Abstract
Modeling and simulation of drug disposition has emerged as an important tool in drug development, clinical study design and regulatory review, and the number of physiologically based pharmacokinetic (PBPK) modeling related publications and regulatory submissions have risen dramatically in recent years. However, the extent of use of PBPK modeling by researchers, and the public availability of models has not been systematically evaluated. This review evaluates PBPK-related publications to 1) identify the common applications of PBPK modeling; 2) determine ways in which models are developed; 3) establish how model quality is assessed; and 4) provide a list of publically available PBPK models for sensitive P450 and transporter substrates as well as selective inhibitors and inducers. PubMed searches were conducted using the terms “PBPK” and “physiologically based pharmacokinetic model” to collect published models. Only papers on PBPK modeling of pharmaceutical agents in humans published in English between 2008 and May 2015 were reviewed. A total of 366 PBPK-related articles met the search criteria, with the number of articles published per year rising steadily. Published models were most commonly used for drug-drug interaction predictions (28%), followed by interindividual variability and general clinical pharmacokinetic predictions (23%), formulation or absorption modeling (12%), and predicting age-related changes in pharmacokinetics and disposition (10%). In total, 106 models of sensitive substrates, inhibitors, and inducers were identified. An in-depth analysis of the model development and verification revealed a lack of consistency in model development and quality assessment practices, demonstrating a need for development of best-practice guidelines.
Introduction
Prediction of disposition characteristics of new drug candidates can identify pharmacokinetic (PK) liabilities such as poor bioavailability, high clearance, potential for drug-drug interactions (DDIs), or the need for dose adjustments in special populations (Obach et al., 1997; Jones et al., 2009; Zhao et al., 2011; Chen et al., 2012; Di et al., 2013; Shardlow et al., 2013). Such predictions can help decision making in relation to development progression, dose selection, and clinical study strategies (Obach et al., 1997; Jones et al., 2009, 2015; Rowland et al., 2011; Zhao et al., 2011; Chen et al., 2012; Di et al., 2013; Shardlow et al., 2013). Various allometric scaling, in vitro-to-in vivo extrapolation (IVIVE), and in silico methods have been developed over the years to enable predictions of human pharmacokinetics prior to first in human dosing. More than 30 different methods exist to predict human volume of distribution (Di et al., 2013), including interspecies scaling (Lombardo et al., 2013a) and in silico methods. Generally, in vivo animal data, log P values, plasma protein binding, and blood-to-plasma ratios are used to predict the human steady-state volume of distribution and tissue-to-plasma partitioning (Poulin and Theil, 2000, 2002; Poulin et al., 2001; Berezhkovskiy, 2004; Rodgers et al., 2005; Rodgers and Rowland, 2006). While interspecies scaling allows predictions of the human volume of distribution, its utility in the prediction of human clearance is limited due to species differences in the expression and substrate specificity of drug metabolizing enzymes and transporters (Obach et al., 1997; Di et al., 2013). Instead, IVIVE tools have been developed to predict hepatic bioavailability and whole organ clearances using in vitro intrinsic clearance, protein binding and permeability data and in vivo blood flows (Houston, 1994; Iwatsubo et al., 1997; Obach et al., 1997; Lombardo et al., 2013b; Cho et al., 2014). While further efforts are needed to improve IVIVE, particularly for transporters and non-P450 enzymes, IVIVE has become an important tool in the process of predicting human exposures and effective dosages.
Quantitative methods to predict pharmacokinetics range in complexity from static mechanistic predictions of specific PK parameters to dynamic physiologically based PK (PBPK) models used to predict plasma concentration-time curves. Static mechanistic methods typically use one or two in vitro parameters to predict specific human PK parameters, and can therefore be easily adopted in screening programs to prioritize and triage compounds based on undesirable pharmacokinetics. Static prediction methods have been used extensively to predict human metabolic (Gillette, 1971; Rowland et al., 1973; Iwatsubo et al., 1997; Obach et al., 1997) and transporter-mediated clearance (Liu and Pang, 2005; Barton et al., 2013; Varma et al., 2013) and drug-drug interactions (Mayhew et al., 2000; Wang et al., 2004; Obach et al., 2007; Fahmi et al., 2008). However, while static models are very useful for predictions of overall drug exposures in humans or the overall magnitude of DDIs, they rely on steady-state assumptions and hence cannot predict the overall shape of the plasma concentration-time curve, time-varying changes in enzyme or transporter inhibition, or the distribution kinetics of new drugs. In contrast, PBPK models provide simulated concentration versus time profiles of a drug and its metabolite(s) in plasma or an organ of interest and simultaneously allow for estimation of maximum plasma concentrations, absorption kinetics, distribution kinetics, and drug elimination. While the simultaneous modeling of drug disposition processes provides multiple advantages (Rostami-Hodjegan and Tucker, 2007; Almond et al., 2009; Fahmi et al., 2009; Jamei et al., 2009a; Rowland et al., 2011; Huang and Rowland, 2012; Di et al., 2013; Shardlow et al., 2013; Galetin, 2014; Tsamandouras et al., 2015; Varma et al., 2015b), it also makes PBPK modeling labor intensive and requires considerably more parameter estimates and more detailed physiologic and drug-specific data than static predictions. The simulated concentration-time profiles can aid in selection of optimal sampling times or dosing strategies in different study populations, including vulnerable subjects (Rowland et al., 2011). They can also aid in design of DDI studies in which the timing of the dosing of the perpetrator drug and the victim drug is critical (Zhao et al., 2009; Shardlow et al., 2013), or in situations where perpetrator concentrations fluctuate over the sampling and dosing interval (Almond et al., 2009; Fahmi et al., 2009; Pang and Durk, 2010; Di et al., 2013). Additionally, the simulated concentrations can be linked to pharmacodynamic endpoints to allow for PK/PD (pharmacokinetic-pharmacodynamic) simulations. Furthermore, because PBPK models account for sequential metabolism and permeability limited processes, they may provide advantages for predicting bioavailability when compared with static models (Fan et al., 2010; Chow and Pang, 2013). This can have important implications for first in human dose selection, particularly for drugs with active or toxic metabolites. In some cases, PBPK models incorporate interindividual variability, thus allowing for the prospective simulation of the population variability in the pharmacokinetics of a given drug. Population variability is not typically accounted for in static models but can provide insight into variability in exposure and drug response in a given population (Rostami-Hodjegan and Tucker, 2007; Jamei et al., 2009a; Cubitt et al., 2011; Brown et al., 2012). Finally, the separation of drug-specific and physiologic parameters within the model can allow a more mechanistic understanding of the sources of interindividual variability than can be provided by population and compartmental modeling techniques (Rostami-Hodjegan and Tucker, 2007; Vinks, 2013; Tsamandouras et al., 2015). However, detailed understanding of physiologic variables in the population of interest is required but not always available, which can hinder the use of PBPK modeling in special populations.
In recent years, the number of publications (Rowland et al., 2011; Rostami-Hodjegan et al., 2012) and regulatory submissions (Zhao et al., 2011; Huang et al., 2013; Sinha et al., 2014) referencing or including PBPK modeling has increased substantially. The development of user-friendly software tools such as Simcyp, GastroPlus, and PK-Sim have made modeling more accessible to those without extensive modeling and/or programming experience (Zhao et al., 2011; Chen et al., 2012; Huang et al., 2013). However, it is possible that many users are not completely familiar with, or aware of, the assumptions made and equations used during model building and implementation. As such, the increased implementation of PBPK modeling has led to a need for comprehensive software and modeling-focused education as well as the need to confirm the sound knowledge of users in ADME principles and fundamental physiology (Jones et al., 2015). A recommendation for the presence of a modeling expert for advice and to review models has also been made to ensure appropriate decision making and interpretation of the modeling (Jones et al., 2015). Advancements in computer science and physiologically based mathematical models have led to the expansion of the potential applications of PBPK modeling. For example, more complex absorption models such as advanced dissolution, absorption, and metabolism (ADAM) models (Jamei et al., 2009b) and advanced compartmental absorption and transit (ACAT) models (Agoram et al., 2001) have been developed that enable the use of PBPK modeling for the simulation of food effects (Shono et al., 2009; Turner et al., 2012; Heimbach et al., 2013; Xia et al., 2013b; Patel et al., 2014; Zhang et al., 2014), the impact of drug properties on absorption kinetics (Kambayashi et al., 2013; Parrott et al., 2014), and intestinal interactions (Fenneteau et al., 2010). The development of sophisticated models that allow for the simulation of multiple inhibitors or inducers, relevant metabolites, and multiple mechanisms of interaction have permitted the prediction of complex DDIs involving enzymes, transporters, and multiple interaction mechanisms (Zhang et al., 2009; Rekić et al., 2011; Varma et al., 2012, 2013; Dhuria et al., 2013; Gertz et al., 2013, 2014; Guo et al., 2013; Kudo et al., 2013; Siccardi et al., 2013; Wang et al., 2013a; Sager et al., 2014; Chen et al., 2015; Shi et al., 2015). Furthermore, the mechanistic understanding of ADME changes that occur in different age groups or disease states has improved, and consequently PBPK modeling has been used to simulate drug disposition in special populations including hepatic (Johnson et al., 2014) and renal impairment populations (Li et al., 2012; Zhao et al., 2012a; Lu et al., 2014; Sayama et al., 2014), children (Leong et al., 2012), and pregnant women (Andrew et al., 2008; Gaohua et al., 2012; Horton et al., 2012; Ke et al., 2012, 2013, 2014; Lu et al., 2012).
In the past 10 years, PBPK modeling has become increasingly accepted by regulatory agencies as a means of informing clinical study strategy, and it has become a useful tool in drug development (Leong et al., 2012; Zhao et al., 2012b; Huang et al., 2013; Sinha et al., 2014). PBPK approaches have been included in regulatory guidance on hepatic impairment [Committee for Medicinal Products for Human Use (CHMP), 2005], pediatrics [Center for Drug Evaluation and Research (CDER), 2014], DDIs (CDER, 2012; CHMP, 2012; Ministry of Health, Labor and Welfare Research Group, 2014), and pharmacogenetics (CHMP, 2011; CDER, 2013) as a means to guide clinical study design and labeling decisions. Hence, in addition to being used to inform internal development decisions (Jones et al., 2009, 2015; Chen et al., 2012; Shardlow et al., 2013), PBPK modeling is increasingly being used in investigating new drugs and new drug applications (Huang et al., 2013; CHMP, 2014; Sinha et al., 2014). The U.S. Food and Drug Administration (FDA) Office of Clinical Pharmacology has been tracking the use of PBPK modeling in regulatory submissions since 2008 (Huang et al., 2013; Pan et al., 2014). Based on 2013 submissions, the models included in regulatory filings were most commonly used for DDI (60%), pediatric (21%), and absorption (6%) predictions (Pan et al., 2014). PBPK models have been used during the review process to inform dose selection and optimal design for clinical studies (Leong et al., 2012), and in some cases to directly inform labeling (Zhao et al., 2012b). For example, cabazitaxel is predicted to cause in vivo CYP3A4 inhibition based on the ratio between its concentration and inhibition constant, ([I]/Ki ratio). However, modeling and simulation suggested minimal risk for DDI in vivo. As a result, the label states that “a post-marketing requirement for the effect of cabazitaxel on the pharmacokinetics of a sensitive CYP3A4 substrate is therefore not necessary” (Huang and Rowland, 2012; Huang et al., 2013; CDER, 2010). Additional examples of PBPK-informed labeling between 2008 and 2014 are included in recent reviews (Zhao et al., 2012b; Huang et al., 2013; Sinha et al., 2014; Jones et al., 2015).
Despite the increasing use of PBPK modeling, there are many challenges that limit the utility of PBPK modeling and simulation. In general, IVIVE using PBPK models requires considerably more experimental and in silico data than static models. Due to the large number of parameters required for PBPK modeling and the limited availability of in vivo data to verify individual parameters, model predictions can be confounded by lack of confidence in individual parameters. For example, for drugs that have not been administered intravenously to humans, distribution and absorption parameters cannot be validated or verified experimentally, which introduces uncertainty into model parameters and output. The application of PBPK modeling to predict the pharmacokinetics in disease populations is hindered by lack of in vivo data in patient populations, poor understanding of the physiologic changes that occur in certain populations, and limited knowledge of tissue-specific changes in enzyme and transporter expression (Edginton and Joshi, 2011; Sjöstedt et al., 2014; Jones et al., 2015). Furthermore, absolute abundances of transporters and non-P450 enzymes in the liver and other tissues are not well established, resulting in poor IVIVE of the kinetics of non-P450 substrates and permeability limited drugs (Edginton and Joshi, 2011; Jones et al., 2012, 2015; Varma et al., 2012; Harwood et al., 2013). Additionally, a lack of selective substrates and inhibitors for some non-P450 enzymes and transporters has prevented model validation against in vivo data (Jones et al., 2015). While efforts are being made to characterize tissue-specific transporter expression, current models of the disposition of transporter substrates rely on the incorporation of empirical scaling factors (Varma et al., 2015b). Although scaling factors have allowed for predictions of the kinetics of a number of uptake transporter substrates (Varma et al., 2012, 2014, 2015a; Kudo et al., 2013; Gertz et al., 2014; Jamei et al., 2014), it is not possible to experimentally verify whether unbound tissue exposures are adequately predicted (Chu et al., 2013; Jones et al., 2015; Varma et al., 2015b). This could have important implications for IVIVE of efflux clearance, metabolism-transporter interplay, and predictions of pharmacological effects. The utility of PBPK modeling in the prediction of therapeutic protein disposition is still relatively limited, as was recently discussed (Jones et al., 2015). While a number of PBPK models have been used to accurately predict the kinetics of monoclonal antibodies (Baxter et al., 1995; Ferl et al., 2005; Shah and Betts, 2012; Cao and Jusko, 2014; Elmeliegy et al., 2014; Li et al., 2014a; Chetty et al., 2015; Zhao et al., 2015), model structures are inconsistent (Chetty et al., 2012; Jones et al., 2015). Limited data on target expression and changes in disease populations result in the risk for overparameterization with PBPK models, and thus there is an effort to move toward reduced PBPK models for therapeutic proteins (Elmeliegy et al., 2014; Li et al., 2014a; Chetty et al., 2015; Diao and Meibohm, 2015).
