Abstract
Ligands that selectively inhibit human α3β2 and α6β2 nicotinic acetylcholine receptor (nAChRs) and not the closely related α3β4 and α6β4 subtypes are lacking. Current α-conotoxins (α-Ctxs) that discriminate among these nAChR subtypes in rat fail to discriminate among the human receptor homologs. In this study, we describe the development of α-Ctx LvIA(N9R,V10A) that is 3000-fold more potent on oocyte-expressed human α3β2 than α3β4 and 165-fold more potent on human α6/α3β2β3 than α6/α3β4 nAChRs. This analog was used in conjuction with three other α-Ctx analogs and patch-clamp electrophysiology to characterize the nAChR subtypes expressed by human adrenal chromaffin cells. LvIA(N9R,V10A) showed little effect on the acetylcholine-evoked currents in these cells at concentrations expected to inhibit nAChRs with β2 ligand-binding sites. In contrast, the β4-selective α-Ctx BuIA(T5A,P6O) inhibited >98% of the acetylcholine-evoked current, indicating that most of the heteromeric receptors contained β4 ligand-binding sites. Additional studies using the α6-selective α-Ctx PeIA(A7V,S9H,V10A,N11R,E14A) indicated that the predominant heteromeric nAChR expressed by human adrenal chromaffin cells is the α3β4* subtype (asterisk indicates the possible presence of additional subunits). This conclusion was supported by polymerase chain reaction experiments of human adrenal medulla gland and of cultured human adrenal chromaffin cells that demonstrated prominent expression of RNAs for α3, α5, α7, β2, and β4 subunits and a low abundance of RNAs for α2, α4, α6, and α10 subunits.
Introduction
Nicotinic acetylcholine receptors (nAChRs) are ligand-gated ion channels composed of five individual subunits and can be classified into two broad categories, homomeric and heteromeric, based on whether the receptors are assembled from one gene product or from two or more gene products, respectively. In humans there are 16 different nAChR genes designated α1–α7, α9, α10, β1-β4, δ, ε, and γ [for a review of nAChRs, see (Albuquerque et al., 2009)]. Highly selective ligands that discriminate between the various human nAChR subtypes are lacking, which makes pharmacological identification of individual subtypes difficult. This is particularly problematic for the more closely related subtypes, such as those that contain α3 or α6 subunits.
Peptides isolated from the venom of marine cone snails, and one subclass in particular, the α-conotoxin (α-Ctx), have been particularly useful in developing ligands that selectively target one nAChR subtype over another. For example, α-Ctx TxIB is highly selective for rat α6/α3β2β3 nAChRs and shows very little activity on other subtypes (Luo et al., 2013). α-Ctx PnIA is a peptide similar in sequence to TxIB and is selective for rat α3β2 and α6β2 nAChRs over the α3β4 and α6/α3β4 subtypes (Luo et al., 1999; Hone et al., 2012a). α-Ctx LvIA is a newly discovered peptide that shows some selectivity for α3β2 over the α3β4 subtype (Luo et al., 2014). The amino acid sequences of these small peptides are all similar, but vary in key positions that are known to be critical for activity. Importantly, they can be modified by substituting one or more of their amino acids to produce analogs with increased potency and selectivity, and several have recently been developed that are highly selective for various rodent nAChR subtypes. For example, PeIA(S9H,V10A,E14N) shows a high degree of selectivity for rat α3β2 and α6β2 nAChRs over the α3β4 and α6β4 subtypes (Hone et al., 2012b), and PeIA(A7V,S9H,V10A,N11R,E14A) selectively inhibits α6-containing nAChRs over other subtypes (Hone et al., 2013). The margins of selectivity of most of these peptides for human receptors, however, are much narrower (unpublished observation), making discrimination among human nAChR subtypes difficult. We took advantage of the modest selectivity of LvIA and made two substitutions in the peptide’s amino acid sequence to further increase its potency and selectivity for human α3β2 and α6β2 nAChRs, and then used this analog to probe for the expression of these two subtypes in human adrenal chromaffin cells.
Chromaffin cells of the adrenal gland are neuroendocrine cells that release catecholamines and other substances into the bloodstream during the flight-or-fight response or as a reaction to a perceived stressful situation. These cells are of neural crest origin and have many of the features of neurons, including the ability to fire action potentials and release neurotransmitters, particularly catecholamines. Activation of nAChRs by acetylcholine (ACh) released from the splanchnic nerve is sufficient to depolarize the adrenal chromaffin cell membrane and activate voltage-gated ion channels, including calcium channels. The influx of calcium ions through nAChRs and voltage-gated calcium channels facilitates the fusion of synaptic vesicles with the plasma membrane to promote the release of vesicular content (Mollard et al., 1995; Perez-Alvarez et al., 2012a). Rodent (Di Angelantonio et al., 2003; Gahring et al., 2014) and bovine (Campos-Caro et al., 1997; Criado et al., 1997) adrenal chromaffin cells have been reported to express a variety of nAChR subtypes, including α7, α3β4*, α3β2*, and α4β2*, whereas in primates the reported receptor subtype(s) has included α7 and α6β4* (Perez-Alvarez et al., 2012a, 2012b; Hernandez-Vivanco et al., 2014). Given the number of nicotinic compounds used clinically, it is critical to identify all of the subtypes expressed by human adrenal chromaffin cells and to assess their pharmacology. In this study, we synthesized a novel α-Ctx analog that is potent and highly selective for human α3β2 and α6β2 nAChRs over the α3β4 and α6β4 subtypes. This analog, LvIA(N9R,V19A), in conjunction with a panel of three other α-Ctxs, allowed us to identify the α3β4* subtype as the predominant nAChR expressed by human adrenal chromaffin cells. Previous studies were inconclusive regarding the presence of nAChRs with β2 ligand-binding sites in these cells, and in the present study we demonstrate that in fact there are likely to be few nAChRs of this type present. Furthermore, we show that the α3β4* nAChRs in these cells contain two α3-β4 ligand-binding sites and suggest that the stoichiometry of the receptor is likely (α3β4)2 with a fifth auxiliary subunit that has yet to be identified. These studies highlight the utility of α-Ctxs for characterizing the possible nAChRs expressed by a given cell type.
Materials and Methods
Reagents.
Acetylcholine chloride, HEPES, amphotericin B, penicillin/streptomycin, protease type XIV, collagenase type I, poly-D-lysine hydrobromide, Red Blood Cell Lysis solution, dimethylsulfoxide, and all salts were purchase from Sigma-Aldrich (St. Louis, MO). Dulbecco’s modified Eagle´s medium and Glutamax were purchased from Life Technologies (Carlsbad, CA). Fetal bovine serum was from LabClinics (Barcelona, Spain), and D-glucose was from Panreac (Barcelona, Spain). Amino acids used in peptide synthesis were obtained from AAPPTec (Louisville, KY).
Peptide Synthesis.
The α-Ctxs BuIA(T5A,P6O) and ArIB(V11L,V16D) were synthesized at the University of Utah DNA/peptide sequencing core facility, according to the procedures described in (Cartier et al., 1996). α-Ctxs LvIA(N9R,V10A) and PeIA(A7V,S9H,V10A,N11R,E14A) were synthesized using an AAPPTec Apex 396 automated peptide synthesizer, according to the procedures described in (Hone et al., 2012b). The peptide masses were verified by matrix-assisted laser desorption ionization/time-of-flight mass spectrometry, which was performed at the Salk Institute for Biologic Studies (La Jolla, CA).
Two-Electrode Voltage-Clamp Electrophysiology of Xenopus laevis Oocytes.
Detailed methods for conducting electrophysiology of Xenopus oocytes and the pharmacological assessment of the activities of α-Ctxs on heterologously expressed nAChRs have been previously described (Hone et al., 2009). Briefly, stage IV–V oocytes were injected with equal ratios of capped cRNA encoding human nAChR subunits prepared using the mMessage mMachine in vitro transcription kit (Ambion, Austin, TX). The α3, α4, β2, β3, and β4 clones were provided by J. Garrett (Cognetix, Salt Lake City, UT). The β4-α3-β4-α3-α5(D) concatemer was prepared, as previously described (George et al., 2012). Coinjection of human α6 subunit cRNA with cRNA for the β2, β3, or β4 subunits results in few or no functional receptors (Kuryatov et al., 2000; McIntosh et al., 2004; Kuryatov and Lindstrom, 2011). Several strategies have been employed to achieve functional expression, including concatenated subunits, as well as chimeras of α6 and other α subunits. We used an α6/α3 construct in which the first 207 amino acids of the α3 subunit are replaced with those of the α6 subunit ligand-binding domain (Kuryatov et al., 2000), and an α6M211L,cytα3 construct in which the Met at position 211 is replaced by a Leu residue and the intracellular loop between transmembranes three and four is replaced with that of the α3 subunit (Ley et al., 2014). For brevity, we use α6β4 constructs throughout the rest of the manuscript to describe data in which both α6M211L,cytα3β4 and α6/α3β4 were used to obtain α-Ctx values. Both of the α6 constructs and the β3-α6-β2-α4-β2 concatemer were prepared as previously described (Kuryatov and Lindstrom, 2011). Human α6/α3 and human β4 were ligated into the digested pSGEM vector by T4 DNA ligase. The ligation reactions were transformed in DH10B chemically competent cells, plated on ampicillin LB plates, and incubated overnight at 37°C. The following day, the cells were incubated in inoculated ampicillin-containing LB at 37°C in a shaker overnight. The DNA for both clones was isolated using the Qiaprep Spin Mini-Prep kit (Qiagen, Valencia, CA) and linearized with NheI restriction enzyme prior to RNA transcription. The linearized DNA was purified using the Qiaquick polymerase chain reaction (PCR) purification kit (Qiagen), and RNA was synthesized using the Ambion T7 mMessage mMachine RNA transcription kit (Life Technologies). The RNA was subsequently purified using the RNeasy mini kit (Qiagen). The concentration of the RNA was determined by UV spectroscopy. Oocytes were subsequently injected with the cRNA, and electrophysiology experiments were conducted 1–5 days after injection. To record ACh-evoked currents, the oocyte membranes were voltage-clamped at a holding potential of −70 mV with a Warner Instruments OC-725 series amplifier (Warner Instruments, Hamden, CT) and then stimulated with 1-second pulses of ACh once every 60 seconds. A concentration of 300 μM ACh was used for all subtypes to elicit a maximal response and to compare the α-Ctx IC50 values obtained in human adrenal chromaffin cells. The α-Ctxs were suspended in extracellular solution (ND96), and applied by perfusion for concentrations ≤1 μM or in a static bath for 5 minutes for concentrations >1 µM.
End-Point PCR Analysis of nAChR Subunit mRNA.
Total RNA was extracted from the adrenal medulla of four donors [age 64 ± 15 years, standard deviation of the mean (SDM)] or from cultured human adrenal chromaffin cells from three donors (age 58 ± 8 years, SDM), according to the manufacturers’ instructions, using an Ambion RNAqueous-4PCR kit (catalogue AM1914), and subsequently treated with DNase I to degrade genomic DNA. The purity of the RNA was assessed by measuring the A260/A280 ratio using a NanoDrop spectrophotometer. cDNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (catalogue 4368814; Applied Biosystems, Foster City, CA), following the manufacturer’s instructions. A quantity amounting to 10 µl RNA was used in a total reaction volume of 20 µl. The cycling conditions for the reverse transcription were as follows: step 1, 25°C for 10 minutes; step 2, 37°C for 120 minutes; and step 3, 85°C for 5 minutes, and were achieved using a PTC-200 peltier thermal cycler (MJ Research, Waltham, MA). In some samples, the reverse transcriptase was omitted to further assess for genomic DNA contamination in the subsequent reverse-transcriptase PCR (RT-PCR) analyses.
