.org/ 10.1158/1078-0432.CCR-07-4756 [DOI] [PubMed] [Google Scholar]
- 18. Schmidt M, Bohm D, von Torne C, Steiner E, Puhl A, Pilch H, Lehr HA, Hengstler JG, Kolbl H, Gehrmann M. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res 2008; 68:5405-13; PMID:18593943; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-5206 [DOI] [PubMed] [Google Scholar]
- 19. Schmidt M, Hengstler JG, von Torne C, Koelbl H, Gehrmann MC. Coordinates in the universe of node-negative breast cancer revisited. Cancer Res 2009; 69:2695-8; PMID:19318558; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-4013 [DOI] [PubMed] [Google Scholar]
- 20. Schmidt M, Victor A, Bratzel D, Boehm D, Cotarelo C, Lebrecht A, Siggelkow W, Hengstler JG, Elsasser A, Gehrmann M, et al. Long-term outcome prediction by clinicopathological risk classification algorithms in node-negative breast cancer–comparison between Adjuvant!, St Gallen, and a novel risk algorithm used in the prospective randomized Node-Negative-Breast Cancer-3 (NNBC-3) trial. Ann Oncol: Off J Eur Soc Med Oncol ESMO 2009; 20:258-64; PMID:18824499; http://dx.doi.org/ 10.1093/annonc/mdn590 [DOI] [PubMed] [Google Scholar]
- 21. Yagita K, Yamaguchi S, Tamanini F, van Der Horst GT, Hoeijmakers JH, Yasui A, Loros JJ, Dunlap JC, Okamura H. Dimerization and nuclear entry of mPER proteins in mammalian cells. Gene Dev 2000; 14:1353-63; PMID:10837028 [PMC free article] [PubMed] [Google Scholar]
- 22. Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 1999; 98:193-205; PMID:10428031 [DOI] [PubMed] [Google Scholar]
- 23. Oster H, Baeriswyl S, Van Der Horst GT, Albrecht U. Loss of circadian rhythmicity in aging mPer1–mCry2– mutant mice. Gene Dev 2003; 17:1366-79; PMID:12782655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oster H, Yasui A, van der Horst GT, Albrecht U. Disruption of mCry2 restores circadian rhythmicity in mPer2 mutant mice. Gene Dev 2002; 16:2633-8; PMID:12381662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell 1997; 91:1055-64; PMID:9428527 [DOI] [PubMed] [Google Scholar]
- 26. Zylka MJ, Shearman LP, Weaver DR, Reppert SM. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 1998; 20:1103-10; PMID:9655499 [DOI] [PubMed] [Google Scholar]
- 27. Okamura H, Miyake S, Sumi Y, Yamaguchi S, Yasui A, Muijtjens M, Hoeijmakers JH, van der Horst GT. Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock. Sci (New York, NY) 1999; 286:2531-4; PMID:10617474 [DOI] [PubMed] [Google Scholar]
- 28. Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature 2002; 418:935-41; PMID:12198538 [DOI] [PubMed] [Google Scholar]
- 29. Yan L, Okamura H. Gradients in the circadian expression of Per1 and Per2 genes in the rat suprachiasmatic nucleus. Eur J Neurosci 2002; 15:1153-62; PMID:11982626 [DOI] [PubMed] [Google Scholar]
- 30. Lee Y, Chen R, Lee HM, Lee C. Stoichiometric relationship among clock proteins determines robustness of circadian rhythms. J Biol Chem 2011; 286:7033-42; PMID:21199878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim JK, Forger DB. A mechanism for robust circadian timekeeping via stoichiometric balance. Mol Syst Biol 2012; 8:630; PMID:23212247; http://dx.doi.org/ 10.1038/msb.2012.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oishi K, Sakamoto K, Okada T, Nagase T, Ishida N. Antiphase circadian expression between BMAL1 and period homologue mRNA in the suprachiasmatic nucleus and peripheral tissues of rats. Biochem Biophys Res Commun 1998; 253:199-203; PMID:9878515 [DOI] [PubMed] [Google Scholar]
- 33. Grundy A, Schuetz JM, Lai AS, Janoo-Gilani R, Leach S, Burstyn I, Richardson H, Brooks-Wilson A, Spinelli JJ, Aronson KJ. Shift work, circadian gene variants and risk of breast cancer. Cancer Epidemiol 2013; 37:606-12; PMID:23725643; http://dx.doi.org/ 10.1016/j.canep.2013.04.006 [DOI] [PubMed] [Google Scholar]
- 34. Climent J, Perez-Losada J, Quigley DA, Kim IJ, Delrosario R, Jen KY, Bosch A, Lluch A, Mao JH, Balmain A. Deletion of the PER3 gene on chromosome 1p36 in recurrent ER-positive breast cancer. J Clin Oncol: Off J Am Soc Clin Oncol 2010; 28:3770-8; PMID:20625127; http://dx.doi.org/ 10.1200/JCO.2009.27.0215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gery S, Virk RK, Chumakov K, Yu A, Koeffler HP. The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene 2007; 26:7916-20; PMID:17599055 [DOI] [PubMed] [Google Scholar]
- 36. Bao AM, Liu RY, van Someren EJ, Hofman MA, Cao YX, Zhou JN. Diurnal rhythm of free estradiol during the menstrual cycle. Eur J Endocrinol Eur Fed Endocr Soc 2003; 148:227-32; PMID:12590642 [DOI] [PubMed] [Google Scholar]
- 37. Rossetti S, Esposito J, Corlazzoli F, Gregorski A, Sacchi N. Entrainment of breast (cancer) epithelial cells detects distinct circadian oscillation patterns for clock and hormone receptor genes. Cell Cycle (Georgetown, Tex) 2012; 11:350-60; PMID:22193044; http://dx.doi.org/ 10.4161/cc.11.2.18792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rabstein S, Harth V, Pesch B, Pallapies D, Lotz A, Justenhoven C, Baisch C, Schiffermann M, Haas S, Fischer HP, et al. Night work and breast cancer estrogen receptor status–results from the German GENICA study. Scand J Work, Env Health 2013; 39:448-55; PMID:23543199; http://dx.doi.org/ 10.5271/sjweh.3360 [DOI] [PubMed] [Google Scholar]
- 39. Kondratov RV, Antoch MP. Circadian proteins in the regulation of cell cycle and genotoxic stress responses. Trends Cell Biol 2007; 17:311-7; PMID:17644383 [DOI] [PubMed] [Google Scholar]
- 40. Mao Y, Fu A, Leaderer D, Zheng T, Chen K, Zhu Y. Potential cancer-related role of circadian gene TIMELESS suggested by expression profiling and in vitro analyses. BMC Cancer 2013; 13:498; PMID:24161199; http://dx.doi.org/ 10.1186/1471-2407-13-498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Borgs L, Beukelaers P, Vandenbosch R, Belachew S, Nguyen L, Malgrange B. Cell “circadian” cycle: new role for mammalian core clock genes. Cell Cycle (Georgetown, Tex) 2009; 8:832-7; PMID:19221497 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.