Another current challenge in the PBPK modeling field is determining how to assess model quality. To date, neither the FDA nor the European Medicines Agency (EMA) have issued a formal guidance regarding model quality assessment during regulatory review. However, the FDA has acknowledged the use of the best-practice methods proposed by the World Health Organization International Program for Chemical Safety (International Programme on Chemical Harmonization Project, 2010; Zhao et al., 2012b). These practices include ensuring the physiologic plausibility of the input parameters, demonstrating the ability of the model to predict the pharmacokinetics in an independent data set, and confirming that sensitivity and uncertainty analysis support model quality. The recommendation to establish guidelines for reporting and qualification of PBPK models was made at the Ministerial Industry Strategy Group (2014) New Technologies Forum on Physiologically Based Modeling and Simulation, and the EMA released a concept paper on the reporting and quality assessment of PBPK models with the goal of publishing a draft guidance in 2015 (CHMP, 2014). However, while some basic guidelines for assessing model quality prior to regulatory review are accepted or in development, no standards exist for how model quality should be evaluated in peer-reviewed publications. Additionally, no formal analysis of the literature has previously been performed to evaluate what quality assessment methods, if any, are typically used in peer-reviewed publications.
Despite the growth of the PBPK modeling field and the well-established use of PBPK models in regulatory submissions, the overall public availability of PBPK models is unclear and the breath of use of PBPK modeling by the research community has not been systematically evaluated. The PBPK models used in regulatory submissions are not publicly available to the outside research community, which prevents the broad use of models that have been accepted by regulatory agencies. Furthermore, the applications of the models in regulatory submissions may be driven primarily by the needs of drug developers and may not reflect how PBPK modeling is used in the larger research community. Identifying and compiling a list of the publicly available models could be beneficial to future research efforts since published models could be used either unchanged or as a starting point in future modeling efforts. Furthermore, determining the common applications of the published PBPK models will provide insight into current modeling interests as well as highlight under-represented applications. This review evaluates recent PBPK publications and identifies the common applications of PBPK modeling, how models are typically developed, and ways in which model quality is assessed, and provides a list of publicly available PBPK models, with a focus on enzyme probes, marker substrates and important perpetrators of DDIs.
Literature Search Strategy
PubMed searches were conducted using the search terms “PBPK” and “physiologically based pharmacokinetic model” within the abstract or title of the manuscript. Papers were selected for review if they were published in English between 2008 and May 20, 2015, and focused on PBPK modeling of pharmaceutical agents in humans. The number of papers referenced is likely an under-representation of the overall body of literature on PBPK modeling due to the strict search criteria and the search terms used. Publications were categorized as a review, commentary, letter to the editor, or an original data paper containing one or more PBPK models. Papers that focused on the development of new modeling software or a modeling strategy were classified as prediction method papers. Original data papers were further categorized by the primary application of the models.
Models for FDA and EMA recommended probe substrates, inhibitors, and inducers (CDER, 2012; CHMP, 2012) were identified within the original data papers. Complete lists of the compounds recognized by the regulatory agencies are shown in Supplemental Tables 1 and 2. Models for these compounds were included in our analysis if 1) they were original published models; 2) enough information was provided to allow for replication of the model in an appropriate software program; and 3) the simulation results were compared with observed in vivo data. A number of models were excluded because they were default library files in a simulation software package, the model input parameters were not reported, or the simulation results were not compared with in vivo data. Compound models were categorized as substrates, inhibitors, and/or inducers based on their classification in the FDA or EMA DDI guidance (CDER, 2012; CHMP, 2012) if the model was built with the clearance pathways or interaction parameters that permitted it to be used for the specified purpose. Models for FDA substrates, inhibitors, and inducers that lacked the appropriate clearance pathways or inhibition/induction parameters to allow them to be used according to their FDA or EMA classification were placed into a category of their own. For each FDA and EMA recommended substrate, inhibitor, or inducer that met the search criteria, information regarding the simulated formulation, genotype, and software used was extracted. Furthermore, the source of the clearance input parameter (in vitro or in vivo), the type of independent quality assessment data set used, and the a priori model acceptance criteria were collected. Finally, the type of model (full or minimal PBPK) was determined. PBPK models were considered to be minimal if the model included no more than five compartments, including the gastrointestinal tract, blood, and liver, and up to two additional compartments. More complex models were considered to be full PBPK.
PBPK Modeling Articles by Year and Application
A total of 366 PBPK-related articles meeting our search criteria were published since 2008. While it is unlikely that the literature search identified all of the papers presenting PBPK modeling in the literature, the search likely provides adequate and representative coverage of the existing models and practices. The number of articles published per year rose steadily with time from nine articles in 2008 to 94 articles published in 2014 (Fig. 1A). Of the papers identified, 74% were original data papers that included one or more PBPK models, while 26% were reviews, commentaries, letters to the editor, or prediction methods papers. The original data papers were analyzed to identify the common applications of PBPK models. The distribution of the model applications is shown in Fig. 1B. The published PBPK models were most commonly used for DDI predictions (28%). The majority of the DDI prediction models were used to simulate P450-mediated DDIs (81%), while the remainder of the models focused on transport DDIs (10%) or a combination of P450 and transporter-mediated interactions (10%). Additionally, models were commonly used to predict interindividual variability and general clinical pharmacokinetics (23%), absorption kinetics (12%), and age-related changes in pharmacokinetics (10%). This distribution of model applications is distinctly different from what has been reported for regulatory submissions to the FDA. The models included in FDA regulatory filings were primarily used for DDI predictions (60%), followed by pediatrics (21%) and absorption (6%) predictions (Pan et al., 2014). Based on this analysis the use of PBPK modeling to evaluate interindividual variability and overall drug disposition characteristics is far more common in the broader research community than in regulatory review. This difference reflects the fact that both the FDA guidance on pediatrics (CDER, 2014) and DDIs (CDER, 2012) includes PBPK modeling as a potentially useful tool for guiding clinical study design; however, PBPK modeling is currently not included in FDA guidance on bioequivalence or first in human studies.
Fig. 1.
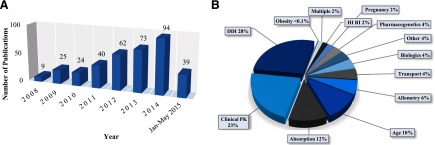
Summary of the PBPK literature analyzed. The number of articles per year that contain one or more PBPK models of pharmaceutical agents in humans is shown in (A). The distribution of the PBPK model applications in the original data papers is shown in (B).
Published Models of FDA and EMA Recommended Substrates, Inhibitors, and Inducers
Each of the 271 original data papers identified included at least one PBPK model of a pharmaceutical agent. The majority of the papers included models of approved drugs, and only 21 papers (8%) used PBPK modeling to simulate the pharmacokinetics of drugs in development. The published PBPK models included default models from software libraries as well as original models. Of the published original models, the models for the FDA and EMA recommended sensitive substrates, inhibitors, and inducers were further evaluated. While these models only represent a fraction of the published PBPK models, these compounds represent a group of drugs for which PBPK models are particularly useful since the models can be used in DDI predictions or to validate altered expression levels or activity of transporters and enzymes in new physiologic models. Fifty-six papers were found that included models for FDA and EMA listed sensitive substrates, inhibitors, and inducers. In these papers, 107 original models representing 61 different compounds were identified. These models were analyzed to gain insight into how peer-reviewed models are commonly developed and how authors assess overall model quality. For each model, information about model development was collected, including the software used, the complexity of the model (full or minimal PBPK), the source of the clearance input value, and the type of dosing simulated. Additionally, information regarding model quality and quality assessment was recorded, including whether the simulated population matched the observed population, if an independent dataset was used to verify model quality, and the type of criteria authors used to determine if a model performance was acceptable. The compounds modeled, the model development methods, and the quality assessment criteria are provided in Tables 1–6, along with references to the original publications.
Table 1.
Summary of PBPK models published for P450 Sensitive Substrates
| Main Clearance Enzyme | Compound | Application | Minimal or Full PBPK | Oral or IV | Clearancea | Simulated genotype specified? | Population matched?b | Verificationc | Acceptance Criteriad | Software | Citation |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CYP1A2 | Caffeine | Allometry | Full | Oral | SF | No | N.S | A,D | 1 | PK Sim | (Thiel et al., 2014) |
| CYP2B6 | Efavirenz | DDI | Minimal | Oral | In vitro | Yes | N.S | A, E | 1 | Simcyp | (Siccardi et al., 2013) |
| Efavirenz | DDI | Full | Oral | In vitro | Yes | Sex | D,E | 1 | Simcyp | (Rekic et al., 2011) | |
| Efavirenz | Absorption | Full | Oral | In vitro | No | N.S | B | 5 | Matlab | (Rajoli et al., 2014) | |
| CYP2C8 | Repaglinide | Diabetes | Full | Oral | In vitro | No | N.S. | B,D | 3 | WinNonlin | (Li et al., 2014b) |
| Repaglinide | RI | Full | Oral | BC | Yes | N.S. | D | 1 | Simcyp | (Zhao et al., 2012a) | |
| Repaglinide | DDI | Minimal | Oral | PE | No | N.S | B, E | 1 | Napp | (Kudo et al., 2013) | |
| Repaglinide | DDI | Full | Oral | BC | No | N.S | C,E | 1 | Simcyp | (Varma et al., 2013) | |
| CYP2C19 | Clobazam | Pediatrics | Full | Oral | In vitro | No | N.S. | B,D,E | 1 | Matlab | (Ogungbenro and Aarons, 2015) |
| Omeprazole | Clinical PK | Minimal | Both | BC | Yes | N.S. | B,E | 1 | Simcyp | (Wu et al., 2014) | |
| CYP2D6 | Metoprolol | Pregnancy | Full | Oral | In vitro, SF | Yes | Sex, PGX | D, E | 2 | Simcyp, Matlab | (Ke et al., 2013b) |
| Dextromethorphan | Pregnancy | Full | Oral | PE | No | Sex, PGX | D, E | 2 | Simcyp, Matlab | (Ke et al., 2013b) | |
| Dextromethorphan | Allometry | Full | Oral | SF | No | N.S | A,D | 1 | PK Sim | (Thiel et al., 2014) | |
| CYP3A4 | Alfentanil | DDI | Full | Oral | BC | No | N.S. | C, E | 5 | Gastroplus | (Baneyx et al., 2014) |
| Alfentanil | DDI | Full | Oral | In vitro | No | N.S. | E | 5 | WinNonlin | (Guo et al., 2013) | |
| Buspirone | DDI | Full | Oral | In vitro | No | N.S. | E | 5 | WinNonlin | (Guo et al., 2013) | |
| Indinavir | Pregnancy | Full | Both | In vitro, SA | No | Sex | C, D, E | 2 | Simcyp, Matlab | (Ke et al., 2012) | |
| Maraviroc | DDI | Minimal | Oral | In vitro | No | N.S. | A,E | 1 | Simcyp | (Hyland et al., 2008) | |
| Midazolam | Pregnancy | Full | Oral | In vitro | No | Sex | A, D | 2 | Simcyp, Matlab | (Ke et al., 2012) | |
| Midazolam | DDI | Full | Oral | BC | No | N.S | C, D | 5 | Gastroplus | (Baneyx et al., 2014) | |
| Midazolam | DDI | Full | Oral | PE | No | N.S | A | 4 | Berkeley Madonna | (Brantley et al., 2014) | |
| Midazolam | Pregnancy | Full | Oral | In vivo | No | Sex | B,D | 3 | Gastroplus | (Xia et al., 2013b) | |
| Midazolam | DDI | Minimal | Oral | In vitro | No | N.S. | E | 5 | WinNonlin | (Wang et al., 2013a) | |
| Midazolam | DDI | Full | Oral | In vitro | No | N.S. | E | 5 | WinNonlin | (Guo et al., 2013) | |
| Midazolam | Allometry | Full | Oral | SF | No | N.S | A, D | 1 | PK Sim | (Thiel et al., 2014) | |
| Quetiapine | Pediatrics | Both | Oral | BC | No | Age, Sex | D, E | 1 | Simcyp | (Johnson et al., 2014) | |
| Sildenafil | RI | Full | Oral | BC | Yes | N.S. | D | 2 | Simcyp | (Zhao et al., 2012a) | |
| Simvastain | DDI | Full | Oral | In vitro | No | N.S. | E | 5 | WinNonlin | (Guo et al., 2013) | |
| Simvastatin | DDI | Minimal | Oral | In vitro | No | N.S. | A, E | 5 | WinNonlin | (Wang et al., 2013a) | |
| Triazolam | DDI | Full | Oral | In vitro | No | N.S. | E | 5 | WinNonlin | (Guo et al., 2013) | |
| Triazolam | DDI | Full | Oral | BC | No | N.S. | C, E | 5 | Gastroplus | (Baneyx et al., 2014) |
BC = back-calculated from in vivo data, PE = parameter estimate, SA = sensitivity analysis, SF = scaling factor from mice.
PGX = genotype, N.S. = not specified.
Data sets used in model verification included: (A) Single dose PK, (B) alternative dosing regimen, (C) alternative formulation, (D) alternative population, (E) DDI.
Acceptance criteria fell into 5 categories: (1) Not specified, (2) Ratio of PK parameter(s) must be within 30% of observed ratio, (3) Ratio of PK parameter(s) must be within 2 fold of observed ratio, (4) PK parameters must be within 30% of observed parameters, (5) PK parameters must be within 2 fold of observed parameters.
Table 6.