End-point RT-PCR analysis was performed using the Qiagen Taq PCR Master Mix Kit (Qiagen) following the manufacturer’s instructions. Briefly, 2.5 µl cDNA template was added to the Master Mix solution containing sense and antisense primers (final concentration of 500 nM for each primer) in a total reaction volume of 50 µl. Negative controls for each reaction were performed by omission of the cDNA template. Several primer sets designed to target Homo sapiens sequences were evaluated (West et al., 2003; Carlisle et al., 2004; Lips et al., 2005; Liu et al., 2009a). Each primer pair was aligned with their respective nicotinic subunit DNA and RNA sequences to verify that the primers spanned introns. All of the primers used for generation of the results presented in this work spanned an intron, with exception of primers for the α9 subunit. The primers were synthesized by the University of Utah Sequencing and Genomics Core Facility (Salt Lake City, UT). The PCR was performed in a PTC-200 thermal cycler using an initial 5-minute denaturation step at 95°C, followed by 35 cycles of denaturation at 94°C for 30 seconds, 55–60°C for 30 seconds for annealing, 72°C for 45 seconds for extension, with a final extension step at 72°C for 10 minutes. Following PCR, the reactions were analyzed by gel (1.5% agarose wt/vol) electrophoresis, stained, and visualized with ethidium bromide. A summary of the primers used to assess the presence of the different nAChR subunit mRNAs and the RT-PCR conditions used is provided in Table 3.
TABLE 3.
Primers for nAChR subunit mRNA, amplicon size, and annealing temperature used in PCR
| Target | Primer Sequence | Size | Temp. | Reference |
|---|---|---|---|---|
| α2 | 5′-CCGGTGGCTTCTGATGA-3′ (s) | 466 bp | 55°C | (West et al., 2003) |
| 5′-CAGATCATTCCAGCTAGG-3′ (as) | ||||
| α3 | 5′-CCATGTCTCAGCTGGTG-3′ (s) | 401 bp | 55°C | (West et al., 2003) |
| 5′-GTCCTTGAGGTTCATGGA-3′ (as) | ||||
| α4 | 5′-GGATGAGAAGAACCAGATGA-3′ (s) | 444 bp | 58°C | (Carlisle et al., 2004) |
| 5′-CTCGTACTTCCTGGTGTTGT-3′ (as) | ||||
| α5 | 5′-GGCCTCTGGACAAGACAA-3′ (s) | 179 bp | 60°C | (Liu et al., 2009a) |
| 5′-AAGATTTTCCTGTGTTCCC-3′ (as) | ||||
| α6 | 5′-TCCATCGTGGTGACTGTGT-3′ (s) | 413 bp | 55°C | (West et al., 2003) |
| 5′-AGGCCACCTCATCAGCAG-3′ (as) | ||||
| α7 | 5′-CTTCACCATCATCTGCACCATC-3′ (s) | 308 bp | 58°C | (Kurzen et al., 2004) |
| 5′-GGTACGGATGTGCCAAGGATAT-3′ (as) | ||||
| α9 | 5′-GTCCAGGGTCTTGTTTGT-3′ (s) | 403 bp | 55°C | (West et al., 2003) |
| 5′-ATCCGCTCTTGCTATGAT-3′ (as) | ||||
| α10 | 5′-CTGTTCCGTGACCTCTTCG-3′ (s) | 388 bp | 60°C | (West et al., 2003) |
| 5′-GAAGGCCGCCACGTCCA-3′ (as) | ||||
| β2 | 5′-CAGCTCATCAGTGTGCA-3′ (s) | 347 bp | 55°C | (West et al., 2003) |
| 5′-GTGCGGTCGTAGGTCCA-3′ (as) | ||||
| β3 | 5′-TGGAGAGTACCTGCTGTTCA-3′ (s) | 439 bp | 58°C | (Carlisle et al., 2004) |
| 5′-CGAGCCTGTTACTGACACTA-3′ (as) | ||||
| β4 | 5′-AGCAAGTCATGCGTGACCAAG-3′ (s) | 210 bp | 60°C | (Liu et al., 2009a) |
| 5′-GCTGACACCTTCTAATGCCTCC-3′ (as) |
Quantitative real-time PCR (qPCR) was performed to quantify the presence of mRNA transcripts. qPCR was performed using inventoried TaqMan Gene Expression Assays (Thermo Fisher Scientific, Waltham, MA), according to manufacturer’s instructions. (See Table 4 for a list of hydrolysis probe assay ID numbers and context sequences representing the human nAChR and reference genes analyzed in this study.) With the exception of GAPDH and ACTB, all probe sets spanned an intron. DNase I–treated human brain reference total RNA pooled from multiple donors and several brain regions was purchased from Ambion (catalogue AM6050). Up to 2 μg total RNA was reverse transcribed into cDNA using the SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific). Each 10 μl qPCR was performed in triplicate and assembled using 5 μl TaqMan 2× Fast Advanced Master Mix [containing AmpliTaq Fast DNA Polymerase, uracil-N glycosylase, dNTPs with dUTP, and ROX dye (passive reference)], 0.5 μl 20× hydrolysis probe set, and 4.5 μl nuclease-free water containing 20 ng cDNA (derived from RNA concentrations). A minus template control was run for each probe set. Samples were prepared in Fast Optical 96-well plates (Thermo Fisher; catalogue 4346906), and the gene targets were amplified using the Applied Biosystems QuantStudio Flex 6 real-time thermocycler. The qPCR thermal cycling program used was as follows: 1 cycle of 50°C for 2 minutes, followed by 95°C for 20 seconds (to activate, and then inactivate uracil-N glycosylase, which degrades any carryover DNA), 40 cycles of 1-second denaturation at 95°C, followed by 20 seconds of annealing and extension at 60°C. Cq values for each reaction were extracted using QuantStudio software to calculate relative gene expression levels and compared with total human brain using the 2−ΔΔCq method (Livak and Schmittgen, 2001). For this study, four reference genes were tested (GAPDH, ACTB, UBC, B2M), and the resulting Cq means were exported to the online reference gene stability analyzer, RefFinder (http://150.216.56.178). Briefly, gene stability was analyzed using the Delta CT (Silver et al., 2006), BestKeeper (Pfaffl et al., 2004), Normfinder (Andersen et al., 2004), and Geonorm (Vandesompele et al., 2002) methods to generate a comprehensive stability ranking of all four reference genes. From this analysis, GAPDH and ACTB produced similar high stability rankings and thus were chosen for geometric mean calculations and qPCR single and double normalization. Single normalization, log10(2−ΔCq), where ΔCq = (mean Cq nAChR gene − geometric mean Cq reference genes), was also employed to assess more directly the relative abundance of each nAChR gene compared with control genes within and between single samples in the absence of brain reference tissue. Statistical analysis of nAChR gene expression was determined using an analysis of variance and a Bonferroni post hoc test. Differences were considered significant if the P value was less than 0.05. All data are reported as the mean ± S.E.M.
TABLE 4.
TaqMan qPCR assay
| Assay ID | Context Sequence | Gene Symbol | Accession Number |
|---|---|---|---|
| Hs00181237_m1 | CTCTACAACAATGCAGATGGGGAGT | CHRNA2 | NM_000742.3 |
| Hs01088199_m1 | ACCTGTGGCTCAAGCAAATCTGGAA | CHRNA3 | NM_001166694.1 |
| Hs00181247_m1 | ACACAGACTTCTCGGTGAAGGAGGA | CHRNA4 | NM_001256573.1 |
| Hs00181248_m1 | AATTGGTGGATGTGGATGAGAAAAA | CHRNA5 | NM_000745.3 |
| Hs00610231_m1 | TCTTTAAAGGCTGTGTGGGCTGTGC | CHRNA6 | NM_001199279.1 |
| Hs01063373_m1 | CTCTATAACAGTGCTGATGAGCGCT | CHRNA7 | NM_001190455.2 |
| Hs00395558_m1 | TGCCCCTGATAGGTAAATACTACAT | CHRNA9 | NM_017581.3 |
| Hs00220710_m1 | TAACAAAGCCGACGCGCAGCCTCCA | CHRNA10 | NM_020402.2 |
| Hs00181267_m1 | ACAACAATGCTGACGGCATGTACGA | CHRNB2 | NM_000748.2 |
| Hs00181269_m1 | TTGAAAATGCTGACGGCCGCTTCGA | CHRNB3 | NM_000749.3 |
| Hs00181269_m1 | CCTTTGCGGGCGCGGGAACTGCCGC | CHRNB4 | NM_001256567.1 |
| Hs00609520_m1 | GACTCATGACCACAGTCCATGCCAT | GAPDH | NM_001256799.1 |
| Hs02758991_g1 | AAGCAGCATCATGGAGGTTTGAAGA | B2M | NM_004048.2 |
| Hs00984230_m1 | GTGATCGTCACTTGACAATGCAGAT | UBC | NM_021009.5 |
| Hs01060665_g1 | CCCAGGCACCAGGGCGTGATGGTGG | ACTB | NM_001101.3 |
Human Adrenal Chromaffin Cell Isolation and Culture.
Adrenal chromaffin cells were isolated from six male, age 58 ± 13 years (SDM), and one 35-year-old female organ donors. To isolate the chromaffin cells, the glands were perfused with a saline solution (154 mM NaCl, 5.6 mM KCl, 3.6 mM NaHCO3, 5.6 mM D-glucose, and 5 mM HEPES; pH was adjusted to 7.4 with NaOH and the observed osmolarity was 325 mOsM) through the suprarenal vein until the solution was clear and blood free, and then with 2 ml saline containing 1 mg/ml protease type XIV. The glands were then placed in a 50-ml conical tube, covered with saline solution, and incubated for 10 minutes at 37°C, followed by a second perfusion with protease and additional 10-minute incubation at 37°C. Following treatment with protease, the glands were placed in a 150-mm petri dish, bisected sagittally, and then opened by a coronal incision. The medullary tissue was scraped out of the gland and transferred to a conical tube partially filled with 30 ml saline solution containing 2 mg/ml collagenase type I. The medullary tissue was then incubated for 60 minutes at 37°C in a water bath with intermittent trituration every 5 minutes with a plastic Pasteur pipette. Following incubation with collagenase, 10 ml saline solution was added to the cell suspension and then filtered first through a 200-μm nylon mesh, followed by an 80-μm mesh. The isolated cells were centrifuged at 200g for 10 minutes, the supernatant was removed, and the cell pellet was suspended in 1 ml Red Blood Cell Lysis Buffer for 60 seconds, after which 39 ml saline was added to terminate the actions of the lysis buffer. Lastly, the cells were centrifuged at 200g for 5 minutes, the supernatant was removed, and the cells were suspended in Dulbecco’s modified Eagle´s medium containing 10% fetal bovine serum, 200 μM Glutamax, 100 U/ml penicillin, and 0.1 mg/ml streptomycin. A total of 500 μl cell suspension was added to each well of a 24-well culture plate containing glass coverslips that had been treated with 0.1 mg/ml poly-D-lysine. The cells were maintained at 37°C in an incubator under an atmosphere of 95% air and 5% CO2 for up to 7 days. The culture medium was changed daily by exchanging approximately 70% of the solution with fresh medium.
Patch-Clamp Electrophysiology of Chromaffin Cells.