Summary of the PBPK models published for compounds that are FDA probe substrates, inhibitors, or inducers but the models were developed for a different purpose than the FDA category.
| Compound | Application | Type | Oral or IV | Simulated genotype specified? | Clearancea | Verificationb | Population matched?c | Acceptance Criteriad | Software | Citation |
|---|---|---|---|---|---|---|---|---|---|---|
| Alprazolam | DDI | Full | Oral | No | In vitro | E | N.S. | 5 | Winnonlin | (Guo et al., 2013) |
| Clopidogrel | DDI | Minimal | Oral | Yes | In vitro | B, E | Sex | 1 | Simcyp | (Tornio et al., 2014) |
| Clopidogrel | Genetics | Full | Oral | Yes | In vitro | B, D, E | N.S. | 1 | Simcyp | (Djebli et al., 2015) |
| Lansoprazole | Absorption | Minimal | Oral | No | In vivo | C | N.S. | 1 | Gastroplus | (Wu et al., 2013) |
| Metformin | Other (diabetes) | Full | Oral | No | in vivo | D | N.S. | 3 | Winnonlin | (Li et al., 2015) |
| Metformin | Pregnancy | Full | Oral | No | In vivo | D | Sex | 5 | Gastroplus | (Xia et al., 2013a) |
| Methadone | Pregnancy | Full | Oral | No | BC | B, D | Sex | 4 | Simcyp, Matlab | (Ke et al., 2013a) |
| Nisoldipine | Other (diabetes) | Full | Oral | No | In vivo | D | N.S. | 3 | Winnonlin | (Li et al., 2015) |
| Oseltamivir | Pediatrics | Full | Both | No | In vitro, SF | C, D, E | N.S. | 1 | Gastroplus | (Parrott et al., 2011) |
| Oseltamivir | Clinical PK | Full | Oral | Yes | In vitro | D | N.S. | 1 | PK-Sim | (Hu et al., 2014) |
| Oseltamivir | RI | Full | Oral | No | In vivo | B | N.S. | 1 | Simcyp | (Hsu et al., 2014) |
| Phenobarbital | DDI | Full | Oral | No | In vivo | B,E | N.S | 5 | WinNonlin | (Guo et al., 2013) |
| Pravastatin | Clinical PK | Full | IV | No | In vitro, SF | C | N.S. | 1 | Berkeley Madonna | (Jones et al., 2012) |
| Propranolol | Formulation | Full | Oral | No | In vivo | B, C | N.S. | 1 | Gastroplus | (Wang et al., 2013b) |
| Rosuvastatin | Clinical PK | Full | IV | No | In vitro, SF | C | N.S. | 1 | Berkeley Madonna | (Jones et al., 2012) |
| Sertraline | DDI | Minimal | Oral | Yes | In vitro | E | N.S. | 1 | Simcyp | (Siccardi et al., 2013) |
| Theophylline | DDI | Minimal | Oral | No | In vivo | B,E | Age, Sex, PGX | 3 | Simcyp | (Xu et al., 2009) |
| Valsartan | Clinical PK | Full | IV | No | In vitro, SF | C | N.S. | 1 | Berkeley Madonna | (Jones et al., 2012) |
| Verapamil | DDI | Full | Oral | No | In vitro | E | N.S. | 3 | Winnonlin | (Guo et al., 2013) |
| Voriconazole | Pediatrics | Full | Both | No | In vitro, SF | C, D | N.S. | 1 | Simcyp | (Zane and Thakker, 2014) |
| Voriconazole | DDI | Minimal | Oral | Yes | In vitro | E | Sex, PGX | 1 | Simcyp | (Damle et al., 2011) |
BC = back calculated from in vivo data, SF = scaling factor.
Data sets used in model verification included: (A) Single dose PK, (B) alternative dosing regimen, (C) alternative formulation, (D) alternative population, (E) DDI.
PGX = genotype, N.S. = not specified.
Acceptance criteria fell into 5 categories: (1) Not specified, (2) Ratio of PK parameter(s) must be within 30% of observed ratio, (3) Ratio of PK parameter(s) must be within 2 fold of observed ratio, (4) PK parameters must be within 30% of observed parameters, (5) PK parameters must be within 2 fold of observed parameters.
Table 2.
Summary of PBPK models published for narrow therapeutic index substrates
| Main Clearance Enzyme | Compound | Application | Model Type | IV or Oral | Clearancea | Simulated genotype specified? | Population matched?b | Verificationc | Acceptance Criteriad | Software | Citation |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CYP1A2 | Theophylline | Pregnancy | Full | Oral | BC | No | Sex | B, D | 2 | Simcyp, Matlab | (Ke et al., 2013b) |
| Theophylline | DDI | Minimal | Both | In vitro | No | N.S. | B, E | 1 | Matlab | (Pan et al., 2011) | |
| CYP2C9 | Phenytoin | Clinical PK | Minimal | Oral | In vitro | Yes | Age, Sex, PGX | B | 1 | Simcyp | (Polasek et al., 2009) |
| Warfarin | DDI | Full | Oral | PE | No | Age, Sex, PGX | A | 4 | Berkeley Madonna | (Brantley et al., 2014) | |
| CYP3A4 | Cyclosporine | Pediatrics | Full | IV | In vivo, SF | No | Age | C | 1 | AdaptII | (Gérard et al., 2010) |
| Cyclosporine | DDI | Full | Oral | In vitro | No | N.S. | E | 5 | WinNonlin | (Guo et al., 2013) | |
| Cyclosporine | DDI | Full | Both | PE | No | N.S. | C, E | 5 | Matlab | (Gertz et al., 2013) | |
| Cyclosporine | Allometry | Full | Oral | SF | No | N.S | A, D | 1 | PK Sim | (Thiel et al., 2014) | |
| Quinidine | DDI | Full | Oral | In vitro | No | N.S. | E | 5 | WinNonlin | (Guo et al., 2013) | |
| Sirolimus | Clinical PK | Full | Oral | In vitro, PE | No | Age, Sex | B, D, E | 1 | Simcyp | (Emoto et al., 2013) | |
| Tacrolimus | DDI | Full | Oral | In vitro | No | N.S. | E | 5 | WinNonlin | (Guo et al., 2013) | |
| Tacrolimus | Clinical PK | Minimal | Oral | BC | Yes | Age, Sex, PGX | D, E | 1 | PKquest | (Gérard et al., 2014) |
BC = back-calculated from in vivo data, PE = parameter estimate, SF = scaling factor.
PGX = genotype, N.S. = not specified.
Data sets used in model verification included: (A) Single dose PK, (B) alternative dosing regimen, (C) alternative formulation, (D) alternative population, (E) DDI.
Acceptance criteria fell into 5 categories: (1) Not specified, (2) Ratio of PK parameter(s) must be within 30% of observed ratio, (3) Ratio of PK parameter(s) must be within 2 fold of observed ratio, (4) PK parameters must be within 30% of observed parameters, (5) PK parameters must be within 2 fold of observed parameters.
Table 3.
PBPK models and model details for recognized P450 inhibitors
| Enzyme Inhibited | Compound | Application | Minimal or Full PBPK | Oral or IV | Clearancea | Additional Inhibition Parameters | Simulated genotype specified? | Population matched?b | Verificationc | Acceptance Criteriad | Software | Citation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strong Inhibitors | ||||||||||||
| CYP2C8 | Gemfibrozil | DDI | Minimal | Oral | PE | CYP3A4 | No | N.S. | A, B, E | 1 | Napp | (Kudo et al., 2013) |
| CYP2D6 | Paroxetine | Pregnancy | Full | Oral | In vitro | CYP3A4 | Yes | Sex, PGX | D, E | 2 | Simcyp, Matlab | (Ke et al., 2013b) |
| Fluoxetine | DDI | Minimal | Oral | In vitro | CYP1A2 | Yes | N.S. | E | 1 | Simcyp | (Siccardi et al., 2013) | |
| CYP2C9 | ||||||||||||
| CYP2C19 | ||||||||||||
| CYP3A4 | Clarithromycin | DDI | Minimal | Oral | In vivo | — | Yes | Age, Sex, PGX | A, B, E | 3 | Simcyp | (Wang, 2010) |
| Clarithromycin | DDI | Minimal | Oral | In vivo | — | No | Age, Sex, PGX | B, E | 3 | Simcyp | (Xu et al., 2009) | |
| Itraconazole | DDI | Minimal | Oral | PE | — | No | N.S. | A, B, E | 1 | Napp | (Kudo et al., 2013) | |
| Ritonavir | DDI | Minimal | Oral | In vitro | CYP2C9 | Yes | N.S. | A, E | 1 | Simcyp | (Siccardi et al., 2013) | |
| CYP2D6 | ||||||||||||
| Ritonavir | Clinical PK | Minimal | Oral | In vitro | CYP3A5 | Yes | N.S. | D | 1 | Simcyp | (Kaspera et al., 2014) | |
| CYP2D6 | ||||||||||||
| CYP2J2 | ||||||||||||
| Telithromycin | RI | Full | Oral | BC | P-gp | Yes | N.S. | D, E | 1 | Simcyp | (Zhao et al., 2012a) | |
| Telithromycin | DDI | Full | Oral | BC | P-gp | No | N.S. | A, B, E | 1 | Simcyp | (Vieira et al., 2012) | |
| CYP3A5 | ||||||||||||
| Moderate Inhibitors | ||||||||||||
| CYP2C9 | Amiodarone | DDI | Full | Both | BC | CYP2D6 | No | N.S. | A, E | 1 | Simcyp | (Chen et al., 2015) |
| CYP3A4 | ||||||||||||
| CYP2C19 | Omeprazole | Clinical PK | Minimal | Both | BC | — | Yes | PGX | B, D, E | 1 | Simcyp | (Wu et al., 2014) |
| CYP3A4 | Diltiazem | DDI | Minimal | Oral | In vivo | — | No | Age, Sex, PGX | B, E | 3 | Simcyp | (Xu et al., 2009) |
| Diltiazem | DDI | Minimal | Oral | In vitro | — | No | N.S. | A, B, E | 1 | WinNonlin | (Zhang et al., 2009) | |
| Diltiazem | DDI | Minimal | Oral | In vivo | CYP2D6 | No | Age, Sex | B | 1 | Simcyp | (Friedman et al., 2011) | |
| Erythromycin | DDI | Minimal | Oral | In vivo | CYP2C8 | No | Age, Sex, PGX | B, E | 3 | Simcyp | (Xu et al., 2009) | |
| Verapamil | DDI | Minimal | Oral | In vitro | CYP2C8 | No | N.S. | A, B, C, E | 5 | WinNonlin | (Wang et al., 2013a) | |
| OATP1B1 | ||||||||||||
| Verapamil | DDI | Full | Oral | BC | — | No | Age, Sex | A, E | 1 | Simcyp | (Neuhoff et al., 2013a) | |
| Verapamil | DDI | Minimal | Oral | In vivo | — | No | Age, Sex, PGX | B, E | 3 | Simcyp | (Xu et al., 2009) | |
| Weak Inhibitors | ||||||||||||
| CYP2C8 | Trimethoprim | DDI | Minimal | Oral | In vivo | — | Yes | N.S. | B, E | 1 | Simcyp | (Yeo et al., 2013) |
PE = parameter estimation from in vivo data, BC = back-calculated from in vivo data.
PGX = genotype, N.S. = not specified.
Data sets used in model verification included: (A) Single dose PK, (B) alternative dosing regimen, (C) alternative formulation, (D) alternative population, (E) DDI.
Acceptance criteria fell into 5 categories: (1) Not specified, (2) Ratio of PK parameter(s) must be within 30% of observed ratio, (3) Ratio of PK parameter(s) must be within 2 fold of observed ratio, (4) PK parameters must be within 30% of observed parameters, (5) PK parameters must be within 2 fold of observed parameters.
Table 4.
PBPK models published for P450 inducers
| Enzyme Induced | Compound | Application | Minimal or Full | IV or Oral | Clearancea | Simulated genotype specified? | Population matched?b | Verificationc | Acceptance Criteriad | Software | Citation |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CYP2B6 and CYP3A4 | Efavirenz | DDI | Minimal | Oral | In vitro | Yes | N.S | A, E | 1 | Simcyp | (Siccardi et al., 2013) |
| Efavirenz | Absorption | Full | Oral | In vitro | No | N.S. | B | 5 | Matlab | (Rajoli et al., 2014) | |
| Efavirenz | DDI | Full | Oral | In vitro | Yes | Sex | D, E | 1 | Simcyp | (Rekic et al., 2011) | |
| CYP3A4 | Carbamazepine | DDI | Full | Oral | In vitro | No | N.S. | B, E | 5 | WinNonlin | (Guo et al., 2013) |
| Etravirine | Absorption | Full | Oral | In vitro | No | N.S. | B | 5 | Matlab | (Rajoli et al., 2014) | |
| Rifampin | DDI | Full | Oral | BC | No | N.S. | E | 5 | Gastroplus | (Baneyx et al., 2014) | |
| Rifampin | DDI | Full | Oral | In vivo | No | N.S. | B, E | 5 | WinNonlin | (Guo et al., 2013) | |
| Rifampin | DDI | Full | Oral | BC | No | Age, sex, PGX | A | 1 | Simcyp | (Neuhoff et al., 2013b) |
BC = back calculated from in vivo data.
PGX = genotype, N.S. = not specified.
Data sets used in model verification included: (A) Single dose PK, (B) alternative dosing regimen, (C) alternative formulation, (D) alternative population, (E) DDI.
Acceptance criteria fell into 5 categories: (1) Not specified, (2) Ratio of PK parameter(s) must be within 30% of observed ratio, (3) Ratio of PK parameter(s) must be within 2 fold of observed ratio, (4) PK parameters must be within 30% of observed parameters, (5) PK parameters must be within 2 fold of observed parameters.
Table 5.