To conduct electrophysiology experiments, the coverslips were placed in a glass-bottom chamber and continuously gravity perfused with extracellular solution (145 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM D-glucose, and 10 mM HEPES; pH adjusted to 7.4 with NaOH; observed osmolarity 325 mOsM) at a flow rate of 1.5 ml/min by means of a polyethylene tube with an inner diameter of 0.58 mm. The outlet of this tube was placed close to the cell of interest to ensure rapid solution exchange. Glass electrodes were pulled from borosilicate glass capillaries (Kimbal Chase, Vineland, NJ; catalogue 3400-99) using a P97 pipette puller (Sutter Instruments, Novato, CA). These electrodes had resistances between 1.5 and 2.5 MΩ when filled with an internal electrode solution composed of 145 mM K-glutamate, 10 mM NaCl, 1 mM MgCl2, 10 mM D-glucose, and 10 mM HEPES (pH adjusted to 7.2 with KOH; observed osmolarity 322 mOsM). To initiate whole-cell recordings, a stock solution of 0.5 mg/ml amphotericin B was prepared daily in dimethysulfoxide, and 5 μl stock solution was added to 500 μl intracellular solution and ultrasonicated immediately before use. All experiments were performed under a sodium lamp for light. A HEKA EPC10 amplifer (HEKA Electronik, Lambrect, Germany) was used to record ACh-evoked responses. Analysis of the α-Ctx activity was performed only on cells where the series resistance obtained was <20 MΩ, and, in general, resistances between 10 and 15 MΩ were achieved and were compensated electronically up to 94% to minimize voltage errors. ACh was applied using a multibarreled pipette that was constructed using polyethylene tubing with an inner diameter of 0.4 mm. These tubes coalesced to a single outlet tube with a 0.28 mm inner diameter and had a flow rate approximately 850 μl/min. The agonist pulses were delivered by gravity and were controlled by a valve controller triggered by the amplifier. The activities of the antagonists were determined by applying 200-ms pulses of ACh once every 3 minutes until a steady baseline response was achieved. The control solution was then switched to one containing the antagonist of interest and was perfused until a steady state level of inhibition was observed. The peak amplitudes of three control responses were averaged, and the level of inhibition by the antagonist was determined by dividing the peak amplitude of the response in the presence of the antagonist by the averaged control response to obtain a percent response value. Human adrenal chromaffin cells are known to express α7-containing nAChRs, but account for <10% of the whole-cell response to ACh (Perez-Alvarez et al., 2012a). In this study, our interest was focused on the predominant heteromeric subtype expressed; therefore, the α7 antagonist α-Ctx ArIB(V11L,V16D) was included in all solutions at a concentration of 100 nM (Whiteaker et al., 2007; Hone et al., 2012a). For simplicity, we refer to heteromeric subtypes throughout the manuscript as those that do not include α7 subunits.
Data Analysis.
All statistical comparisons of antagonist data, concentration-response analyses, and qPCR-determined gene expression levels were performed using GraphPad Prism (La Jolla, CA). Current traces were rendered using IGOR Pro (WaveMetrics, Lake Oswego, OR). Images for gel electrophoresis were processed using ImageJ (National Institutes of Health, Bethesda, MD).
Results
α-Ctx LvIA(N9R,V10A) Is a Potent and Selective Ligand for Human α3β2- and α6β2-Containing nAChRs Expressed in X. laevis Oocytes.
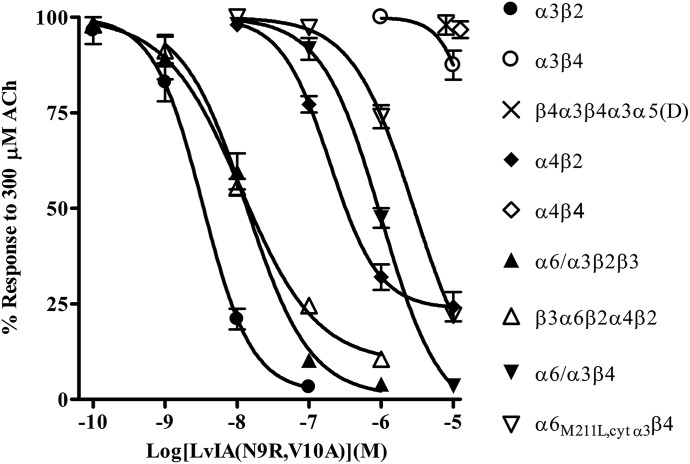
As previously discussed, α-Ctxs TxIB, PnIA, and LvIA show some degree of selectivity for particular nAChR subtypes. Table 1 shows the sequences of these peptides and compares their selectivity profiles. Note that many of the amino acids are strictly conserved, whereas others are highly variable. The residues that vary most likely account for the observed differences in the peptides’ selectivity profiles. Indeed, we have previously shown that specific positions in the sequence of α-Ctx PeIA are important for determining potency and selectivity for β2- over β4-containing subtypes and can be substituted with other amino acids to generate analogs with more favorable selectivity profiles. For example, substituting the Val in the tenth position of PeIA with Ala, the residue found in this position in PnIA, increases the selectivity for α3β2 over α3β4 by >1000-fold and by >100-fold for α6/α3β2β3 over α6/α3β4 nAChRs (Hone et al., 2012b). Substituting Ser9 with Arg, the residue found in this position in TxIB, results in a 1800-fold increase in selectivity for α6/α3β2β3 over α6/α3β4 nAChRs (Hone et al., 2013). Thus, taking cues from the natural diversity found in the α-Ctxs shown in Table 1 and from our studies with PeIA, we synthesized a LvIA analog that was expected to show greater selectivity for human α3β2 and α6β2 nAChRs over the α3β4 and α6β4 subtypes. Two substitutions in the amino acid sequence of LvIA were made to generate LvIA(N9R,V10A). Verification of the peptide synthesis was confirmed by mass spectrometry. The calculated monoisotopic mass for LvIA(N9R,V10A) is 1693.64 Da, and the observed monoisotopic mass was determined to be 1693.97 Da. The peptide was then tested on a panel of human nAChRs expressed in Xenopus oocytes that included α3β2, α3β4, β4α3β4α3α5(D), α4β2, α4β4, α6/α3β2β3, β3α6β2α4β2, α6/α3β4, and α6M211L,cytα3β4. As shown in Fig. 1, this analog was >3000-fold more potent on α3β2 than α3β4, and was between 85- and 245-fold more potent on α6β2-containing nAChRs over the α6β4 constructs. In addition, the analog showed some activity on the α4β2 subtype, but very little activity on the α4β4 subtype (Fig. 1). Thus, when used at a concentration of 100 nM, >90% of responses mediated by α3β2 or α6β2 nAChRs were inhibited while inhibiting <5% of the responses from β4-containing nAChRs.
TABLE 1.
Sequence comparison of select α-Ctxs and their selectivity profiles
| Amino Acid Sequence | nAChR Potency | Reference | |
|---|---|---|---|
| α-Conotoxin | |||
| LvIA | GCCSHPACNVDHPEIC | α3β2 > α3β4; α6/α3β2β3 = α6/α3β4 | (Luo et al., 2014) |
| TxIB | GCCSDPPCRNKHPDLC | α6/α3β2β3 > >α3β2,α3β4,α6/α3β4 | (Luo et al., 2013) |
| PnIA | GCCSLPPCAANNPDYC | α3β2 > >α3β4; α6/α3β2β3 > α6/α3β4 | (Luo et al., 1999) |
| LvIA(N9R,V10A) | GCCSHPACRADHPEIC | α3β2 > >>α3β4; α6/α3β2β3 > >α6/α3β4 | This work |
>, 2- to 99-fold; > >, 100- to 500-fold; > >>, >1000-fold.
Residues shaded are conserved between the four peptides, and residues bolded are those substituted from TxIB and PnIA.
Fig. 1.
Pharmacological profile of LvIA(N9R,V10A) for inhibition of human nAChRs expressed in Xenopus oocytes. Oocytes expressing different nAChR subtypes were subjected to two-electrode voltage-clamp electrophysiology, as described in Materials and Methods. LvIA(N9R,V10A) inhibited α3β2, α6/α3β2β3, and β3α6β2α4β2 nAChRs with IC50 values of 3.3 (2.4–4.7) nM (n = 4), 13.5 (8.6–21.2) nM (n = 3), and 11.4 (8.1–16.0) nM (n = 4), respectively. The α6/α3β4 and α6M211L,cytα3β4 constructs were inhibited with IC50 values of 1.0 (0.8–1.3) μM (n = 4) and 2.8 (2.5–3.4) µM, respectively. The peptide also inhibited α4β2 nAChRs with an IC50 of 195 (133–284) nM (n = 4). For α3β4, β4α3β4α3α5(D), and α4β4 nAChRs, the average response after a 5-minute static bath exposure to 10 µM LvIA(N9R,V10A) was 88 ± 4% (n = 4), 98 ± 3 (n = 4), and 97 ± 1% (n = 4), respectively. The error bars denote the S.E.M., and the values in parentheses denote the 95% confidence interval.
The α-Ctx Analog BuIA(T5A,P6O) Discriminates between Human β2 and β4 Subunit-Containing nAChRs.
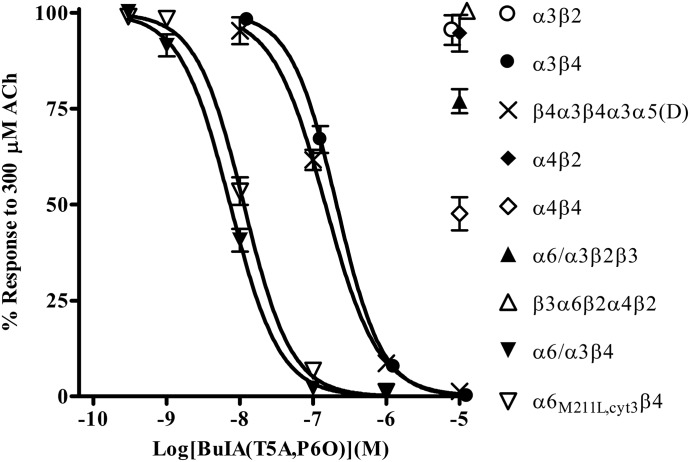
To dissect the responses mediated by β2-containing nAChRs from those mediated by β4-containing nAChRs, a ligand is needed that discriminates between the different nAChRs that contain these subtypes. We therefore assessed the activity of α-Ctx BuIA(T5A,P6O), which has been shown to selectively inhibit rat and mouse α6β4 and α3β4 nAChRs over other subtypes (Azam et al., 2010). When tested on human nAChRs expressed in Xenopus oocytes, we found that this analog was >1000-fold more potent on the α6β4 constructs than on α3β2, α4β2, α4β4, α6/α3β2β3, and β3α6β2α4β2 nAChRs (Fig. 2). Additionally, BuIA(T5A,P6O) was >20-fold more potent on the α6β4 constructs than on α3β4 and β4α3β4α3α5(D) nAChRs (Fig. 2). Thus, this ligand can be used to selectively inhibit α3β4 and α6β4 nAChRs while only minimally inhibiting those that contain the β2 subunit.
Fig. 2.
Pharmacological profile of α-Ctx BuIA(T5A,P6O) for inhibition of human nAChRs expressed in Xenopus oocytes. Oocytes expressing different nAChR subtypes were subjected to two-electrode voltage-clamp electrophysiology, as described in Materials and Methods. BuIA(T5A,P6O) inhibited α6/α3β4 nAChRs with an IC50 of 7.4 (6.5–8.3) nM (n = 4), α6M211L,cytα3β4 with an IC50 of 11.3 (10.1–12.7) nM (n = 4), α3β4 nAChRs with an IC50 of 166 (141–196) nM (n = 4), and β4α3β4α5(D) with an IC50 of 147 (127–171) nM (n = 3). For other subtypes, the average response after a 5-minute static bath exposure to 10 µM BuIA(T5A,P6O) was 96 ± 4% (n = 4) for α3β2, 95 ± 5% (n = 4) for α4β2, 48 ± 4% (n = 3) for α4β4, 77 ± 3% (n = 4) for α6/α3β2β3, and 101 ± 2% (n = 4) for β3α6β2α4β2. The error bars denote the S.E.M., and the values in parentheses denote the 95% confidence interval. For clarity, data for α3β4 and β3α6β2α4β2 have been nudged to the right to avoid overlap.
α-Ctxs BuIA(T5A,P6O) and LvIA(N9R,V10A) Demonstrate That the Predominant Functional Heteromeric nAChRs Expressed in Human Adrenal Chromaffin Cells Only Contain β4 Ligand-Binding Sites.