Summary of the PBPK models published for transporter substrates, inhibitors and inducers
| Compound Modeled (Relevant Transporter) | Application | Minimal or Full | Oral or IV | Clearancea | Simulated genotype specified? | Population matched?b | Verificationc | Acceptance Criteriad | Software | Citation |
|---|---|---|---|---|---|---|---|---|---|---|
| Inducers | ||||||||||
| Rifampin (P-gP) | Transport | Minimal | Oral | BC | No | A,S | A, E | 1 | Simcyp | (Neuhoff et al., 2013b) |
| Inhibitors | ||||||||||
| Cyclosporine ( OATP1B1) | Transport | Full | Oral | In vivo | Yes | N.S. | A, E | 1 | Simcyp | (Varma et al., 2012) |
| Cyclosporine (OATP1B1, OATP1B3 BCRP) | Transport | Full | Oral | BC | No | Age, sex, PGX | B | 1 | Simcyp | (Jamei et al., 2014) |
| Cyclosporine (OATP1B1, OATP1B3 BCRP) | DDI | Full | Both | PE | No | N.S. | C, E | 5 | Matlab | (Gertz et al., 2013) |
| Gemfibrozil ( OATP1B1) | DDI | Minimal | Oral | PE | No | N.S. | A, B, E | 1 | Napp | (Kudo et al., 2013) |
| Gemfibrozil (OATP1B1) | Transport | Full | Oral | BC | Yes | N.S. | E | 1 | Simcyp | (Varma et al., 2015a)* updated Varma et al 2012 |
| Probenecid (OAT1, OAT3) | DDI | Full | Both | BC | No | N.S. | C | 1 | Simcyp | (Hsu et al., 2014) |
| Verapamil (P-gp) | DDI | Full | Oral | BC | No | Age, sex | A, E | 1 | Simcyp | (Neuhoff et al., 2013b) |
| Substrates | ||||||||||
| Atorvastatin (OATP1B1) | Absorption | Full | Oral | In vitro | No | Age, sex | D | 1 | Simcyp | (Darwich et al., 2013) |
| Bosentan (OATP1B1) | Transport | Full | IV | In vitro, SF | No | N.S. | None | 1 | Berkeley Madonna | (Jones et al., 2012) |
| Dabigatran (P-gp) | DDI | Full | Both | In vitro | No | N.S. | A, B, E | 1 | PK Sim | (Zhao and Hu, 2014) |
| Digoxin (P-gp) | Transport | Full | Both | BC | No | Age, sex | A, B | 4 | Simcyp | (Neuhoff et al., 2013b) |
| Fluvastatin (OATP1B1) | Transport | Full | IV | In vitro, SF | No | N.S. | None | 1 | Berkeley Madonna | (Jones et al., 2012) |
| Glyburide (OATP1B1) | DDI | Full | Both | In vitro | Yes | PDX | B, C, E | 1 | Simcyp | (Varma et al., 2014) |
| Glyburide (OATP1B1) | Pregnancy | Full | Oral | BC | No | sex | B, D, E | 2 | Simcyp, Matlab | (Ke et al., 2013a) |
| Pravastatin (OATP1B1, OAT3) | Clinical PK | Full | Both | In vitro, SF | No | N.S. | C | 1 | Matlab, PK Sim | (Meyer et al., 2012) |
| Pravastatin (OATP1B1) | Transport | Full | Both | In vitro, SF | No | N.S. | A, B, E | 1 | Simcyp | (Varma et al., 2012) |
| Repaglinide (OATP1B1) | Transport | Full | IV | In vitro, SF | No | N.S. | None | 1 | Berkeley Madonna | (Jones et al., 2012) |
| Repaglinide (OATP1B1) | RI | Full | Oral | BC | Yes | N.S. | D | 1 | Simcyp | (Zhao et al., 2012a) |
| Repaglinide (OATP1B1) | DDI | Minimal | Oral | In vivo | No | N.S. | E | 1 | Napp | (Kudo et al., 2013) |
| Repaglinide (OATP1B1) | DDI | Full | Both | BC | No | N.S. | C, E | 1 | Simcyp | (Varma et al., 2013) |
| Rosuvastatin (BCRP OATP1B1, OATP1B3) | Transport | Full | Oral | BC, PE, SA | No | Age, sex | B, E | 1 | Simcyp | (Jamei et al., 2014) |
| Rosuvastatin (OATP1B1, OATP1B3) | Transport | Full | Both | In vitro, SF | No | N.S. | C | 1 | ASCLX | (Bosgra et al., 2014) |
| Telmisartan (OATP1B3) | Transport | Full | Both | In vitro, SF | No | N.S. | C | 1 | Matlab | (Li et al., 2014c) |
BC = back calculated from in vivo data, PE = parameter estimation, SA= sensitivity analysis, SF = scaling factor.
PGX = genotype, N.S. = not specified.
Data sets used in model verification included: (A) Single dose PK, (B) alternative dosing regimen, (C) alternative formulation, (D) alternative population, (E) DDI.
Acceptance criteria fell into 5 categories: (1) Not specified, (2) Ratio of PK parameter(s) must be within 30% of observed ratio, (3) Ratio of PK parameter(s) must be within 2 fold of observed ratio, (4) PK parameters must be within 30% of observed parameters, (5) PK parameters must be within 2 fold of observed parameters.
How Were the Models Developed?
PBPK models can vary in complexity from full PBPK models, where all of the distribution organs and tissues are represented as separate perfused compartments, to more simplified, minimal PBPK models in which tissues with similar kinetics are lumped (Nestorov et al., 1998; Leahy, 2003; Parrott et al., 2005; Bois et al., 2010; Pilari and Huisinga, 2010; Cao and Jusko, 2012; Tsamandouras et al., 2015). The majority of the models for the FDA and EMA substrates, inhibitors, and inducers listed in Tables 1–6 were full PBPK models (72%) as opposed to minimal PBPK models (27%). Full PBPK models will typically fit the experimental data better than minimal models due to the larger number of parameters used, which increases the degrees of freedom. However, confidence in any individual parameter is decreased when moving from a minimal to a full PBPK model. Minimal PBPK models can be used to reduce model complexity while still allowing for mechanistic simulations in only the compartments of interest (Nestorov et al., 1998; Pilari and Huisinga, 2010; Cao and Jusko, 2012; Tsamandouras et al., 2015). One advantage of full PBPK modeling is the ability to simulate the exposure of a drug or its metabolites in specific tissues that are not accessible to clinical sampling. This can be particularly important if the pharmacological or toxicological effects are driven by the concentrations in that tissue (Tsamandouras et al., 2015). However, none of the models listed in Tables 1–6 and only 13 of the 271 original data papers used simulated tissue concentrations to address pharmacology and toxicology questions (Table 7). Instead, full PBPK models were generally used to enable the systematic prediction of distribution kinetics in order to simulate plasma concentration-time profiles. All of the models that were used to simulate kinetics in special populations in which distribution kinetics can be highly altered, such as pediatrics and pregnancy, incorporated full PBPK models. Full PBPK modeling was also used in all but two of the models for transporter substrates and inhibitors due to the need to capture permeability rate–limited processes.
Table 7.
List of compounds for which Full PBPK models were used to address pharmacological and toxicological questions
| Compound | Model purpose | A priori criteria? | Model Quality Assessment | Conclusions | Citation |
|---|---|---|---|---|---|
| Acetaminophen | Assessing various calibration strategies for linking PBPK models to toxicodynamic models of hepatotoxicity | No | Qualitative discussion of the agreement between simulated and observed Cmax, and metabolite ratios | Predicted liver toxicity in agreement with observed | (Péry et al., 2013) |
| Cyclosporine | Simulation of receptor occupancy in accute graft-versus-host organs and kidneys after intermittent or continuous infusion | No | Mann-Whitney test to compare means, chi-square test to compare proportions, bias and precision, number of simulations within 2-fold of the observed, weighted residuals | A greater therapeutic index was predicted following continuous infusion | (Gérard et al., 2010) |
| Cyclosporine | Establish a connection between the likelihood and severity of graft-versus-host disease and cyclosporine exposures in circulation, graft-versus-host target organs and lymphoid tissues | No | Student's t-test to compare means, chi-square test to compare proportions, Akaike information criterion for model selection | Blood cyclosporine levels can be used as an indicator of therapeutic efficacy | (Gérard et al., 2011) |
| Efalizumab | Develop a PD linked PBPK model to predict efficacy of efalizumab | Yes | Observed data within the predicted 5th and 95th centile | The model predicted the efficacy of efalizumab in treatment of psoriasis | (Chetty et al., 2015) |
| Formamide | Evaluate the relationship between dose and hepatic exposure | No | — | 40mg/day dose was proposed bases on a safety index | (Yan et al., 2012) |
| Levofloxacin | Exploratory study to predict the extent of tissue exposure of levofloxacin in humans as a basis for future PK/PD work. | Yes | Fold error in PK parameters less than 2 | Levofloxacin penetrated well into tissues, including the liver kidneys and spleen | (Zhu et al., 2015a) |
| Moxifloxacin | Simulate tissue concentrations versus time in patients with intra-abdominal infections | Yes | Fold error in PK parameters less than 2 | Concentrations in intra-abdominal tissues were predicted to be higher than the minimum inhibitory concentration for common pathogens | (Zhu et al., 2015b) |
| Moxifoxacin | Evaluate the effect of macrophages on tissue concentrations of moxifloxacin to enhance understanding of the effects of disease on PK/PD | No | Simulated concentration versus time profiles were evaluated for bias and precision | Macrophage concentrations are predicted to effect tissue concentration of moxifloxacin | (Edginton et al., 2009) |
| Nicotine | Develop a PBPK model to describe nicotine exposure and receptor binding in the brain | No | Qualitative discussion of the agreement of the predicted and observed data | PK/PD modeling allowed for prediction of nicotine receptor occupancy in the brain | (Teeguarden et al., 2013) |
| “S1” | Predicting brain extracellular fluid concentrations as a starting point for PK-PD modeling | No | — | Unclear whether the PBPK model would accurately predict PD | (Ball et al., 2014) |
| Temozolomide | PD linked PBPK model for simulating the brain concentration of temozolomide and the levels DNA brain adducts | No | — | Predictions were in close agreement with observed data and parameter estimates had low coefficients of variation | (Ballesta et al., 2014) |
| Zidovudine | Model intra-cellular concentrations of zidovudine in peripheral blood mononuclear cells and establish efficacy and toxicity following various dosing regimens | No | PK features are well represented by the predictions | 100mg 4 times daily is predicted to be the safest and most efficacious dosing scheme | (von Kleist and Huisinga, 2009) |
| Theoretical Compounds | Proof of concept study to evaluate mechanisms for differences in unbound plasma and tissue concentrations | No | This approach can be used to predict free tissue concentrations of various classes of drugs | (Poulin, 2015) |
PBPK models are comprised of system- and drug-specific parameters. System-specific parameters include blood flow, organ volumes, enzyme and transporter expression, and plasma protein concentrations (Jamei et al., 2009a; Rowland et al., 2011; Galetin, 2014). Drug-specific parameters include intrinsic clearances, volume of distribution, solubility and physicochemical parameters, tissue partitioning, plasma protein binding affinity, and membrane permeability. As a result, drug-dependent parameters are independent of the system parameters, allowing for mechanistic extrapolation of human pharmacokinetics from in vitro and in silico data in a bottom-up approach (Jamei et al., 2009a; Rowland et al., 2011; Rostami-Hodjegan et al., 2012; Galetin, 2014; Tsamandouras et al., 2015). While bottom-up approaches are generally considered to be more mechanistic, in many cases sufficient in vitro data and characterization of all drug elimination pathways are not available to allow bottom-up predictions, or existing in vitro data do not predict in vivo disposition well enough. Similarly, in many cases, the knowledge of the biologic system is too limited to allow for bottom-up predictions of disposition kinetics in the population of interest. The bottom-up approach is usually not the method of choice in situations where PBPK models are built to specifically evaluate the disposition characteristics of a drug that has been administered to humans or to a special population. In these situations, the model is built to fit the data rather than for predictive IVIVE purposes and a combination of top-down and bottom-up approaches is often used. Several reviews have provided excellent discussions of the utility and setbacks of these combination or middle-out approaches to model development (Jamei et al., 2009a; Li et al., 2014b; Tsamandouras et al., 2015). In general, when using middle-out approaches, in vitro intrinsic clearances are back-calculated from in vivo clearance by assigning the fractions of the in vivo clearance associated with each clearance pathway, or scaling factors are assigned to the in vitro or in vivo clearance value(s) to accurately predict the observed data. Parameter estimation methods and sensitivity analysis can also be used in instances where in vitro data are unavailable and in vivo fm values are not known.
For the models shown in Tables 1–6, in vitro clearance values (bottom-up approach) were used for clearance parameters in 35% of the cases. The most common alternative to IVIVE was back-calculating in vitro intrinsic clearance data from in vivo clearance (21%). Because this approach incorporates the fractional contribution of individual enzymes into the model, models developed using this technique can potentially be used to simulate pharmacokinetics in situations where enzyme expression levels or activity are altered. However, the reliability of the back-calculations to capture the true intrinsic clearances requires knowledge of the fractional contribution of each enzyme to in vivo clearance and an understanding of the true systemic clearance and bioavailability, which may not be available. In 18% of the models, in vivo clearance was used as an input parameter. While this can be a reliable way to ensure that the total body clearance is captured, no specific elimination pathways are accounted for, and thus the model is not useful for predicting the effects of an inhibitor or inducer or the consequences of changes in enzyme or transporter expression levels. In 17% of the models, a scaling factor was applied to the in vitro or in vivo clearance value(s) to accurately predict the observed data. Scaling factors were particularly common for transporter substrates, likely due to the current limitations in IVIVE of transporter-mediated clearance (Harwood et al., 2013; Li et al., 2014b). Finally, parameter estimation methods and sensitivity analysis were performed to determine the in vitro clearance values required to capture the true in vivo clearance for 9% of the models. While these approaches can permit extrapolation to observed in vivo clearance, caution should be exercised when estimating input parameters. In cases where in vitro parameter values and their variability are well understood, low prediction success could indicate that the model is lacking a critical PK process (Jones et al., 2015; Tsamandouras et al., 2015).
What Makes a Good Model and How Is Model Quality Assessed?
Best practices for model assessment have been proposed by the World Health Organization (International Programme on Chemical Harmonization Project, 2010) and have been discussed in the context of regulatory review (Caldwell et al., 2012; Zhao et al., 2012b; CHMP, 2014; Ministerial Industry Strategy Group, 2014). However, no requirements or guidelines exist regarding how to determine the quality of a PBPK model in general research applications and prior to publication. In regulatory guidance the criteria for assessing model validity are often presented in the context of whether the model meets the performance requirements for its specific purpose. However, in the research literature the specific goal or purpose for the model is often not specified, and PBPK modeling is frequently used to explain observed clinical findings or to support a particular mechanistic hypothesis rather than to predict drug disposition in a specific population or clinical situation. To establish the scope of current practices in the PBPK models that have been published for various purposes and applications, an evaluation of the current state of model development and quality assessment was conducted. The compound models listed in Tables 1–6 were assessed 1) to identify the criteria that were typically used to determine if a model was adequate and 2) to determine if models were tested against multiple in vivo data sets.
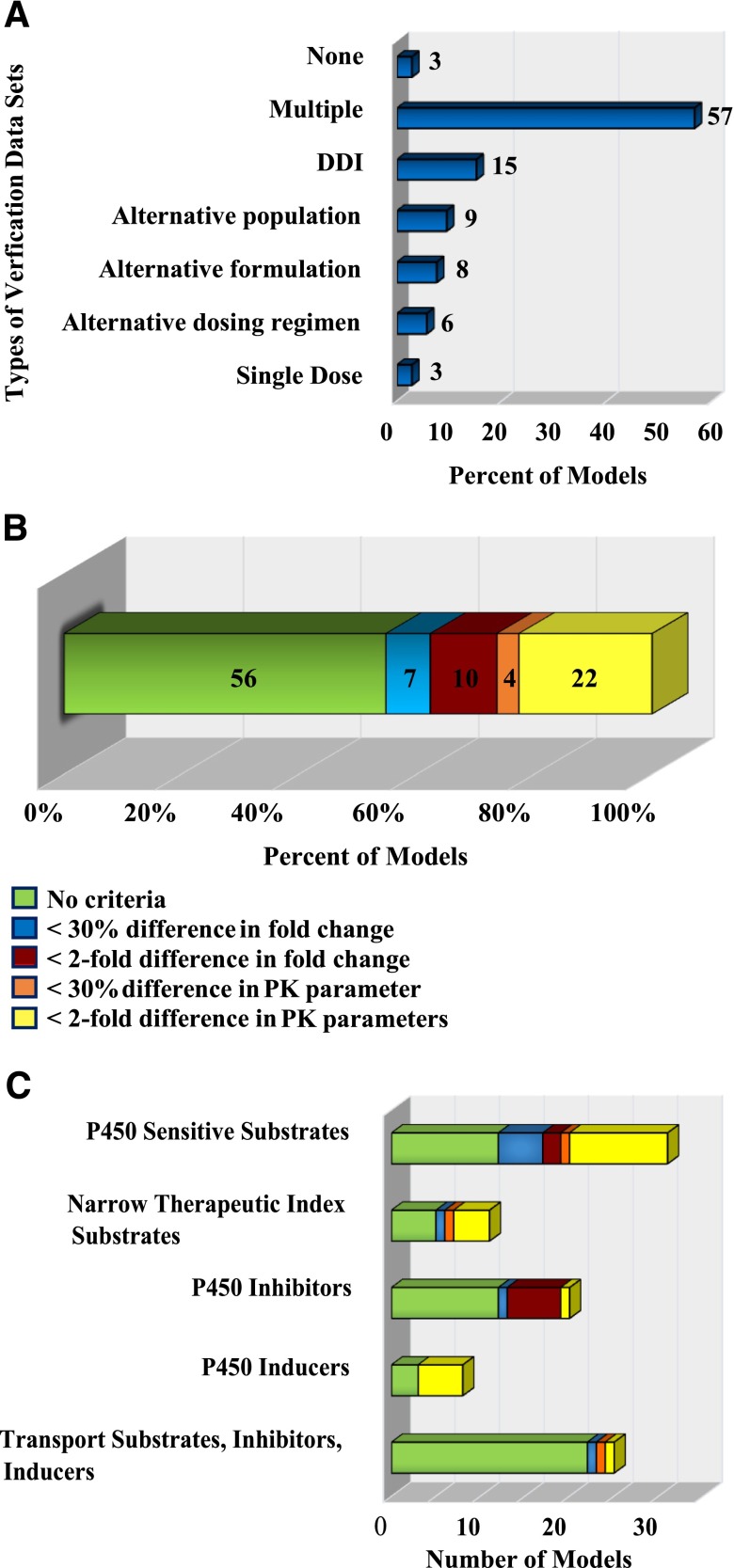
It is considered good practice to assess the quality of a model against in vivo data that were not used in the model development process and in situations where one of the parameters is altered, such as in a DDI or an alternative genotype population (International Programme on Chemical Harmonization Project, 2010; McLanahan et al., 2012; Zhao et al., 2012b; Sinha et al., 2014; Jones et al., 2015). Our analysis revealed that the PK simulations of 97% of the models were compared with pharmacokinetics in independent study populations. When an independent data set was used to test a model, the data set typically described the pharmacokinetics after a single dose or DDI, or for an alternative population, formulation, or dosing regimen. The distribution of the types of in vivo data sets used to assess the quality of the models is shown in Fig. 2A. Most of the models were assessed using multiple types of data sets (57%), DDI data (15%), or PK data from alternative populations (9%). Only 3% of the models were not compared with an independent data set. However, despite the fact that most models were assessed against data that were not used in model development, the simulated populations were rarely matched with the population demographics of the clinical study subjects, or the population demographics used in the simulations were not reported (Tables 1–6). The simulated age, gender, and genotypes were reported to match the observed population for only 32% of the models. Additionally, the simulated genotypes were only specified for 21% of the models. It is possible that the demographics of the clinical study and the simulated population were matched in many of the papers but were not reported. However, reporting the strategy for how the simulated populations were made to reflect the observed would provide greater confidence for the reader that the simulated population was reasonably representative of the true observed population. The population-specific parameters used in PBPK models (such as enzyme and transporter abundance, organ volume, blood flow, plasma protein binding, and glomerular filtration rate) are dependent on the population demographics (such as age, gender, genotype, and disease state). Similarly, the interindividual variability in the physiologic parameters is dependent on the population demographics. Thus, ensuring that the demographics of the simulated population match those of the observed population may improve the accuracy of both the mean PK parameters (Steere et al., 2015) and the predicted population variability. More careful reporting of the simulated and observed study populations would also be critically important when model performance is assessed. As has been highlighted in the literature (Abduljalil et al., 2014), PBPK simulations are often compared to clinical studies with small study populations, and the true inter- and intraindividual variability of the observed PK parameters of the compound of interest are not known. This can lead to a situation in which one clinical study does not accurately predict the PK parameters observed in another study with the same compound (Abduljalil et al., 2014). In such situations, a PBPK model cannot simultaneously meet the common acceptance criteria for both studies. However, the simulated population variability was rarely compared with the observed variability in the literature evaluated, and we found no papers in our analysis in which a priori model acceptance criteria were driven by knowledge of the variability in the PK parameters of the drug of interest in the target population. However, the 90% confidence interval is generally shown in simulated plasma concentration-time curves (Jones et al., 2015), and several studies used the simulated 90% confidence interval of the plasma concentration curves as a criterion for model acceptance (Sager et al., 2014; Bui et al., 2015; Chetty et al., 2015).
Fig. 2.
Summary of the verification criteria and alternative datasets used for the PBPK models in the literature evaluated. The types of in vivo datasets used to verify the quality of the models are shown in (A). The distribution of the acceptance criteria used in PBPK models of FDA probe substrates and inhibitors is shown in (B). The distribution of the model acceptance criteria used specifically for each compound class is shown in (C).
Determination of model performance was inconsistent and largely subjective in a majority of the papers. In 56% of the published models in Tables 1–6, the authors did not specify a priori a criterion by which they would decide if their model was successful or not (Fig. 2B). A recent publication from the International Consortium for Innovation & Quality in Pharmaceutical Development (IQ) IQ PBPK working group suggests that criteria should be predefined regarding whether a model fits the purpose (Jones et al., 2015). However, there is no consensus on what criteria should be applied for different modeling purposes. The IQ working group suggested that for drugs with a broad therapeutic window, common 2-fold criteria for the model would be acceptable, but for drugs with a narrow therapeutic index more stringent criteria would be appropriate (Jones et al., 2015). On the other hand, for PBPK models used for risk assessment the IQ proposed that acceptance criteria should reflect the effect of accuracy on dose selection. However, these recommendations are not consistent with the methods used to evaluate model performance in the literature. Overall, in the papers (Tables 1–6) in which the acceptance criteria were specified a priori, four standard choices were employed for model acceptance. For 22% of the models, the authors specified that predicted ADME characteristics [i.e., the area under the plasma concentration-time curve (AUC), the steady state average or the maximum concentration in plasma (Css,avg, and Cmax)] in a given population must be within 2-fold of the observed value in order for the model to be considered acceptable. In 7% of the cases, predicted mean PK parameters were required to be within 25%–30% of the observed mean. In addition, for 10% of the models, the predicted fold change in the AUC or Cmax between different simulated populations or study conditions had to be within 2-fold of the observed fold change in order for the model to be acceptable. Finally, for 4% of the models, the authors specified that the predicted fold change in the AUC and Cmax needed to be within 30% of the observed fold change. When the acceptance criteria were analyzed according to the types of applications, a more striking discrepancy with the proposed guidance was observed (Fig. 2C). For models built for narrow therapeutic index drugs only 17% (two papers) used a 30% difference as the standard for model acceptance; 50% (six papers) of the papers had no criteria; and 33% (four papers) considered a 2-fold difference to be an acceptable criterion for these drugs. Similarly, for P450-sensitive substrates, which are expected to clinically report less than 2-fold changes in clearance, only 3% (one paper) used <30% difference in fold change as an acceptance criterion for the PBPK models, and 42% (13 papers) considered less than 2-fold difference between predicted and observed values or in fold changes acceptable. Only 16% of the papers (five papers) used the <30% difference observed and predicted values as acceptance criteria for P450-sensitive substrates. For P450 inducers, there were no models that required a <30% difference in PK parameters, and for transporter substrates, inhibitors, and inducers nearly all papers (84%) had no specified acceptance criteria. Taken together, these data suggest that there is a lack of consistency in model quality assessment, which does not reflect the different purposes for which the models are developed. The data also suggest that there is a need for more rigorous evaluation of model quality assessment during peer review. The issue of the lack of strict peer-review requirements for published models has been discussed previously in the literature (McLanahan et al., 2012) but it has not been formally addressed by the larger research community.
Based on the analysis of the PBPK models used to simulate drug absorption, more stringent criteria of model assessment were used in this field, likely adapted from bioequivalence standards. For some absorption models, model performance was determine to be high if error was <25%, medium if error was 25%–50%, low if error was 50%–100%, and inaccurate if error was greater than 2-fold (Sjögren et al., 2013). Importantly, many of the absorption models systematically evaluated model performance in terms of the plasma concentration-time curves rather than specific PK parameters using a similarity factor (f1 or f2) to calculate the percent difference between the simulated and measured plasma concentrations at each measured time point (Shono et al., 2009; Wagner et al., 2012; Fei et al., 2013; Kambayashi et al., 2013; Wang et al., 2013a). In addition, many absorption models were evaluated using statistical criteria such as linear regression between observed and predicted parameters or concentrations and method of residuals (Shono et al., 2009; Turner et al., 2012; Kambayashi et al., 2013). In some studies evaluating PBPK models of biologics (Kletting et al., 2010; Cao et al., 2013) model performances and discrimination between different models was achieved using statistical criteria that account for the added degrees of freedom in the model. The Akaike information criterion and correlation analyses were used to specifically differentiate between developed PBPK models and to identify the model that best fit the observed data (Kletting et al., 2010; Cao et al., 2013). Adaptation of some of these methods and criteria into PBPK modeling in other research areas may provide good standardization of model acceptance criteria.
Conclusions
PBPK modeling is increasingly being used in peer-reviewed publications to provide mechanistic predictions of pharmacokinetics and disposition in diverse populations and dosing regimens. Since 2008, 106 models of sensitive substrates, inhibitors, and inducers have been published, with applications ranging from DDIs to pregnancy. However, there is a relative lack of consistency in how models are developed and how model quality is assessed. Published models use bottom-up, top-down, and middle-out approaches to estimate clearance input values, and they vary in complexity. While model performance was found to be tested against model-independent data sets 97% of the time, model acceptance criteria and the extent to which the simulated populations reflect the observed population were not always specified. Thorough and consistent reporting of model development techniques and quality assessment could increase reader confidence and result in more widespread acceptance of published models. Thus, the development of best-practice guidelines for peer-review submissions might be beneficial. Table 8 includes suggestions for the information that should be included in peer-reviewed publications containing PBPK models. These suggestions are consistent with best-practice guidelines for regulatory review (International Programme on Chemical Harmonization Project, 2010; Zhao et al., 2012b; CHMP, 2014; Ministerial Industry Strategy Group, 2014) but also acknowledge that guidelines for peer-reviewed models may not require the same degree of reporting detail as has been proposed for regulatory submissions.
TABLE 8.