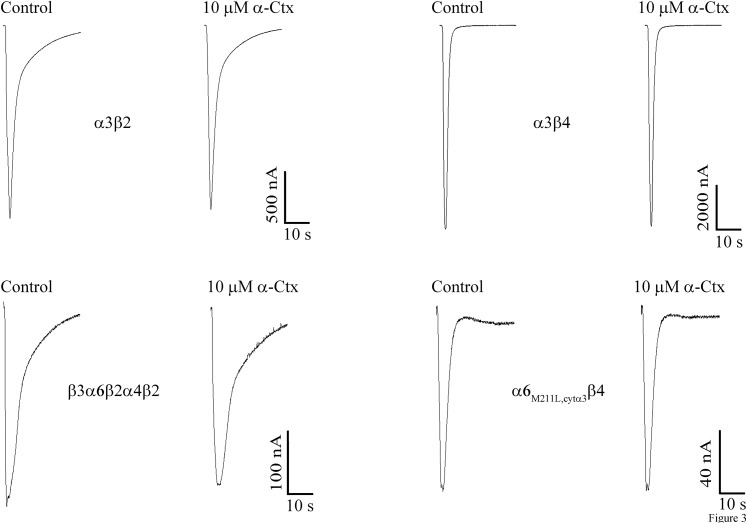
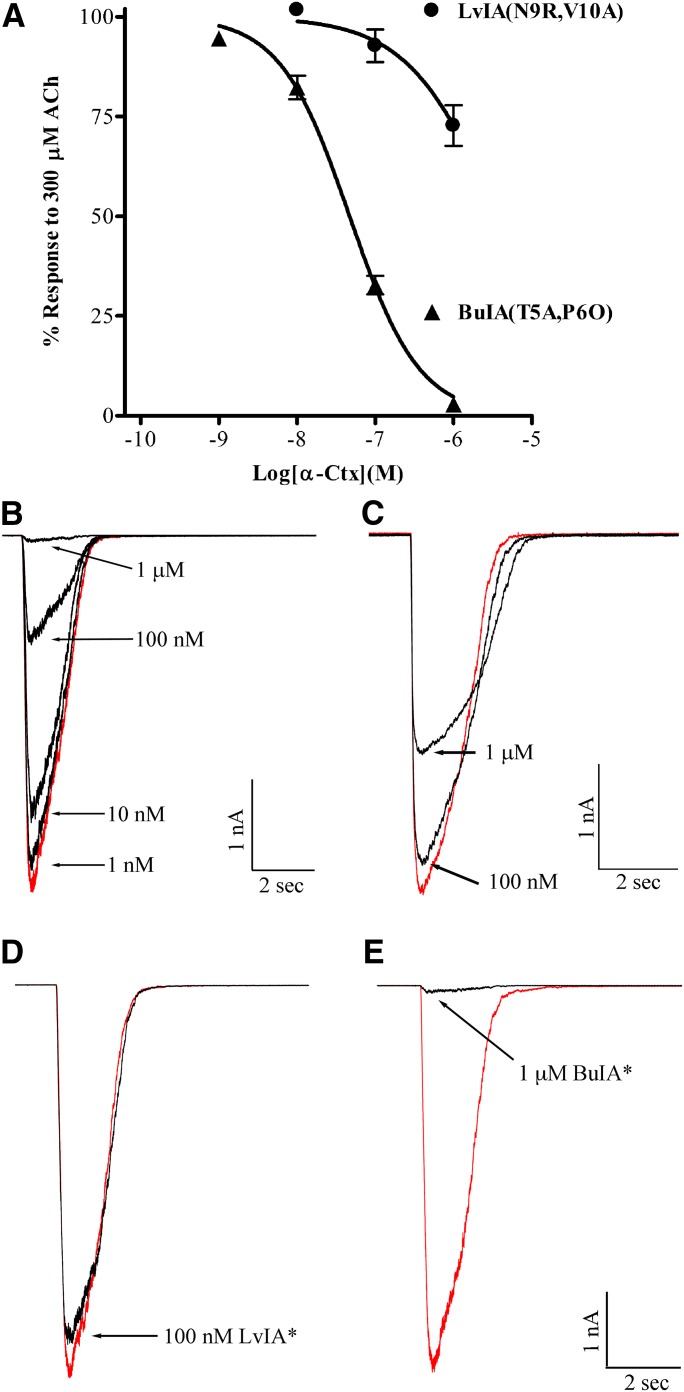
A series of experiments were conducted using the α-Ctx analogs presented in Figs. 1 and 2 to assess the presence of β2 or β4 subunits at the α(+)β(−) subunit interface. We included α-Ctx ArIB(V11L,V16D) in the perfusion solution to ensure that only non-α7 nAChRs were being assessed. This α7 antagonist has previously been shown to be a potent inhibitor of human α7 nAChRs (Innocent et al., 2008). However, because this α-Ctx had not been tested on heteromeric human subtypes, we first tested it on oocyte-expressed nAChRs and found that as for rat nAChRs, ArIB(V11L,V16D) showed very little activity on non-α7 human subtypes even at a concentration of 10 µM (Fig. 3). Therefore, in the presence of 100 nM ArIB(V11L,V16D), any inhibition by BuIA(T5A,P6O) would be attributed to the presence of α3β4 or α6β4 nAChRs. BuIA(T5A,P6O) was then tested in human adrenal chromaffin cells and found to inhibit the ACh-evoked responses with an IC50 value of ∼50 nM (Fig. 4A). At 1 µM, the responses were inhibited by 97 ± 0.6% (n = 7), indicating that most of the heteromeric nAChRs present contained β4 ligand-binding sites (Fig. 4A).
Fig. 3.
Human non-α7 nAChRs are insensitive to α-Ctx ArIB(V11L,V16D). The indicated nAChR subtypes were expressed in X. laevis oocytes and subjected to two-electrode voltage clamp, as described in Materials and Methods. Current traces showing the inhibition of ACh-evoked currents after a 5-minute static bath exposure to 10 µM α-Ctx. The average responses for α3β2, α3β4, β3α6β2α4β2, and α6M211L,cytα3β4 were 93 ± 2% (n = 4), 95 ± 3% (n = 4), 91 ± 11% (n = 3), and 88 ± 2% (n = 4), respectively. Values are the S.E.M.; C, control response to 300 µM ACh prior to application of ArIB(V11L,V16D).
Fig. 4.
α-Ctx BuIA(T5A,P6O), but not LvIA(N9R,V10A), potently inhibits ACh-evoked currents in human adrenal chromaffin cells. Chromaffin cells were isolated from human adrenal glands and subjected to perforated patch-clamp electrophysiology, as described in Materials and Methods. (A) BuIA(T5A,P6O) inhibited ACh-evoked currents with an IC50 of 46.8 (39.8–55.1) nM (n = 7); the Hill slope was −0.98 (0.85–1.1). LvIA(N9R,V10A) inhibited ACh-evoked currents with an IC50 value >1 μM (n = 4). The error bars denote the S.E.M., and the values in parentheses denote the 95% confidence intervals. (B) Representative traces for the inhibition of ACh-evoked currents by increasing concentrations of BuIA(T5A,P6O) and (C) LvIA(N9R,V10A). (D and E) Representative traces showing the inhibition of ACh-evoked currents by both α-Ctxs when tested in the same cell. The LvIA analog inhibited ACh-evoked currents by 7 ± 2% (n = 6), and after washout the BuIA analog inhibited the currents by 98.0 ± 0.3% (n = 6). Values are S.E.M. In (D and E), the LvIA and BuIA analogs are denoted with an asterisk for brevity. Control responses to ACh in the absence of α-Ctxs are shown in red.
Next we used LvIA(N9R,V10A) to determine whether receptors containing β2 ligand-binding sites were present. As shown in Fig. 4A, significant inhibition of the ACh-evoked current by LvIA(N9R,V10A) was only observed at concentrations greater than those required for inhibition of β2-containing nAChRs (>100 nM). Examples of current traces showing the inhibition produced by the two α-Ctxs are shown in Fig. 4, B and C. Most heteromeric human nAChR subtypes, with the exception of the α9α10 subtype, fall into two categories that contain either β2 or β4 subunits, but mixed subtypes that contain both β2 and β4 ligand-binding sites have been shown to be present in several brain regions in rodents (Turner and Kellar, 2005; Azam and McIntosh, 2006; Grady et al., 2009; Whiteaker et al., 2009). Binding of a single α-Ctx molecule to a nAChR subunit interface is sufficient to block the allosteric transitions required for ion channel opening (Talley et al., 2006), and binding to a target site can occur with high affinity without regard to composition of other binding sites in the receptor complex. Examples include inhibition of one of five sites in an α7 homopentamer, binding to the α-ε or α-δ interface in a muscle type nAChR, or binding to the α6-β2 interface in an α6β2α4β2β3 nAChR (Groebe et al., 1995; Sine et al., 1995; Jacobsen et al., 1999; Gotti et al., 2005; Teichert et al., 2005). Thus, the affinities of the LvIA and BuIA analogs for their respective binding would be expected to be similar for a mixed β2 and β4 nAChR subtype. We tested the LvIA and the BuIA analogs on the same cells to probe for nAChR with mixed β2 and β4 ligand-binding sites and observed very little inhibition by LvIA(N9R,V10A) at 100 nM (Fig. 4D). After washout, the responses were nearly completely inhibited by 1 µM BuIA(T5A,P6O) (Fig. 4E). These experiments suggest that human adrenal chromaffin cells are unlikely to express significant levels of functional heteromeric nAChRs on the cell surface that contains β2 ligand-binding sites. These experiments, however, do not exclude the possibility of a β2 subunit in the auxiliary fifth position of the α3β4* nAChR complex.
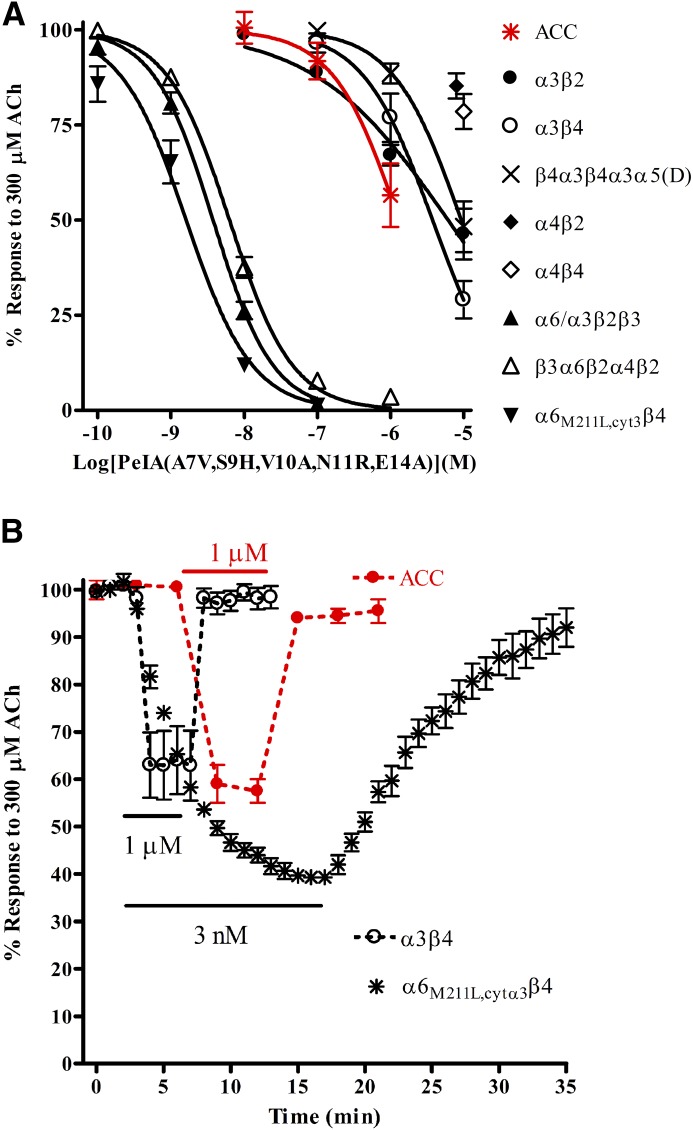
Human adrenal chromaffin cells have been reported to express α6β4* nAChRs (Perez-Alvarez et al., 2012b), yet there are some inconsistencies between α-Ctx IC50 values for inhibition of human adrenal chromaffin cell nAChRs and values for human α6/α3β4 nAChRs heterologously expressed in Xenopus oocytes (Hernandez-Vivanco et al., 2014). Similarly, during the course of the testing of BuIA(T5A,P6O) and LvIA(N9R,V10A), we observed that the IC50 values we obtained in human chromaffin cells were also somewhat different than the values we obtained using the α6β4 constructs expressed in Xenopus oocytes (Figs. 1, 2, and 5). Thus, we chose an additional α-Ctx, namely PeIA(A7V,S9H,V10A,N11R,E14A) (Hone et al., 2013), which has been demonstrated to be 275-fold more potent on rat α6β4 than α3β4 nAChRs expressed in Xenopus oocytes and thus offers superior selectivity for α6β4 over α3β4 compared with BuIA(T5A,P6O). The activity of this ligand on human nAChR subtypes had also not previously been assessed; therefore, we tested the peptide on a panel of human nAChRs expressed in Xenopus oocytes to ensure that this peptide retained its selectivity for human α6-containing subtypes. PeIA(A7V,S9H,V10A,N11R,E14A) potently inhibited human α6M211L,cytα3β4 nAChRs with an IC50 value of 1.6 nM, a value >2900-fold lower than the IC50 value for the α3β4 subtype (Fig. 5A). The peptide also inhibited α6/α3β2β3 and β3α6β2α4β2 receptors with low nM potencies, but μM concentrations were required to inhibit the α3β2 subtype (Fig. 5A), confirming that the peptide retained its selectivity for human α6-containing subtypes. When we tested this PeIA analog for inhibition of the ACh-evoked currents in human adrenal chromaffin cells, inhibition was observed only at concentrations of 100 nM or higher, suggesting that there were few α6β4 nAChRs present (Fig. 5A). In addition to the difference in IC50 values between oocyte-expressed human α3β4 and α6M211L,cytα3β4 nAChRs, the kinetics for inhibition were strikingly different. Inhibition equilibrium of α3β4 nAChRs by IC50 concentrations of PeIA(A7V,S9H,V10A,N11R,E14A) required ∼1 minute of toxin exposure, whereas inhibition of α6M211L,cytα3β4 nAChRs required ∼15 minutes to reach steady state equilibrium (Fig. 5B). Furthermore, recovery from inhibition of α6M211L,cytα3β4 nAChRs was markedly slower than for α3β4 nAChRs (Fig. 5B). The kinetics of inhibition and recovery from inhibition of the ACh-evoked responses in human adrenal chromaffin cells closely matched the time frames observed for α3β4 nAChRs expressed in Xenopus oocytes (Fig. 5B). A comparison of the α-Ctx potencies for the various human nAChR subtypes expressed in Xenopus oocytes as well as their activities in human adrenal chromaffin cells is shown in Table 2.