List of relevant details to report for publication of PBPK models.
| Modeling workflow | Suggested information |
|---|---|
| Objectives | What is the purpose of the model? |
| Model acceptance criteria | What criteria are being used to determine if a model is fit-for-purpose? |
| What is the clinical relevance of these criteria? | |
| What independent data sets are used for model testing? | |
| Model development | Was the model built using a PBPK software package? If not, information regarding the model structure, the source of parameters, and their physiologic context should be reported. |
| What input parameters were used? For recommended parameters to include, see Zhao et al., 2012b | |
| What parameters, if any, were estimated when using parameter estimation or sensitivity analysis? | |
| Are the estimated parameters physiologically plausible? | |
| Are the parameters within the range of previously reported values (if applicable)? | |
| Population demographics (Do the simulated and observed populations and study sizes match?) | |
| Model outcomes | Comparison of the predicted and observed PK |
| Do the predictions meet the predetermined model specification criteria? | |
| Model performance | Was sensitivity analysis performed to assess whether model output parameters are sensitive to specific input parameters? (Yes/No) |
| What are the verified applications of the model? | |
| What is the level of uncertainty in the model components? |
Supplementary Material
Abbreviations
- AUC
Area under plasma concentration time curve
- CDER
Center for Drug Evaluation and Research
- CHMP
Committee for Medicinal Products for Human Use
- DDI
drug-drug interaction
- EMA
European Medicines Agency
- FDA
Food and Drug Administration
- IVIVE
in vitro-to-in vivo extrapolation
- IQ
innovation and quality in pharmaceutical development
- PBPK
physiologically based pharmacokinetic
- PD
Pharmacodynamics
- PK
pharmacokinetic
Authorship Contributions
Participated in research design: Sager, Yu, Ragueneau-Majlessi, Isoherranen.
Performed data analysis: Sager.
Wrote or contributed to the writing of this manuscript: Sager, Yu, Ragueneau-Majlessi, Isoherranen.
Footnotes
This work was supported in part by the National Institutes of Health National Institute of General Medical Sciences [Grant T32-GM007750] and National Institute of Drug Abuse [Grant P01 DA032507].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Abduljalil K, Cain T, Humphries H, Rostami-Hodjegan A. (2014) Deciding on success criteria for predictability of pharmacokinetic parameters from in vitro studies: an analysis based on in vivo observations. Drug Metab Dispos 42:1478–1484. [DOI] [PubMed] [Google Scholar]
- Agoram B, Woltosz WS, Bolger MB. (2001) Predicting the impact of physiological and biochemical processes on oral drug bioavailability. Adv Drug Deliv Rev 50 (Suppl 1):S41–S67. [DOI] [PubMed] [Google Scholar]
- Almond LM, Yang J, Jamei M, Tucker GT, Rostami-Hodjegan A. (2009) Towards a quantitative framework for the prediction of DDIs arising from cytochrome P450 induction. Curr Drug Metab 10:420–432. [DOI] [PubMed] [Google Scholar]
- Andrew MA, Hebert MF, Vicini P. (2008). Physiologically based pharmacokinetic model of midazolam disposition during pregnancy. Conf Proc IEEE Eng Med Biol Soc 2008:5454–5457. [DOI] [PubMed] [Google Scholar]
- Ball K, Bouzom F, Scherrmann JM, Walther B, Declèves X. (2014) Comparing translational population-PBPK modelling of brain microdialysis with bottom-up prediction of brain-to-plasma distribution in rat and human. Biopharm Drug Dispos 35:485–499. [DOI] [PubMed] [Google Scholar]
- Ballesta A, Zhou Q, Zhang X, Lv H, Gallo JM. (2014) Multiscale design of cell-type-specific pharmacokinetic/pharmacodynamic models for personalized medicine: application to temozolomide in brain tumors. CPT Pharmacometrics Syst Pharmacol 3:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baneyx G, Parrott N, Meille C, Iliadis A, Lavé T. (2014) Physiologically based pharmacokinetic modeling of CYP3A4 induction by rifampicin in human: influence of time between substrate and inducer administration. Eur J Pharm Sci 56:1–15. [DOI] [PubMed] [Google Scholar]
- Barton HA, Lai Y, Goosen TC, Jones HM, El-Kattan AF, Gosset JR, Lin J, Varma MV. (2013) Model-based approaches to predict drug-drug interactions associated with hepatic uptake transporters: preclinical, clinical and beyond. Expert Opin Drug Metab Toxicol 9:459–472. [DOI] [PubMed] [Google Scholar]
- Baxter LT, Zhu H, Mackensen DG, Butler WF, Jain RK. (1995) Biodistribution of monoclonal antibodies: scale-up from mouse to human using a physiologically based pharmacokinetic model. Cancer Res 55:4611–4622. [PubMed] [Google Scholar]
- Berezhkovskiy LM. (2004) Volume of distribution at steady state for a linear pharmacokinetic system with peripheral elimination. J Pharm Sci 93:1628–1640. [DOI] [PubMed] [Google Scholar]
- Bois FY, Jamei M, Clewell HJ. (2010) PBPK modelling of inter-individual variability in the pharmacokinetics of environmental chemicals. Toxicology 278:256–267. [DOI] [PubMed] [Google Scholar]
- Bosgra S, van de Steeg E, Vlaming ML, Verhoeckx KC, Huisman MT, Verwei M, Wortelboer HM. (2014) Predicting carrier-mediated hepatic disposition of rosuvastatin in man by scaling from individual transfected cell-lines in vitro using absolute transporter protein quantification and PBPK modeling. Eur J Pharm Sci 65:156–166. [DOI] [PubMed] [Google Scholar]
- Brantley SJ, Gufford BT, Dua R, Fediuk DJ, Graf TN, Scarlett YV, Frederick KS, Fisher MB, Oberlies NH, Paine MF. (2014) Physiologically based pharmacokinetic modeling framework for quantitative prediction of an herb-drug interaction. CPT Pharmacometrics Syst Pharmacol 3:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J, Crison J, Timmins P. (2012) Predicting feasibility and characterizing performance of extended-release formulations using physiologically based pharmacokinetic modeling. Ther Deliv 3:1047–1059. [DOI] [PubMed] [Google Scholar]
- Bui KH, Zhou D, Agbo F, Guo J. (2015). Effect of multiple intravenous doses of lanicemine (AZD6765) on the pharmacokinetics of midazolam in healthy subjects. J Clin Pharmacol 55:1024–1030. [DOI] [PubMed] [Google Scholar]
- Caldwell JC, Evans MV, Krishnan K. (2012) Cutting edge PBPK models and analyses: providing the basis for future modeling efforts and bridges to emerging toxicology paradigms. J Toxicol 2012:852384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Balthasar JP, Jusko WJ. (2013) Second-generation minimal physiologically-based pharmacokinetic model for monoclonal antibodies. J Pharmacokinet Pharmacodyn 40:597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Jusko WJ. (2012) Applications of minimal physiologically-based pharmacokinetic models. J Pharmacokinet Pharmacodyn 39:711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Jusko WJ. (2014) Survey of monoclonal antibody disposition in man utilizing a minimal physiologically-based pharmacokinetic model. J Pharmacokinet Pharmacodyn 41:571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center for Drug Evaluation and Research (CDER) (2010) Drug approval package for cabazitaxel. U.S. Food and Drug Administration, Silver Spring, MD. [Google Scholar]
- Center for Drug Evaluation and Research (CDER) (2012). Guidance for industry: Drug interaction studies–study design, data analysis, implications for dosing, and labeling recommendations. U.S. Food and Drug Administration, Silver Spring, MD. [Google Scholar]
- Center for Drug Evaluation and Research (CDER) (2013). Guidance for industry: Clinical pharmacogenomics: Premarket evaluation in early-phase clinical studies and recommendations for labeling. U.S. Food and Drug Administration, Silver Spring, MD. [Google Scholar]
- Center for Drug Evaluation and Research (CDER) (2014). General clinical pharmacology considerations for pediatric studies for drugs and biological products. U.S. Food and Drug Administration, Silver Spring, MD. [Google Scholar]
- Chen Y, Jin JY, Mukadam S, Malhi V, Kenny JR. (2012) Application of IVIVE and PBPK modeling in prospective prediction of clinical pharmacokinetics: strategy and approach during the drug discovery phase with four case studies. Biopharm Drug Dispos 33:85–98. [DOI] [PubMed] [Google Scholar]
- Chen Y, Mao J, Hop CECA. (2015) Physiologically based pharmacokinetic modeling to predict drug-drug interactions involving inhibitory metabolite: a case study of amiodarone. Drug Metab Dispos 43:182–189. [DOI] [PubMed] [Google Scholar]
- Chetty M, Li L, Rose R, Machavaram K, Jamei M, Rostami-Hodjegan A, Gardner I. (2015) Prediction of the pharmacokinetics, pharmacodynamics, and efficacy of a monoclonal antibody, using a physiologically based pharmacokinetic FcRn model. Front Immunol 5:670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetty M, Mattison D, Rostami-Hodjegan A. (2012) Sex differences in the clearance of CYP3A4 substrates: exploring possible reasons for the substrate dependency and lack of consensus. Curr Drug Metab 13:778–786. [DOI] [PubMed] [Google Scholar]
- Cho HJ, Kim JE, Kim DD, Yoon IS. (2014) In vitro-in vivo extrapolation (IVIVE) for predicting human intestinal absorption and first-pass elimination of drugs: principles and applications. Drug Dev Ind Pharm 40:989–998. [DOI] [PubMed] [Google Scholar]
- Chow EC, Pang KS. (2013) Why we need proper PBPK models to examine intestine and liver oral drug absorption. Curr Drug Metab 14:57–79. [PubMed] [Google Scholar]
- Chu X, Korzekwa K, Elsby R, Fenner K, Galetin A, Lai Y, Matsson P, Moss A, Nagar S, Rosania GR, et al. International Transporter Consortium (2013) Intracellular drug concentrations and transporters: measurement, modeling, and implications for the liver. Clin Pharmacol Ther 94:126–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Committee for Medicinal Products for Human Use (CHMP) (2005). Guideline on the evaluation of the pharmacokinetics of medicinal products in patients with impaired hepatic function. CPMP/EWP/2339/02, European Medicines Agency, London.
- Committee for Medicinal Products for Human Use (CHMP) (2011). Guideline on the use of pharmacogenetic methodologies in the pharmacokinetic evaluation of medicinal products. EMA/CHMP/37646/2009, European Medicines Agency, London.
- Committee for Medicinal Products for Human Use (CHMP) (2012). Guideline on the investigation of drug-drug interactions. European Medicines Agency, London. [Google Scholar]
- Committee for Medicinal Products for Human Use (CHMP) (2014). Concept paper on qualification and reporting of physiologically-based pharmacokinetic (pbpk) modeling and analyses. EMA/CHMP/211243/2014, European Medicines Agency, London.
- Cubitt HE, Yeo KR, Howgate EM, Rostami-Hodjegan A, Barter ZE. (2011) Sources of interindividual variability in IVIVE of clearance: an investigation into the prediction of benzodiazepine clearance using a mechanistic population-based pharmacokinetic model. Xenobiotica 41:623–638. [DOI] [PubMed] [Google Scholar]
- Damle B, Varma MV, Wood N. (2011) Pharmacokinetics of voriconazole administered concomitantly with fluconazole and population-based simulation for sequential use. Antimicrob Agents Chemother 55:5172–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwich AS, Pade D, Rowland-Yeo K, Jamei M, Asberg A, Christensen H, Ashcroft DM, Rostami-Hodjegan A. (2013) Evaluation of an in silico PBPK post-bariatric surgery model through simulating oral drug bioavailability of atorvastatin and cyclosporine. CPT Pharmacometrics Syst Pharmacol 2:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhuria S, Einolf H, Mangold J, Sen S, Gu H, Wang L, Cameron S. (2013) Time-dependent inhibition and induction of human cytochrome P4503A4/5 by an oral IAP antagonist, LCL161, in vitro and in vivo in healthy subjects. J Clin Pharmacol 53:642–653. [DOI] [PubMed] [Google Scholar]
- Di L, Feng B, Goosen TC, Lai Y, Steyn SJ, Varma MV, Obach RS. (2013) A perspective on the prediction of drug pharmacokinetics and disposition in drug research and development. Drug Metab Dispos 41:1975–1993. [DOI] [PubMed] [Google Scholar]
- Diao L, Meibohm B. (2015) Tools for predicting the PK/PD of therapeutic proteins. Expert Opin Drug Metab Toxicol 11:1115–1125. [DOI] [PubMed] [Google Scholar]
- Djebli N, Fabre D, Boulenc X, Fabre G, Sultan E, Hurbin F. (2015). Physiologically based pharmacokinetic modeling for sequential metabolism: effect of CYP2C19 genetic polymorphism on clopidogrel and clopidogrel active metabolite pharmacokinetics. Drug Metab Dispos 43:510–522. [DOI] [PubMed] [Google Scholar]
- Edginton AN, Ahr G, Willmann S, Stass H. (2009) Defining the role of macrophages in local moxifloxacin tissue concentrations using biopsy data and whole-body physiologically based pharmacokinetic modelling. Clin Pharmacokinet 48:181–187. [DOI] [PubMed] [Google Scholar]
- Edginton AN, Joshi G. (2011) Have physiologically-based pharmacokinetic models delivered? Expert Opin Drug Metab Toxicol 7:929–934. [DOI] [PubMed] [Google Scholar]
- Elmeliegy M, Lowe P, Krzyzanski W. (2014) Simplification of complex physiologically based pharmacokinetic models of monoclonal antibodies. AAPS J 16:810–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto C, Fukuda T, Cox S, Christians U, Vinks AA. (2013) Development of a physiologically-based pharmacokinetic model for sirolimus: predicting bioavailability based on intestinal CYP3A content. CPT Pharmacometrics Syst Pharmacol 2:e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahmi OA, Hurst S, Plowchalk D, Cook J, Guo F, Youdim K, Dickins M, Phipps A, Darekar A, Hyland R, et al. (2009) Comparison of different algorithms for predicting clinical drug-drug interactions, based on the use of CYP3A4 in vitro data: predictions of compounds as precipitants of interaction. Drug Metab Dispos 37:1658–1666. [DOI] [PubMed] [Google Scholar]
- Fahmi OA, Maurer TS, Kish M, Cardenas E, Boldt S, Nettleton D. (2008) A combined model for predicting CYP3A4 clinical net drug-drug interaction based on CYP3A4 inhibition, inactivation, and induction determined in vitro. Drug Metab Dispos 36:1698–1708. [DOI] [PubMed] [Google Scholar]
- Fan J, Chen S, Chow EC, Pang KS. (2010) PBPK modeling of intestinal and liver enzymes and transporters in drug absorption and sequential metabolism. Curr Drug Metab 11:743–761. [DOI] [PubMed] [Google Scholar]
- Fei Y, Kostewicz ES, Sheu MT, Dressman JB. (2013) Analysis of the enhanced oral bioavailability of fenofibrate lipid formulations in fasted humans using an in vitro-in silico-in vivo approach. Eur J Pharm Biopharm 85:1274–1284. [DOI] [PubMed] [Google Scholar]
- Fenneteau F, Poulin P, Nekka F. (2010) Physiologically based predictions of the impact of inhibition of intestinal and hepatic metabolism on human pharmacokinetics of CYP3A substrates. J Pharm Sci 99:486–514. [DOI] [PubMed] [Google Scholar]
- Ferl GZ, Wu AM, DiStefano JJ., 3rd (2005) A predictive model of therapeutic monoclonal antibody dynamics and regulation by the neonatal Fc receptor (FcRn). Ann Biomed Eng 33:1640–1652. [DOI] [PubMed] [Google Scholar]
- Friedman EJ, Fraser IP, Wang YH, Bergman AJ, Li CC, Larson PJ, Chodakewitz J, Wagner JA, Stoch SA. (2011) Effect of different durations and formulations of diltiazem on the single-dose pharmacokinetics of midazolam: how long do we go? J Clin Pharmacol 51:1561–1570. [DOI] [PubMed] [Google Scholar]
- Galetin A. (2014) Rationalizing underprediction of drug clearance from enzyme and transporter kinetic data: From in vitro tools to mechanistic modeling, in Enzyme Kinetics in Drug Metabolism: Fundamentals and Applications (Nagar S, Argikar UA, Tweedie DJ, eds), pp 255–288, Springer-Verlag; New York. [DOI] [PubMed] [Google Scholar]
- Gaohua L, Abduljalil K, Jamei M, Johnson TN, Rostami-Hodjegan A. (2012) A pregnancy physiologically based pharmacokinetic (p-PBPK) model for disposition of drugs metabolized by CYP1A2, CYP2D6 and CYP3A4. Br J Clin Pharmacol 74:873–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérard C, Bleyzac N, Girard P, Freyer G, Bertrand Y, Tod M. (2010) Influence of dosing schedule on organ exposure to cyclosporin in pediatric hematopoietic stem cell transplantation: analysis with a PBPK model. Pharm Res 27:2602–2613. [DOI] [PubMed] [Google Scholar]
- Gérard C, Bleyzac N, Girard P, Freyer G, Bertrand Y, Tod M. (2011) Links between cyclosporin exposure in tissues and graft-versus-host disease in pediatric bone marrow transplantation: analysis by a PBPK model. Pharm Res 28:531–539. [DOI] [PubMed] [Google Scholar]
- Gérard C, Stocco J, Hulin A, Blanchet B, Verstuyft C, Durand F, Conti F, Duvoux C, Tod M. (2014) Determination of the most influential sources of variability in tacrolimus trough blood concentrations in adult liver transplant recipients: a bottom-up approach. AAPS J 16:379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz M, Cartwright CM, Hobbs MJ, Kenworthy KE, Rowland M, Houston JB, Galetin A. (2013) Cyclosporine inhibition of hepatic and intestinal CYP3A4, uptake and efflux transporters: application of PBPK modeling in the assessment of drug-drug interaction potential. Pharm Res 30:761–780. [DOI] [PubMed] [Google Scholar]
- Gertz M, Tsamandouras N, Säll C, Houston JB, Galetin A. (2014) Reduced physiologically-based pharmacokinetic model of repaglinide: impact of OATP1B1 and CYP2C8 genotype and source of in vitro data on the prediction of drug-drug interaction risk. Pharm Res 31:2367–2382. [DOI] [PubMed] [Google Scholar]
- Gillette JR. (1971) Factors affecting drug metabolism. Ann N Y Acad Sci 179:43–66. [DOI] [PubMed] [Google Scholar]
- Guo H, Liu C, Li J, Zhang M, Hu M, Xu P, Liu L, Liu X. (2013) A mechanistic physiologically based pharmacokinetic-enzyme turnover model involving both intestine and liver to predict CYP3A induction-mediated drug-drug interactions. J Pharm Sci 102:2819–2836. [DOI] [PubMed] [Google Scholar]
- Harwood MD, Neuhoff S, Carlson GL, Warhurst G, Rostami-Hodjegan A. (2013) Absolute abundance and function of intestinal drug transporters: a prerequisite for fully mechanistic in vitro-in vivo extrapolation of oral drug absorption. Biopharm Drug Dispos 34:2–28. [DOI] [PubMed] [Google Scholar]
- Heimbach T, Xia B, Lin TH, He H. (2013) Case studies for practical food effect assessments across BCS/BDDCS class compounds using in silico, in vitro, and preclinical in vivo data. AAPS J 15:143–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton S, Tuerk A, Cook D, Cook J, Dhurjati P. (2012) Maximum recommended dosage of lithium for pregnant women based on a PBPK model for lithium absorption. Adv Bioinforma 2012:352729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston JB. (1994) Utility of in vitro drug metabolism data in predicting in vivo metabolic clearance. Biochem Pharmacol 47:1469–1479. [DOI] [PubMed] [Google Scholar]
- Hsu V, de LT Vieira M, Zhao P, Zhang L, Zheng JH, Nordmark A, Berglund EG, Giacomini KM, Huang SM. (2014) Towards quantitation of the effects of renal impairment and probenecid inhibition on kidney uptake and efflux transporters, using physiologically based pharmacokinetic modelling and simulations. Clin Pharmacokinet 53:283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu ZY, Edginton AN, Laizure SC, Parker RB. (2014) Physiologically based pharmacokinetic modeling of impaired carboxylesterase-1 activity: effects on oseltamivir disposition. Clin Pharmacokinet 53:825–836. [DOI] [PubMed] [Google Scholar]
- Huang SM, Abernethy DR, Wang Y, Zhao P, Zineh I. (2013) The utility of modeling and simulation in drug development and regulatory review. J Pharm Sci 102:2912–2923. [DOI] [PubMed] [Google Scholar]
- Huang SM, Rowland M. (2012) The role of physiologically based pharmacokinetic modeling in regulatory review. Clin Pharmacol Ther 91:542–549. [DOI] [PubMed] [Google Scholar]
- Hyland R, Dickins M, Collins C, Jones H, Jones B. (2008) Maraviroc: in vitro assessment of drug-drug interaction potential. Br J Clin Pharmacol 66:498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Programme on Chemical Harmonization Project (2010). Characterization and application of physiologically based pharmacokinetic models in risk assessment. Harmonization Project Document No. 9, World Health Organization, Geneva, Switzerland.
- Iwatsubo T, Hirota N, Ooie T, Suzuki H, Shimada N, Chiba K, Ishizaki T, Green CE, Tyson CA, Sugiyama Y. (1997) Prediction of in vivo drug metabolism in the human liver from in vitro metabolism data. Pharmacol Ther 73:147–171. [DOI] [PubMed] [Google Scholar]
- Jamei M, Bajot F, Neuhoff S, Barter Z, Yang J, Rostami-Hodjegan A, Rowland-Yeo K. (2014) A mechanistic framework for in vitro-in vivo extrapolation of liver membrane transporters: prediction of drug-drug interaction between rosuvastatin and cyclosporine. Clin Pharmacokinet 53:73–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamei M, Dickinson GL, Rostami-Hodjegan A. (2009a) A framework for assessing inter-individual variability in pharmacokinetics using virtual human populations and integrating general knowledge of physical chemistry, biology, anatomy, physiology and genetics: A tale of ‘bottom-up’ vs ‘top-down’ recognition of covariates. Drug Metab Pharmacokinet 24:53–75. [DOI] [PubMed] [Google Scholar]
- Jamei M, Turner D, Yang J, Neuhoff S, Polak S, Rostami-Hodjegan A, Tucker G. (2009b) Population-based mechanistic prediction of oral drug absorption. AAPS J 11:225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TN, Zhou D, Bui KH. (2014) Development of physiologically based pharmacokinetic model to evaluate the relative systemic exposure to quetiapine after administration of IR and XR formulations to adults, children and adolescents. Biopharm Drug Dispos 35:341–352. [DOI] [PubMed] [Google Scholar]
- Jones HM, Barton HA, Lai Y, Bi YA, Kimoto E, Kempshall S, Tate SC, El-Kattan A, Houston JB, Galetin A, et al. (2012) Mechanistic pharmacokinetic modeling for the prediction of transporter-mediated disposition in humans from sandwich culture human hepatocyte data. Drug Metab Dispos 40:1007–1017. [DOI] [PubMed] [Google Scholar]
- Jones HM, Chen Y, Gibson C, Heimbach T, Parrott N, Peters SA, Snoeys J, Upreti VV, Zheng M, Hall SD. (2015) Physiologically based pharmacokinetic modeling in drug discovery and development: a pharmaceutical industry perspective. Clin Pharmacol Ther 97:247–262. [DOI] [PubMed] [Google Scholar]
- Jones HM, Gardner IB, Watson KJ. (2009) Modelling and PBPK simulation in drug discovery. AAPS J 11:155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambayashi A, Blume H, Dressman J. (2013) Understanding the in vivo performance of enteric coated tablets using an in vitro-in silico-in vivo approach: case example diclofenac. Eur J Pharm Biopharm 85:1337–1347. [DOI] [PubMed] [Google Scholar]
- Kaspera R, Kirby BJ, Sahele T, Collier AC, Kharasch ED, Unadkat JD, Totah RA. (2014) Investigating the contribution of CYP2J2 to ritonavir metabolism in vitro and in vivo. Biochem Pharmacol 91:109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke AB, Nallani SC, Zhao P, Rostami-Hodjegan A, Isoherranen N, Unadkat JD. (2013) A physiologically based pharmacokinetic model to predict disposition of CYP2D6 and CYP1A2 metabolized drugs in pregnant women. Drug Metab Dispos 41:801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke AB, Nallani SC, Zhao P, Rostami-Hodjegan A, Unadkat JD. (2012) A PBPK model to predict disposition of CYP3A-metabolized drugs in pregnant women: verification and discerning the site of CYP3A induction. CPT Pharmacometrics Syst Pharmacol 1:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke AB, Nallani SC, Zhao P, Rostami-Hodjegan A, Unadkat JD. (2014) Expansion of a PBPK model to predict disposition in pregnant women of drugs cleared via multiple CYP enzymes, including CYP2B6, CYP2C9 and CYP2C19. Br J Clin Pharmacol 77:554–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kletting P, Kull T, Bunjes D, Mahren B, Luster M, Reske SN, Glatting G. (2010) Radioimmunotherapy with anti-CD66 antibody: improving the biodistribution using a physiologically based pharmacokinetic model. J Nucl Med 51:484–491. [DOI] [PubMed] [Google Scholar]
- Kudo T, Hisaka A, Sugiyama Y, Ito K. (2013) Analysis of the repaglinide concentration increase produced by gemfibrozil and itraconazole based on the inhibition of the hepatic uptake transporter and metabolic enzymes. Drug Metab Dispos 41:362–371. [DOI] [PubMed] [Google Scholar]
- Leahy DE. (2003) Progress in simulation modelling for pharmacokinetics. Curr Top Med Chem 3:1257–1268. [DOI] [PubMed] [Google Scholar]
- Leong R, Vieira MLT, Zhao P, Mulugeta Y, Lee CS, Huang SM, Burckart GJ. (2012) Regulatory experience with physiologically based pharmacokinetic modeling for pediatric drug trials. Clin Pharmacol Ther 91:926–931. [DOI] [PubMed] [Google Scholar]