Fig. 5.
α-Ctx PeIA(A7V,S9H,V10A,N11R,E14A) is selective for human α6M211L,α3cytβ4 over α3β4 nAChRs expressed in Xenopus oocytes and identifies the α3β4* subtype as the main nAChR subtype expressed by human adrenal chromaffin cells. Oocytes expressing different nAChR subtypes were subjected to two-electrode voltage-clamp electrophysiology, as described in Materials and Methods. (A) PeIA(A7V,S9H,V10A,N11R,E14A) inhibited α6M211L,cytα3β4 nAChRs with an IC50 of 1.6 (1.1–2.2) nM (n = 4), α6/α3β2β3 with an IC50 of 3.8 (3.2–4.5) nM (n = 4), and β3α6β2α4β2 with an IC50 of 6.3 (5.6–7.1) nM (n = 4). The α3β2, α3β4, and β4α3β4α3α5(D) subtypes were inhibited with IC50 values of 6.1 (3.6–10.3) µM, 4.7 (3.5–6.2) µM, and 9.2 (6.1–13.4) µM, respectively (n = 4 for all). For α4β2 and α4β4, the average responses after a 5-minute static bath exposure to 10 µM peptide were 85 ± 2% (n = 4) and 79 ± 5% (n = 4), respectively. (A) Human adrenal chromaffin cells were subjected to patch-clamp electrophysiology, as described in Materials and Methods. The cells were perfused with increasing concentrations of the PeIA analog, and the ACh-evoked currents were monitored for inhibition. The IC50 value for inhibition of these currents was estimated to be greater than the maximal concentration tested (1 µM) (n = 6). (B) Oocytes expressing α3β4 or α6M211L,cytα3β4 nAChRs were perfused with 1 µM and 3 nM PeIA(A7V,S9H,V10A,N11R,E14A), respectively, until steady state inhibition was observed and then perfused with ND96 only, and the responses were monitored for recovery. Complete inhibition equilibrium of α3β4 nAChRs was obtained in ∼1, and complete recovery occurred in <2 minutes. In contrast, steady state inhibition of α6M211L,cytα3β4 required ∼15 minutes, and recovery from inhibition required >15 minutes. The error bars denote the S.E.M., and the values in parentheses denote the 95% confidence interval; ACC, human adrenal chromaffin cells.
TABLE 2.
IC50 values for inhibition of human nAChRs expressed in Xenopus oocytes and adrenal chromaffin cell nAChRs
| BuIA(T5A,P6O) | LvIA(N9R,V10A) | PeIA(A7V,S9H,V10A,N11R,E14A) | |
|---|---|---|---|
| α3β2 | >10 μM | 3.3 (2.4–4.7) nM | 6.1 (3.6–10.3) µM |
| α3β4 | 166 (141–196) nM | >10 μM | 3.7 (2.4–5.8) µM |
| β4α3β4α3α5(D) | 147 (125–173) nM | >10 μM | 9.2 (6.4–13.4) µM |
| α4β2 | >10 μM | 195 (133–284) nM | >10 μM |
| α4β4 | >10 μM | >10 μM | >10 μM |
| α6/α3β2β3 | >10 μM | 13.5 (8.6–21.2) nM | 3.8 (3.2–4.5) nM |
| β3α6β2α4β2 | >10 μM | 11.4 (8.1–16.0) nM | 6.3 (5.6–7.1) nM |
| α6/α3β4 | 7.4 (6.5–8.3) nM | 1.0 (7.5–13.3) µM | N.D. |
| α6M211L,cytα3β4 | 11.3 (10.1–12.7) nM | 2.8 (2.5–3.3) μM | 1.6 (1.2–2.2) nM |
| ACC | 46.7 (39.8–55.1) nM | >1 μM | >1 μM |
ACC, adrenal chromaffin cells.
Values in parentheses indicate the 95% confidence interval.
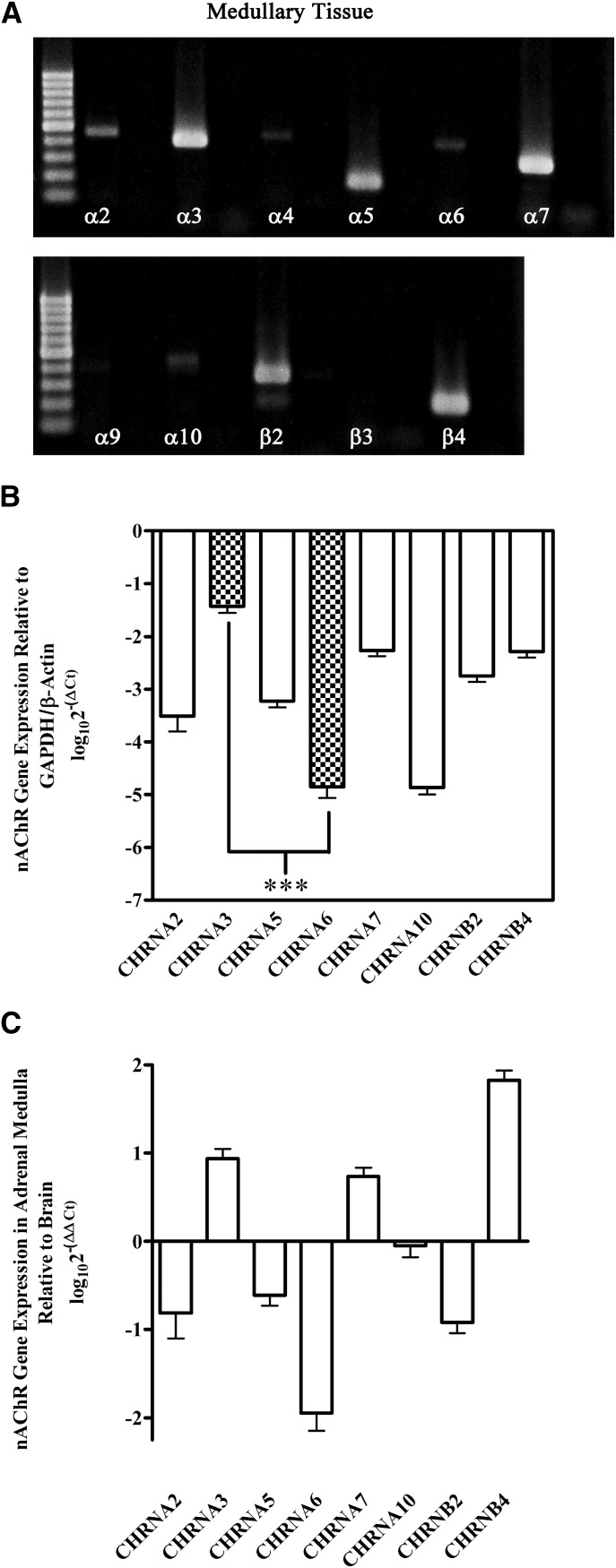
Human Adrenal Medulla Gland and Cultured Adrenal Chromaffin Cells Predominantly Express mRNAs for α3, α5, α7, β2, and β4 Subunits.
The pharmacology experiments suggested that the predominant nAChR expressed by human adrenal chromaffin cells was the α3β4 subtype and that nAChRs containing the α6 subunit were likely few in number. As an additional confirmation, we assessed human adrenal gland tissue for the expression of nAChR subunit mRNAs using both end-point and quantitative real-time PCR methodologies. Total RNA extracted from pieces of adrenal medulla tissue was reverse transcribed into cDNA and subjected to end-point PCR to qualitatively probe for the expression of nAChR subunit mRNAs, as described in Materials and Methods. Figure 6A shows the presence of mRNAs for multiple nAChR subunits, including α2, α3, α4, α5, α6, α7, α10, β2, and β4 subunits. However, signals for α3, α5, α7, β2, and β4 were relatively stronger than the signals for the other subunits. qPCR was used to quantify and extend the results presented in Fig. 6A. Similar to the end-point RT-PCR results, transcripts for α3, α7, and β4 subunits were found to be the most abundant species present (Fig. 6B). Transcripts for α5 and β2 were somewhat less abundant, whereas those for α2, α6, and α10 were nearly absent (Fig. 6B). Transcripts for α4, α9, and β3 were detected infrequently (data not shown). Internal controls were also performed by comparing the expression levels of α2, α3, α4, α5, α6, α7, α10, β2, and β4 subunits in adrenal medullary tissue to human brain. These experiments indicated that, in adrenal gland, transcripts for α3 were more abundant compared with α6, whereas in human brain α6 were more abundant than α3 (Fig. 6C).
Fig. 6.
PCR analysis of human adrenal medulla tissue demonstrates the presence of mRNAs for multiple nAChR subunits. (A) Strong signals for nAChR subunit transcripts were detected for α3, α5, α7, β2, and β4 subunits and relatively weak signals for α2, α4, α6, and α10 subunits. Transcripts for α9 and β3 were not detected under the conditions used in this study. Negative control results for reactions performed in the absence of cDNA template are shown in the lane immediately to the right of each respective subunit. Equal volumes of all reactions were loaded on the gel. The molecular weight ladder is shown and denotes size of the amplicon in numbers of base pairs (100-bp increments). Expected sizes for each amplicon are listed in Table 3. The data shown were obtained from one adrenal medulla (n = 3). (B) qPCR analysis quantitatively confirmed and extended the results presented in (A). Transcripts for α3, α7, and β4 were the three most abundant mRNA species present, followed by α5 and β2. Transcripts for α2, α6, and α10 were weakly detected, whereas those for α4, α9, and β3 were infrequently detected (data not shown). Transcripts for α3 were on average 3.4 ± 0.2 (n = 4) orders of magnitude more abundant than α6. Data were normalized to the expression levels of reference genes using the log10(2−ΔCq) method, as described in Materials and Methods. (C) Comparison of the relative abundance of mRNA transcripts in adrenal medulla with human brain. Transcripts for α3 were more abundant than those for α6 in adrenal medulla, whereas in brain tissue α6 was the more abundant species. Data were normalized to the expression levels in human brain using the log10(2−ΔΔCq) method, as described in Materials and Methods. In (B and C), the error bars represent the S.E.M. of four experiments using adrenal glands from four individual donors. Statistical significance was determined using an analysis of variance and Bonferroni test (***P < 0.001).
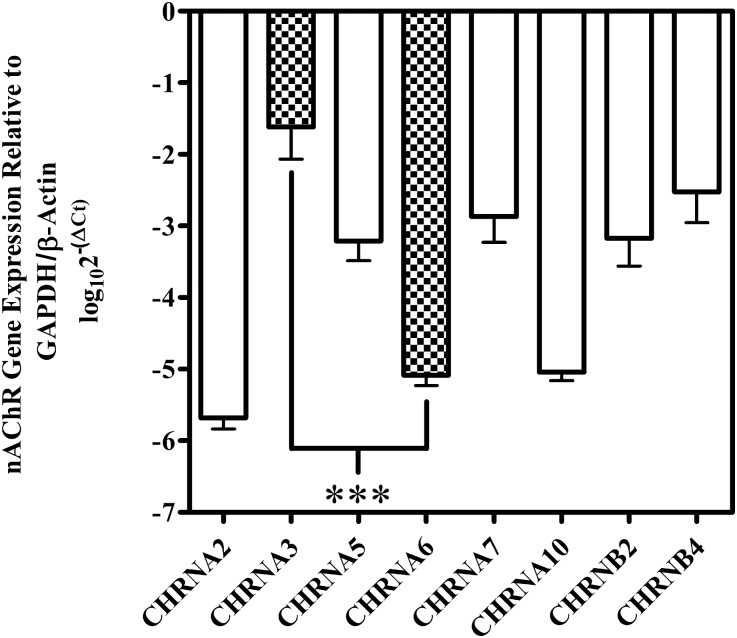
Changes in gene expression may contribute to the differences in mRNAs present in medullary tissue versus functionally expressed nAChRs in cultured adrenal chromaffin cells. Additionally, immune cells, which are known to express different nAChR subunit mRNAs (Peng et al., 2004; Mikulski et al., 2010), may have been present in the medullary tissue that we assessed. Thus, we reassessed the PCR results by performing the experiments on adrenal chromaffin cells isolated and cultured according to the methods used to obtain cells for electrophysiology experiments. In some cultures, 10 µM AraC was added to inhibit proliferating cells. On day 2, the cells were washed extensively with extracellular solution to remove any nonadhering cells before harvesting the mRNA. qPCR was then performed, and, similar to the results found in adrenal medulla, the two most abundant mRNA species were α3 and β4 (Fig. 7). Transcripts for α5, α7, and β2 were also abundant, whereas those for α2, α6, and α10 were nearly absent. Transcripts for α4, α9, and β3 were detected infrequently (data not shown). These experiments corroborate the electrophysiology experiments identifying α3β4* as the predominant heteromeric nAChR subtype expressed by human adrenal chromaffin cells.
Fig. 7.
qPCR analysis of cultured human adrenal chromaffin cells. Transcripts for α3, α7, and β4 were the three most highly abundant mRNA species present in cultured adrenal chromaffin cells, followed by α5 and β2. Transcripts for α2, α6, and α10 were weakly detected, whereas those for α4, α9, and β3 were infrequently detected (data not shown). Transcripts for α3 were on average 3.5 ± 0.5 (n = 3) orders of magnitude more abundant than α6. Data were normalized to the expression levels of reference genes using the log10(2−ΔCq) method, as described in Materials and Methods. The error bars represent the S.E.M. of three experiments using cell cultures of adrenal glands from three individual donors. Statistical significance was determined using an analysis of variance and Bonferroni test (***P < 0.001).
Discussion
α-Ctxs are widely used as pharmacological tools to study nAChRs. We have observed that some α-Ctxs that distinguish well among rat α3β2, α6β2, and α6β4 nAChRs fail to distinguish among the equivalent human subtypes. We took advantage of the unique sequence of the recently discovered α-Ctx LvIA, and, based on our previous work with α-Ctx PeIA, synthesized a new analog that is a potent and highly selective antagonist of human α3β2 and α6β2 nAChRs. This analog, LvIA(N9R,V10A), is particularly selective (>3000-fold) for α3β2 over α3β4 nAChRs, but also has excellent selectivity (∼165-fold) for α6β2-containing nAChRs over the α6β4 nAChR constructs (Fig. 1). Thus, LvIA(N9R,V10A) should prove to be a highly useful tool for identifying human α3β2 and α6β2 nAChRs in cells that potentially express both β2 and β4 subunit-containing subtypes.
In conjunction with three other α-Ctx analogs, LvIA(N9R,V10A) facilitated the identification of α3β4* as the predominant functional nAChR subtype expressed by human adrenal chromaffin cells. We arrived at this conclusion based on results obtained using a combination of pharmacology and molecular biology. BuIA(T5A,P6O) is a selective antagonist of oocyte-expressed human α3β4 and α6β4 nAChR constructs and is essentially inactive on all other subtypes at concentrations ≤1 µM (Fig. 2). The fact that across all experiments BuIA(T5A,P6O) (1 μM) inhibited 97 ± 1% (n = 13) of the ACh-evoked current in human chromaffin cells suggests that these cells express few nAChRs with β2 ligand-binding sites only (Fig. 4). This conclusion is supported by the observation that the currents were insensitive to inhibition by LvIA(N9R,V10A) at concentrations selective for α3β2 and α6β2 subtypes (Fig. 4A). Furthermore, when both α-Ctxs were sequentially tested on the same cells, the LvIA analog inhibited the ACh-evoked currents by only 7 ± 2% (n = 6), whereas the BuIA analog inhibited the currents by 98.0 ± 0.3% (n = 6) (Fig. 4, D and E). These data suggest that nAChRs with both β2 and β4 ligand-binding sites are also likely to be few in number.
We previously showed that α-Ctx PeIA(A7V,S9H,V10A,N11R,E14A) was >200-fold more potent on rat α6β4 nAChRs than the α3β4 subtype (Hone et al., 2013). Similarly, we found that this peptide was ∼600-fold more potent on human α6M211L,α3cytβ4 than α3β4 nAChRs (Fig. 5A). Thus, the expectation was that it would also be a potent inhibitor of the ACh-evoked currents in human adrenal chromaffin cells because α6β4* nAChRs were previously reported to be present (Perez-Alvarez et al., 2012b). However, substantial inhibition of the ACh-evoked current by PeIA(A7V,S9H,V10A,N11R,E14A) was only observed at 1 μM, a concentration >625-fold higher than the IC50 value for inhibition of α6M211L,α3cytβ4 nAChRs expressed in oocytes (Fig. 5A). This coupled with the absence of significant inhibition by the LvIA analog suggests that the predominant nAChR subtype contains α3-β4 ligand-binding sites only.
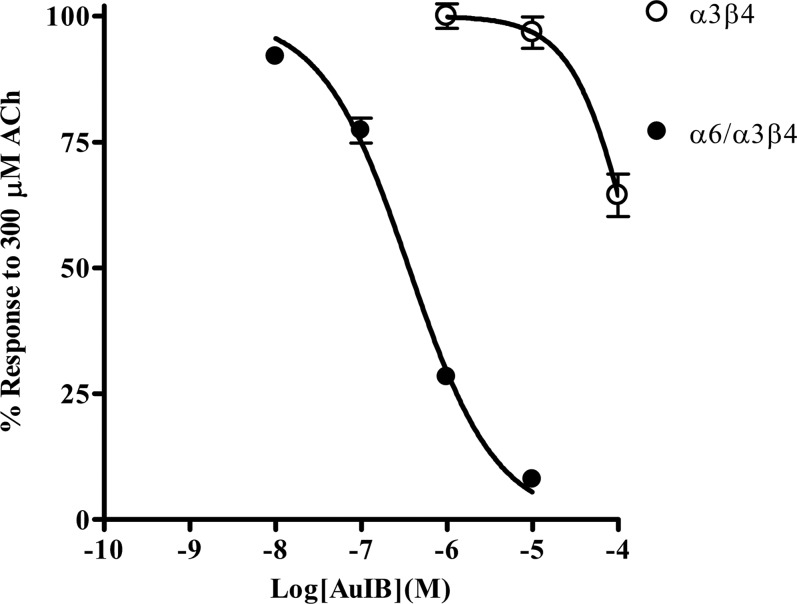
In Pérez-Alvarez et al. (2012b), evidence for the of α6β4* nAChRs was based on the observation that α-Ctxs selective for rat α6-containing nAChRs (McIntosh et al., 2004; Azam et al., 2008) inhibited the ACh-evoked currents in human adrenal chromaffin cells with similar IC50 values. Additionally, the IC50 value for the α3β4 nAChR antagonist α-Ctx AuIB was estimated to be >10 μM, which is inconsistent with the presence of a large population of α3β4 nAChRs based on the IC50 value (750 nM) reported for rat α3β4 nAChRs (Luo et al., 1998). We found that human α3β4 nAChRs are relatively insensitive to inhibition by AuIB, yet human α6/α3β4 nAChRs are 20-fold more sensitive to inhibition (Fig. 9) than rat α6/α3β4 nAChRs (Smith et al., 2013). The lack of human α3β4 nAChR sensitivity to AuIB in oocytes is consistent with the results observed in human adrenal chromaffin cells and further supports our conclusion that these cells predominantly express α3β4* nAChRs. Other α-Ctxs have also been found to be more potent on human β4-containing nAChRs than the rat homologs (Hernandez-Vivanco et al., 2014). In the present study, PeIA(A7V,S9H,V10A,N11R,E14A) was found to be ∼27-fold more potent on human α6M211L,cytα3β4 nAChRs (Fig. 5A) compared with the value reported for rat α6β4 nAChRs (Hone et al., 2013). BuIA(T5A,P6O) is also more potent on both human α3β4 and the α6β4 nAChR constructs (Fig. 2) than the equivalent rat receptors (Azam et al., 2010). A comparison of the IC50 values for inhibition of rat and human α3β4 and α6β4 nAChRs by these α-Ctxs is presented in Table 5.
Fig. 9.
α-Ctx AuIB inhibits human α6/α3β4 nAChRs more potently that α3β4 nAChRs. Oocytes expressing α6/α3β4 or α3β4 nAChR subtypes were subjected to two-electrode voltage-clamp electrophysiology, as described in Materials and Methods. AuIB inhibited α6/α3β4 nAChRs with an IC50 value of 360 (305–424) nM (n = 4) and α3β4 nAChRs with an IC50 value >10 μM (n = 4). The error bars denote the S.E.M., and the values in parentheses denote the 95% confidence interval.
TABLE 5.
Comparison of α-Ctx IC50 values for inhibition of rat versus human nAChRs expressed in Xenopus oocytes
| rα3β4 | hα3β4 | Ratio | rα6β4 | hα6β4 | Ratio | |
|---|---|---|---|---|---|---|
| BuIA(T5A,P6O) | 1.2 μMa | 166 nMb | 7 | 58 nMa | 7 nMb | 8 |
| MII[H9A,L15A] | 7.8 μMa | 1.4 μMc | 6 | 269 nMa | 13 nMc | 21 |
| PeIA(A7V,S9H,V10A,N11R,E14A) | >10 μMd | 3.7 μMb | 3 | 44 nMd | 1.6 nMb | 28 |
| AuIB | 750 nMe | >10 μMb | −3 | 7.3 μMf | 360 nMb | 20 |
h, human; r, rat; ratios compare rat/human.
This work.
Smith et al., 2014.
Species differences in the amino acid sequences of rat α3 and α6 subunits have been shown to influence α-Ctx on- and off-rate kinetics, potencies, and selectivity profiles (Azam et al., 2008; Hone et al., 2013). In the α6 subunit, these differing residues include Glu152, Asp184, and Thr195, which in the α3 subunit are Lys, Glu, and Gln, respectively. Of these three residues, only Glu152 is conserved in the human α3 subunit, whereas the other two residues at positions 184 and 195 are Asp and Pro, respectively (Fig. 8). Interestingly, human and rat α6 subunits also have an Asp at position 184 and thus, in this aspect, the human α3 subunit is more like rat α6 than rat α3. Similar observations have been made regarding the α4 subunit. α-Ctxs in general, including BuIA and its analogs, show very little activity on α4-containing nAChRs. This lack of activity has been shown to be due to the presence of specific residues in the α4 ligand-binding pocket that are not present in other subtypes and that prevent efficient α-Ctx binding (Everhart et al., 2003; Beissner et al., 2012; Kim and McIntosh, 2012). These observations highlight the importance of continued work developing selective ligands that discriminate among the different human nAChR subtypes.
Fig. 8.
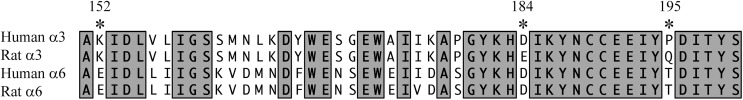
Residues in the ligand-binding domains of rat and human α3 and α6 subunits are only partially conserved between the two species. A sequence comparison of amino acids 150–200 of rat and human α3 and α6 ligand-binding domains was performed. Residues marked with the asterisk and numbered 152, 184, and 195 are important for α-Ctx binding. Note that residue 152 is conserved between rat and human α3 subunits, but not residues 184 and 195.
PCR experiments revealed that human medullary tissue and cultured human adrenal chromaffin cells predominantly express mRNA for the α3 subunit and express relatively low levels of mRNA for the α6 subunit (Figs. 6 and 7). Quantification of the transcripts in medullary tissue and cultured adrenal chromaffin cells by qPCR revealed that α3 transcripts are ∼3.5 orders of magnitude more abundant than α6 transcripts (Figs. 6B and 7). Noteworthily, the concordance across tissue versus cell culture and PCR methods was exceptionally high. Thus, together with the pharmacology experiments, converging lines of evidence strongly suggest that most heteromeric nAChRs in human chromaffin cells are of the α3β4* subtype.
nAChRs composed of α and β subunits contain two putative ligand-binding sites composed of the primary site, contributed by the α subunit, and the complementary site, contributed by the β subunit. Hence, in a receptor composed of α3 and β4 subunits, there are expected to be two pharmacologically similar ligand-binding sites. However, some receptor subtypes, such as those found on nigrostriatal dopaminergic terminals composed of α4, α6, and β2 subunits, contain two ligand-binding sites that are pharmacologically distinct (Gotti et al., 2005; Quik et al., 2005). This does not appear to be the case with the α3β4* nAChRs in human adrenal chromaffin cells. We conclude that the α3β4* nAChRs in these cells possess two α3-β4 ligand-binding interfaces. However, signals for both β2 and α5 subunits were detected in PCR experiments (Figs. 6 and 7), suggesting that these subunits may be present in the α3β4* nAChR complex. The α5 subunit has been detected in bovine adrenal chromaffin cells, where it has been suggested to form α3β4α5 nAChRs (Campos-Caro et al., 1997) and rat adrenal chromaffin cells have been reported to express α3β2 nAChRs (Di Angelantonio et al., 2003). If β2 subunits are present in human adrenal chromaffin cell α3β4* nAChRs, they are likely to be restricted to the fifth or auxiliary position because no pharmacological evidence for an α3-β2 ligand-binding site was found. Additionally, β2 subunits may assemble with α7 subunits to form α7β2 nAChRs, which have recently been identified in rodent and human basal forebrain as well as mouse hippocampal interneurons (Liu et al., 2009b, 2012; Moretti et al., 2014).
In conclusion, this study describes the development of a novel pharmacological tool, LvIA(N9R,V10A), that can be used to selectively target human α3β2 and α6β2 nAChRs. We demonstrated its utility by using it in conjunction with a panel of α-Ctxs to characterize the nAChR subtypes expressed by a cell that was thought to potentially express multiple subtypes. Data from previous studies suggested that human adrenal chromaffin cells may express both α3β4 and α6β4 nAChRs as well as subtypes with β2 ligand-binding sites (Perez-Alvarez et al., 2012b). In this study, we present evidence that human adrenal chromaffin cells in fact predominantly express nAChRs with only α3-β4 ligand-binding sites. Thus, these studies not only clarify the nAChR expression in human adrenal chromaffin cells, but also suggest that the stoichiometry of the predominant nAChR is likely (α3β4)2 with an auxiliary fifth subunit that is yet to be identified. Future experiments are clearly warranted to identify this fifth subunit because auxiliary subunits can play an important role in determining agonist sensitivity and calcium permeability (Gerzanich et al., 1998; Brown et al., 2007; Sciaccaluga et al., 2015). Combined approaches using selective immunoprecipitation, radioligand binding, and immunohistochemistry may be particularly helpful in this regard (Whiteaker et al., 2000; Gotti et al., 2005; Grady et al., 2009; Lomazzo et al., 2010; Marks et al., 2011).
Acknowledgments
The authors thank Bob Shackman at the DNA/Peptide Synthesis Core Facility (University of Utah), Joanna Gajewiak (University of Utah) for assistance with peptide synthesis, Sean Christensen (University of Utah) for assistance with the peptide folding, and William Lowe at the Salk Institute for performing the matrix-assisted laser desorption ionization/time-of-flight mass spectrometry mass spectrometry.
Abbreviations
- α-Ctx
α-conotoxin
- ACh
acetylcholine
- nAChR
nicotinic acetylcholine receptor
- PCR
polymerase chain reaction
- qPCR
quantitative real-time PCR
- RT-PCR
reverse-transcription PCR
- SEM
standard error of the mean
- SDM
standard deviation of the mean
Authorship Contributions
Participated in research design: Hone, McIntosh, Albillos.
Conducted experiments: Hone, Azam, Lucero.
Contributed new reagents or analytic tools: Lindstrom, Whiteaker, Passas, Blázquez.
Performed data analysis: Hone, Lucero.
Wrote or contributed to the writing of the manuscript: Hone, McIntosh, Lucero, Whiteaker, Albillos.
Footnotes
A.J.H. holds a Marie Curie International Fellowship from the European Commission [NRHACC-329966]. This work was also supported by Spanish Ministerio de Ciencia y Tecnología [Grant BFU2012-30997 to A.A.]; National Institutes of Health [Grants GM-103801 and GM-48677 to J.M.M., Grant DA030929 to J.L., and Grants R21 DA026627, R21 DA027070S, and R01 DA012242 to P.W.]; and Barrow Neurological Foundation [to P.W.].
References
- Albuquerque EX, Pereira EF, Alkondon M, Rogers SW. (2009) Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol Rev 89:73–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Ørntoft TF. (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250. [DOI] [PubMed] [Google Scholar]
- Azam L, Maskos U, Changeux JP, Dowell CD, Christensen S, De Biasi M, McIntosh JM. (2010) α-Conotoxin BuIA[T5A;P6O]: a novel ligand that discriminates between α6ß4 and α6ß2 nicotinic acetylcholine receptors and blocks nicotine-stimulated norepinephrine release. FASEB J 24:5113–5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam L, McIntosh JM. (2006) Characterization of nicotinic acetylcholine receptors that modulate nicotine-evoked [3H]norepinephrine release from mouse hippocampal synaptosomes. Mol Pharmacol 70:967–976. [DOI] [PubMed] [Google Scholar]
- Azam L, Yoshikami D, McIntosh JM. (2008) Amino acid residues that confer high selectivity of the alpha6 nicotinic acetylcholine receptor subunit to alpha-conotoxin MII[S4A,E11A,L15A]. J Biol Chem 283:11625–11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissner M, Dutertre S, Schemm R, Danker T, Sporning A, Grubmüller H, Nicke A. (2012) Efficient binding of 4/7 α-conotoxins to nicotinic α4β2 receptors is prevented by Arg185 and Pro195 in the α4 subunit. Mol Pharmacol 82:711–718. [DOI] [PubMed] [Google Scholar]
- Brown RW, Collins AC, Lindstrom JM, Whiteaker P. (2007) Nicotinic alpha5 subunit deletion locally reduces high-affinity agonist activation without altering nicotinic receptor numbers. J Neurochem 103:204–215. [DOI] [PubMed] [Google Scholar]
- Campos-Caro A, Smillie FI, Domínguez del Toro E, Rovira JC, Vicente-Agulló F, Chapuli J, Juíz M, Sala S, Sala F, Ballesta JJ, et al. (1997) Neuronal nicotinic acetylcholine receptors on bovine chromaffin cells: cloning, expression, and genomic organization of receptor subunits. J Neurochem 68:488–497. [DOI] [PubMed] [Google Scholar]
- Carlisle DL, Hopkins TM, Gaither-Davis A, Silhanek MJ, Luketich JD, Christie NA, Siegfried JM. (2004) Nicotine signals through muscle-type and neuronal nicotinic acetylcholine receptors in both human bronchial epithelial cells and airway fibroblasts. Respir Res 5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier GE, Yoshikami D, Gray WR, Luo S, Olivera BM, McIntosh JM. (1996) A new alpha-conotoxin which targets alpha3beta2 nicotinic acetylcholine receptors. J Biol Chem 271:7522–7528. [DOI] [PubMed] [Google Scholar]
- Criado M, Domínguez del Toro E, Carrasco-Serrano C, Smillie FI, Juíz JM, Viniegra S, Ballesta JJ. (1997) Differential expression of alpha-bungarotoxin-sensitive neuronal nicotinic receptors in adrenergic chromaffin cells: a role for transcription factor Egr-1. J Neurosci 17:6554–6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Angelantonio S, Matteoni C, Fabbretti E, Nistri A. (2003) Molecular biology and electrophysiology of neuronal nicotinic receptors of rat chromaffin cells. Eur J Neurosci 17:2313–2322. [DOI] [PubMed] [Google Scholar]
- Everhart D, Reiller E, Mirzoian A, McIntosh JM, Malhotra A, Luetje CW. (2003) Identification of residues that confer alpha-conotoxin-PnIA sensitivity on the alpha 3 subunit of neuronal nicotinic acetylcholine receptors. J Pharmacol Exp Ther 306:664–670. [DOI] [PubMed] [Google Scholar]
- Gahring LC, Myers E, Palumbos S, Rogers SW. (2014) Nicotinic receptor Alpha7 expression during mouse adrenal gland development. PLoS One 9:e103861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George AA, Lucero LM, Damaj MI, Lukas RJ, Chen X, Whiteaker P. (2012) Function of human α3β4α5 nicotinic acetylcholine receptors is reduced by the α5(D398N) variant. J Biol Chem 287:25151–25162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerzanich V, Wang F, Kuryatov A, Lindstrom J. (1998) alpha 5 Subunit alters desensitization, pharmacology, Ca++ permeability and Ca++ modulation of human neuronal alpha 3 nicotinic receptors. J Pharmacol Exp Ther 286:311–320. [PubMed] [Google Scholar]
- Gotti C, Moretti M, Clementi F, Riganti L, McIntosh JM, Collins AC, Marks MJ, Whiteaker P. (2005) Expression of nigrostriatal alpha 6-containing nicotinic acetylcholine receptors is selectively reduced, but not eliminated, by beta 3 subunit gene deletion. Mol Pharmacol 67:2007–2015. [DOI] [PubMed] [Google Scholar]
- Grady SR, Moretti M, Zoli M, Marks MJ, Zanardi A, Pucci L, Clementi F, Gotti C. (2009) Rodent habenulo-interpeduncular pathway expresses a large variety of uncommon nAChR subtypes, but only the alpha3beta4* and alpha3beta3beta4* subtypes mediate acetylcholine release. J Neurosci 29:2272–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groebe DR, Dumm JM, Levitan ES, Abramson SN. (1995) alpha-Conotoxins selectively inhibit one of the two acetylcholine binding sites of nicotinic receptors. Mol Pharmacol 48:105–111. [PubMed] [Google Scholar]
- Hernández-Vivanco A, Hone AJ, Scadden ML, Carmona-Hidalgo B, McIntosh JM, Albillos A. (2014) Monkey adrenal chromaffin cells express α6β4* nicotinic acetylcholine receptors. PLoS One 9:e94142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hone AJ, Meyer EL, McIntyre M, McIntosh JM. (2012a) Nicotinic acetylcholine receptors in dorsal root ganglion neurons include the α6β4* subtype. FASEB J 26:917–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hone AJ, Ruiz M, Scadden M, Christensen S, Gajewiak J, Azam L, McIntosh JM. (2013) Positional scanning mutagenesis of α-conotoxin PeIA identifies critical residues that confer potency and selectivity for α6/α3β2β3 and α3β2 nicotinic acetylcholine receptors. J Biol Chem 288:25428–25439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hone AJ, Scadden M, Gajewiak J, Christensen S, Lindstrom J, McIntosh JM. (2012b) α-Conotoxin PeIA[S9H,V10A,E14N] potently and selectively blocks α6β2β3 versus α6β4 nicotinic acetylcholine receptors. Mol Pharmacol 82:972–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hone AJ, Whiteaker P, Christensen S, Xiao Y, Meyer EL, McIntosh JM. (2009) A novel fluorescent alpha-conotoxin for the study of alpha7 nicotinic acetylcholine receptors. J Neurochem 111:80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocent N, Livingstone PD, Hone A, Kimura A, Young T, Whiteaker P, McIntosh JM, Wonnacott S. (2008) Alpha-conotoxin Arenatus IB[V11L,V16D] [corrected] is a potent and selective antagonist at rat and human native alpha7 nicotinic acetylcholine receptors. J Pharmacol Exp Ther 327:529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen RB, DelaCruz RG, Grose JH, McIntosh JM, Yoshikami D, Olivera BM. (1999) Critical residues influence the affinity and selectivity of alpha-conotoxin MI for nicotinic acetylcholine receptors. Biochemistry 38:13310–13315. [DOI] [PubMed] [Google Scholar]
- Kim HW, McIntosh JM. (2012) α6 nAChR subunit residues that confer α-conotoxin BuIA selectivity. FASEB J 26:4102–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuryatov A, Lindstrom J. (2011) Expression of functional human α6β2β3* acetylcholine receptors in Xenopus laevis oocytes achieved through subunit chimeras and concatamers. Mol Pharmacol 79:126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuryatov A, Olale F, Cooper J, Choi C, Lindstrom J. (2000) Human alpha6 AChR subtypes: subunit composition, assembly, and pharmacological responses. Neuropharmacology 39:2570–2590. [DOI] [PubMed] [Google Scholar]
- Kurzen H, Berger H, Jäger C, Hartschuh W, Näher H, Gratchev A, Goerdt S, Deichmann M. (2004) Phenotypical and molecular profiling of the extraneuronal cholinergic system of the skin. J Invest Dermatol 123:937–949. [DOI] [PubMed] [Google Scholar]
- Ley CK, Kuryatov A, Wang J, Lindstrom JM. (2014) Efficient expression of functional (α6β2)2β3 AChRs in Xenopus oocytes from free subunits using slightly modified α6 subunits. PLoS One 9:e103244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lips KS, Brüggmann D, Pfeil U, Vollerthun R, Grando SA, Kummer W. (2005) Nicotinic acetylcholine receptors in rat and human placenta. Placenta 26:735–746. [DOI] [PubMed] [Google Scholar]
- Liu J, McGlinn AM, Fernandes A, Milam AH, Strang CE, Andison ME, Lindstrom JM, Keyser KT, Stone RA. (2009a) Nicotinic acetylcholine receptor subunits in rhesus monkey retina. Invest Ophthalmol Vis Sci 50:1408–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Huang Y, Shen J, Steffensen S, Wu J. (2012) Functional α7β2 nicotinic acetylcholine receptors expressed in hippocampal interneurons exhibit high sensitivity to pathological level of amyloid β peptides. BMC Neurosci 13:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Huang Y, Xue F, Simard A, DeChon J, Li G, Zhang J, Lucero L, Wang M, Sierks M, et al. (2009b) A novel nicotinic acetylcholine receptor subtype in basal forebrain cholinergic neurons with high sensitivity to amyloid peptides. J Neurosci 29:918–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. [DOI] [PubMed] [Google Scholar]
- Lomazzo E, MacArthur L, Yasuda RP, Wolfe BB, Kellar KJ. (2010) Quantitative analysis of the heteromeric neuronal nicotinic receptors in the rat hippocampus. J Neurochem 115:625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Kulak JM, Cartier GE, Jacobsen RB, Yoshikami D, Olivera BM, McIntosh JM. (1998) alpha-conotoxin AuIB selectively blocks alpha3 beta4 nicotinic acetylcholine receptors and nicotine-evoked norepinephrine release. J Neurosci 18:8571–8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Nguyen TA, Cartier GE, Olivera BM, Yoshikami D, McIntosh JM. (1999) Single-residue alteration in alpha-conotoxin PnIA switches its nAChR subtype selectivity. Biochemistry 38:14542–14548. [DOI] [PubMed] [Google Scholar]
- Luo S, Zhangsun D, Schroeder CI, Zhu X, Hu Y, Wu Y, Weltzin MM, Eberhard S, Kaas Q, Craik DJ, et al. (2014) A novel α4/7-conotoxin LvIA from Conus lividus that selectively blocks α3β2 vs. α6/α3β2β3 nicotinic acetylcholine receptors. FASEB J 28:1842–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Zhangsun D, Wu Y, Zhu X, Hu Y, McIntyre M, Christensen S, Akcan M, Craik DJ, McIntosh JM. (2013) Characterization of a novel α-conotoxin from conus textile that selectively targets α6/α3β2β3 nicotinic acetylcholine receptors. J Biol Chem 288:894–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks MJ, McClure-Begley TD, Whiteaker P, Salminen O, Brown RW, Cooper J, Collins AC, Lindstrom JM. (2011) Increased nicotinic acetylcholine receptor protein underlies chronic nicotine-induced up-regulation of nicotinic agonist binding sites in mouse brain. J Pharmacol Exp Ther 337:187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JM, Azam L, Staheli S, Dowell C, Lindstrom JM, Kuryatov A, Garrett JE, Marks MJ, Whiteaker P. (2004) Analogs of alpha-conotoxin MII are selective for alpha6-containing nicotinic acetylcholine receptors. Mol Pharmacol 65:944–952. [DOI] [PubMed] [Google Scholar]
- Mikulski Z, Hartmann P, Jositsch G, Zasłona Z, Lips KS, Pfeil U, Kurzen H, Lohmeyer J, Clauss WG, Grau V, et al. (2010) Nicotinic receptors on rat alveolar macrophages dampen ATP-induced increase in cytosolic calcium concentration. Respir Res 11:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollard P, Seward EP, Nowycky MC. (1995) Activation of nicotinic receptors triggers exocytosis from bovine chromaffin cells in the absence of membrane depolarization. Proc Natl Acad Sci USA 92:3065–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti M, Zoli M, George AA, Lukas RJ, Pistillo F, Maskos U, Whiteaker P, Gotti C. (2014) The novel α7β2-nicotinic acetylcholine receptor subtype is expressed in mouse and human basal forebrain: biochemical and pharmacological characterization. Mol Pharmacol 86:306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Ferris RL, Matthews T, Hiel H, Lopez-Albaitero A, Lustig LR. (2004) Characterization of the human nicotinic acetylcholine receptor subunit alpha (alpha) 9 (CHRNA9) and alpha (alpha) 10 (CHRNA10) in lymphocytes. Life Sci 76:263–280. [DOI] [PubMed] [Google Scholar]
- Pérez-Alvarez A, Hernández-Vivanco A, Alonso Y, Gregorio S, Tabernero A, McIntosh JM, Albillos A. (2012a) Pharmacological characterization of native α7 nicotinic ACh receptors and their contribution to depolarization-elicited exocytosis in human chromaffin cells. Br J Pharmacol 165:908–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Alvarez A, Hernández-Vivanco A, McIntosh JM, Albillos A. (2012b) Native α6β4* nicotinic receptors control exocytosis in human chromaffin cells of the adrenal gland. FASEB J 26:346–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol Lett 26:509–515. [DOI] [PubMed] [Google Scholar]
- Quik M, Vailati S, Bordia T, Kulak JM, Fan H, McIntosh JM, Clementi F, Gotti C. (2005) Subunit composition of nicotinic receptors in monkey striatum: effect of treatments with 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine or L-DOPA. Mol Pharmacol 67:32–41. [DOI] [PubMed] [Google Scholar]
- Sciaccaluga M, Moriconi C, Martinello K, Catalano M, Bermudez I, Stitzel JA, Maskos U, and Fucile S (2015) Crucial role of nicotinic alpha5 subunit variants for Ca2+ fluxes in ventral midbrain neurons. FASEB J. 29:3389-3398. [DOI] [PMC free article] [PubMed]
- Silver N, Best S, Jiang J, Thein SL. (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol 7:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sine SM, Kreienkamp HJ, Bren N, Maeda R, Taylor P. (1995) Molecular dissection of subunit interfaces in the acetylcholine receptor: identification of determinants of alpha-conotoxin M1 selectivity. Neuron 15:205–211. [DOI] [PubMed] [Google Scholar]
- Smith NJ, Hone AJ, Memon T, Bossi S, Smith TE, McIntosh JM, Olivera BM, Teichert RW. (2013) Comparative functional expression of nAChR subtypes in rodent DRG neurons. Front Cell Neurosci 7:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talley TT, Olivera BM, Han KH, Christensen SB, Dowell C, Tsigelny I, Ho KY, Taylor P, McIntosh JM. (2006) Alpha-conotoxin OmIA is a potent ligand for the acetylcholine-binding protein as well as alpha3beta2 and alpha7 nicotinic acetylcholine receptors. J Biol Chem 281:24678–24686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichert RW, Rivier J, Torres J, Dykert J, Miller C, Olivera BM. (2005) A uniquely selective inhibitor of the mammalian fetal neuromuscular nicotinic acetylcholine receptor. J Neurosci 25:732–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JR, Kellar KJ. (2005) Nicotinic cholinergic receptors in the rat cerebellum: multiple heteromeric subtypes. J Neurosci 25:9258–9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, and Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034. [DOI] [PMC free article] [PubMed]
- West KA, Brognard J, Clark AS, Linnoila IR, Yang X, Swain SM, Harris C, Belinsky S, Dennis PA. (2003) Rapid Akt activation by nicotine and a tobacco carcinogen modulates the phenotype of normal human airway epithelial cells. J Clin Invest 111:81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteaker P, Christensen S, Yoshikami D, Dowell C, Watkins M, Gulyas J, Rivier J, Olivera BM, McIntosh JM. (2007) Discovery, synthesis, and structure activity of a highly selective alpha7 nicotinic acetylcholine receptor antagonist. Biochemistry 46:6628–6638. [DOI] [PubMed] [Google Scholar]
- Whiteaker P, Jimenez M, McIntosh JM, Collins AC, Marks MJ. (2000) Identification of a novel nicotinic binding site in mouse brain using [(125)I]-epibatidine. Br J Pharmacol 131:729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteaker P, Wilking JA, Brown RW, Brennan RJ, Collins AC, Lindstrom JM, Boulter J. (2009) Pharmacological and immunochemical characterization of alpha2* nicotinic acetylcholine receptors (nAChRs) in mouse brain. Acta Pharmacol Sin 30:795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]