- Li GF, Wang K, Chen R, Zhao HR, Yang J, Zheng QS. (2012) Simulation of the pharmacokinetics of bisoprolol in healthy adults and patients with impaired renal function using whole-body physiologically based pharmacokinetic modeling. Acta Pharmacol Sin 33:1359–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Guo HF, Liu C, Zhong Z, Liu L, Liu XD. (2015) Prediction of drug disposition in diabetic patients by means of a physiologically based pharmacokinetic model. Clin Pharmacokinet 54:179–193. [DOI] [PubMed] [Google Scholar]
- Li L, Gardner I, Dostalek M, Jamei M. (2014a) Simulation of monoclonal antibody pharmacokinetics in humans using a minimal physiologically based model. AAPS J 16:1097–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Barton HA, Yates PD, Ghosh A, Wolford AC, Riccardi KA, Maurer TS. (2014b) A “middle-out” approach to human pharmacokinetic predictions for OATP substrates using physiologically-based pharmacokinetic modeling. J Pharmacokinet Pharmacodyn 41:197–209. [DOI] [PubMed] [Google Scholar]
- Li R, Ghosh A, Maurer TS, Kimoto E, Barton HA. (2014c) Physiologically based pharmacokinetic prediction of telmisartan in human. Drug Metab Dispos 42:1646–1655. [DOI] [PubMed] [Google Scholar]
- Liu L, Pang KS. (2005) The roles of transporters and enzymes in hepatic drug processing. Drug Metab Dispos 33:1–9. [DOI] [PubMed] [Google Scholar]
- Lombardo F, Waters NJ, Argikar UA, Dennehy MK, Zhan J, Gunduz M, Harriman SP, Berellini G, Liric Rajlic I, Obach RS. (2013a) Comprehensive assessment of human pharmacokinetic prediction based on in vivo animal pharmacokinetic data, part 1: volume of distribution at steady state. J Clin Pharmacol 53:167–177. [DOI] [PubMed] [Google Scholar]
- Lombardo F, Waters NJ, Argikar UA, Dennehy MK, Zhan J, Gunduz M, Harriman SP, Berellini G, Liric Rajlic I, Obach RS. (2013b) Comprehensive assessment of human pharmacokinetic prediction based on in vivo animal pharmacokinetic data, part 2: clearance. J Clin Pharmacol 53:178–191. [DOI] [PubMed] [Google Scholar]
- Lu C, Suri A, Shyu WC, Prakash S. (2014) Assessment of cytochrome P450-mediated drug-drug interaction potential of orteronel and exposure changes in patients with renal impairment using physiologically based pharmacokinetic modeling and simulation. Biopharm Drug Dispos 35:543–552. [DOI] [PubMed] [Google Scholar]
- Lu G, Abduljalil K, Jamei M, Johnson TN, Soltani H, Rostami-Hodjegan A. (2012) Physiologically-based pharmacokinetic (PBPK) models for assessing the kinetics of xenobiotics during pregnancy: achievements and shortcomings. Curr Drug Metab 13:695–720. [DOI] [PubMed] [Google Scholar]
- Mayhew BS, Jones DR, Hall SD. (2000) An in vitro model for predicting in vivo inhibition of cytochrome P450 3A4 by metabolic intermediate complex formation. Drug Metab Dispos 28:1031–1037. [PubMed] [Google Scholar]
- McLanahan ED, El-Masri HA, Sweeney LM, Kopylev LY, Clewell HJ, Wambaugh JF, Schlosser PM. (2012) Physiologically based pharmacokinetic model use in risk assessment—Why being published is not enough. Toxicol Sci 126:5–15. [DOI] [PubMed] [Google Scholar]
- Meyer M, Schneckener S, Ludewig B, Kuepfer L, Lippert J. (2012) Using expression data for quantification of active processes in physiologically based pharmacokinetic modeling. Drug Metab Dispos 40:892–901. [DOI] [PubMed] [Google Scholar]
- Ministerial Industry Strategy Group (2014) MISG New technologies forum on physiologically-based pharmacokinetic (PBPK) modelling and simulation. Association of the British Pharmaceutical Industry, London. [Google Scholar]
- Ministry of Health, Labour and Welfare Research Group (2014). Drug interaction guideline for drug development and labeling recommendations (draft for public comment). Japanese Ministry of Health, Labour and Welfare, Tokyo. [Google Scholar]
- Nestorov IA, Aarons LJ, Arundel PA, Rowland M. (1998) Lumping of whole-body physiologically based pharmacokinetic models. J Pharmacokinet Biopharm 26:21–46. [DOI] [PubMed] [Google Scholar]
- Neuhoff S, Yeo KR, Barter Z, Jamei M, Turner DB, Rostami-Hodjegan A. (2013a) Application of permeability-limited physiologically-based pharmacokinetic models: part I—digoxin pharmacokinetics incorporating P-glycoprotein-mediated efflux. J Pharm Sci 102:3145–3160. [DOI] [PubMed] [Google Scholar]
- Neuhoff S, Yeo KR, Barter Z, Jamei M, Turner DB, Rostami-Hodjegan A. (2013b) Application of permeability-limited physiologically-based pharmacokinetic models: part II—prediction of P-glycoprotein mediated drug-drug interactions with digoxin. J Pharm Sci 102:3161–3173. [DOI] [PubMed] [Google Scholar]
- Obach RS, Baxter JG, Liston TE, Silber BM, Jones BC, MacIntyre F, Rance DJ, Wastall P. (1997) The prediction of human pharmacokinetic parameters from preclinical and in vitro metabolism data. J Pharmacol Exp Ther 283:46–58. [PubMed] [Google Scholar]
- Obach RS, Walsky RL, Venkatakrishnan K. (2007) Mechanism-based inactivation of human cytochrome p450 enzymes and the prediction of drug-drug interactions. Drug Metab Dispos 35:246–255. [DOI] [PubMed] [Google Scholar]
- Ogungbenro K, Aarons L, CRESim & Epi-CRESim Project Groups (2015) A physiologically based pharmacokinetic model for clobazam and stiripentol in adults and children. Pharm Res 32:144–157. [DOI] [PubMed] [Google Scholar]
- Pan X, Wang P, Hu N, Liu L, Liu X, Xie L, Wang G. (2011) A physiologically based pharmacokinetic model characterizing mechanism-based inhibition of CYP1A2 for predicting theophylline/antofloxacin interaction in both rats and humans. Drug Metab Pharmacokinet 26:387–398. [DOI] [PubMed] [Google Scholar]
- Pan Y, Grillo J, Hsu V, Zhang L, Sinha V, Huang S. (2014). Application of the FDA PBPK knowledgebase in evaluating model predictability for drug-drug interactions. Clin Pharmacol Ther 95 S102. [Google Scholar]
- Pang KS, Durk MR. (2010) Physiologically-based pharmacokinetic modeling for absorption, transport, metabolism and excretion. J Pharmacokinet Pharmacodyn 37:591–615. [DOI] [PubMed] [Google Scholar]
- Parrott N, Davies B, Hoffmann G, Koerner A, Lave T, Prinssen E, Theogaraj E, Singer T. (2011) Development of a physiologically based model for oseltamivir and simulation of pharmacokinetics in neonates and infants. Clin Pharmacokinet 50:613–623. [DOI] [PubMed] [Google Scholar]
- Parrott N, Hainzl D, Scheubel E, Krimmer S, Boetsch C, Guerini E, Martin-Facklam M. (2014) Physiologically based absorption modelling to predict the impact of drug properties on pharmacokinetics of bitopertin. AAPS J 16:1077–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott N, Jones H, Paquereau N, Lavé T. (2005) Application of full physiological models for pharmaceutical drug candidate selection and extrapolation of pharmacokinetics to man. Basic Clin Pharmacol Toxicol 96:193–199. [DOI] [PubMed] [Google Scholar]
- Patel N, Polak S, Jamei M, Rostami-Hodjegan A, Turner DB. (2014) Quantitative prediction of formulation-specific food effects and their population variability from in vitro data with the physiologically-based ADAM model: a case study using the BCS/BDDCS class II drug nifedipine. Eur J Pharm Sci 57:240–249. [DOI] [PubMed] [Google Scholar]
- Péry ARR, Brochot C, Zeman FA, Mombelli E, Desmots S, Pavan M, Fioravanzo E, Zaldívar JM. (2013) Prediction of dose-hepatotoxic response in humans based on toxicokinetic/toxicodynamic modeling with or without in vivo data: a case study with acetaminophen. Toxicol Lett 220:26–34. [DOI] [PubMed] [Google Scholar]
- Pilari S, Huisinga W. (2010) Lumping of physiologically-based pharmacokinetic models and a mechanistic derivation of classical compartmental models. J Pharmacokinet Pharmacodyn 37:365–405. [DOI] [PubMed] [Google Scholar]
- Polasek TM, Polak S, Doogue MP, Rostami-Hodjegan A, Miners JO. (2009) Assessment of inter-individual variability in predicted phenytoin clearance. Eur J Clin Pharmacol 65:1203–1210. [DOI] [PubMed] [Google Scholar]
- Poulin P. (2015) A paradigm shift in pharmacokinetic-pharmacodynamic (PKPD) modeling: rule of thumb for estimating free drug level in tissue compared with plasma to guide drug design. J Pharm Sci 104:2359–2368. [DOI] [PubMed] [Google Scholar]
- Poulin P, Schoenlein K, Theil FP. (2001) Prediction of adipose tissue: plasma partition coefficients for structurally unrelated drugs. J Pharm Sci 90:436–447. [DOI] [PubMed] [Google Scholar]
- Poulin P, Theil FP. (2000) A priori prediction of tissue:plasma partition coefficients of drugs to facilitate the use of physiologically-based pharmacokinetic models in drug discovery. J Pharm Sci 89:16–35. [DOI] [PubMed] [Google Scholar]
- Poulin P, Theil FP. (2002) Prediction of pharmacokinetics prior to in vivo studies. 1. Mechanism-based prediction of volume of distribution. J Pharm Sci 91:129–156. [DOI] [PubMed] [Google Scholar]
- Rajoli RKR, Back DJ, Rannard S, Freel Meyers CL, Flexner C, Owen A, Siccardi M. (2015) Physiologically based pharmacokinetic modelling to inform development of intramuscular long-acting nanoformulations for HIV. Clin Pharmacokinet 54:639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekić D, Röshammar D, Mukonzo J, Ashton M. (2011) In silico prediction of efavirenz and rifampicin drug-drug interaction considering weight and CYP2B6 phenotype. Br J Clin Pharmacol 71:536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers T, Leahy D, Rowland M. (2005) Physiologically based pharmacokinetic modeling 1: predicting the tissue distribution of moderate-to-strong bases. J Pharm Sci 94:1259–1276. [DOI] [PubMed] [Google Scholar]
- Rodgers T, Rowland M. (2006) Physiologically based pharmacokinetic modelling 2: predicting the tissue distribution of acids, very weak bases, neutrals and zwitterions. J Pharm Sci 95:1238–1257. [DOI] [PubMed] [Google Scholar]
- Rostami-Hodjegan A, Tamai I, Pang KS. (2012) Physiologically based pharmacokinetic (PBPK) modeling: it is here to stay! Biopharm Drug Dispos 33:47–50. [DOI] [PubMed] [Google Scholar]
- Rostami-Hodjegan A, Tucker GT. (2007) Simulation and prediction of in vivo drug metabolism in human populations from in vitro data. Nat Rev Drug Discov 6:140–148. [DOI] [PubMed] [Google Scholar]
- Rowland M, Benet LZ, Graham GG. (1973) Clearance concepts in pharmacokinetics. J Pharmacokinet Biopharm 1:123–136. [DOI] [PubMed] [Google Scholar]
- Rowland M, Peck C, Tucker G. (2011) Physiologically-based pharmacokinetics in drug development and regulatory science. Annu Rev Pharmacol Toxicol 51:45–73. [DOI] [PubMed] [Google Scholar]
- Sager JE, Lutz JD, Foti RS, Davis C, Kunze KL, Isoherranen N. (2014) Fluoxetine- and norfluoxetine-mediated complex drug-drug interactions: in vitro to in vivo correlation of effects on CYP2D6, CYP2C19, and CYP3A4. Clin Pharmacol Ther 95:653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayama H, Takubo H, Komura H, Kogayu M, Iwaki M. (2014) Application of a physiologically based pharmacokinetic model informed by a top-down approach for the prediction of pharmacokinetics in chronic kidney disease patients. AAPS J 16:1018–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah DK, Betts AM. (2012) Towards a platform PBPK model to characterize the plasma and tissue disposition of monoclonal antibodies in preclinical species and human. J Pharmacokinet Pharmacodyn 39:67–86. [DOI] [PubMed] [Google Scholar]
- Shardlow CE, Generaux GT, Patel AH, Tai G, Tran T, Bloomer JC. (2013) Impact of physiologically based pharmacokinetic modeling and simulation in drug development. Drug Metab Dispos 41:1994–2003. [DOI] [PubMed] [Google Scholar]
- Shi JG, Fraczkiewicz G, Williams WV, Yeleswaram S. (2015) Predicting drug-drug interactions involving multiple mechanisms using physiologically based pharmacokinetic modeling: a case study with ruxolitinib. Clin Pharmacol Ther 97:177–185. [DOI] [PubMed] [Google Scholar]
- Shono Y, Jantratid E, Janssen N, Kesisoglou F, Mao Y, Vertzoni M, Reppas C, Dressman JB. (2009) Prediction of food effects on the absorption of celecoxib based on biorelevant dissolution testing coupled with physiologically based pharmacokinetic modeling. Eur J Pharm Biopharm 73:107–114. [DOI] [PubMed] [Google Scholar]
- Siccardi M, Marzolini C, Seden K, Almond L, Kirov A, Khoo S, Owen A, Back D. (2013) Prediction of drug-drug interactions between various antidepressants and efavirenz or boosted protease inhibitors using a physiologically based pharmacokinetic modelling approach. Clin Pharmacokinet 52:583–592. [DOI] [PubMed] [Google Scholar]
- Sinha V, Zhao P, Huang SM, Zineh I. (2014) Physiologically based pharmacokinetic modeling: from regulatory science to regulatory policy. Clin Pharmacol Ther 95:478–480. [DOI] [PubMed] [Google Scholar]
- Sjögren E, Westergren J, Grant I, Hanisch G, Lindfors L, Lennernäs H, Abrahamsson B, Tannergren C. (2013) In silico predictions of gastrointestinal drug absorption in pharmaceutical product development: application of the mechanistic absorption model GI-Sim. Eur J Pharm Sci 49:679–698. [DOI] [PubMed] [Google Scholar]
- Sjöstedt N, Kortejärvi H, Kidron H, Vellonen KS, Urtti A, Yliperttula M. (2014) Challenges of using in vitro data for modeling P-glycoprotein efflux in the blood-brain barrier. Pharm Res 31:1–19. [DOI] [PubMed] [Google Scholar]
- Steere B, Baker JAR, Hall SD, Guo Y. (2015) Prediction of in vivo clearance and associated variability of CYP2C19 substrates by genotypes in populations utilizing a pharmacogenetics-based mechanistic model. Drug Metab Dispos 43:870–883. [DOI] [PubMed] [Google Scholar]
- Teeguarden JG, Housand CJ, Smith JN, Hinderliter PM, Gunawan R, Timchalk CA. (2013) A multi-route model of nicotine-cotinine pharmacokinetics, pharmacodynamics and brain nicotinic acetylcholine receptor binding in humans. Regul Toxicol Pharmacol 65:12–28. [DOI] [PubMed] [Google Scholar]
- Thiel C, Schneckener S, Krauss M, Ghallab A, Hofmann U, Kanacher T, Zellmer S, Gebhardt R, Hengstler JG, Kuepfer L. (2015) A systematic evaluation of the use of physiologically based pharmacokinetic modeling for cross-species extrapolation. J Pharm Sci 104:191–206. [DOI] [PubMed] [Google Scholar]
- Tornio A, Filppula AM, Kailari O, Neuvonen M, Nyrönen TH, Tapaninen T, Neuvonen PJ, Niemi M, Backman JT. (2014) Glucuronidation converts clopidogrel to a strong time-dependent inhibitor of CYP2C8: a phase II metabolite as a perpetrator of drug-drug interactions. Clin Pharmacol Ther 96:498–507. [DOI] [PubMed] [Google Scholar]
- Tsamandouras N, Rostami-Hodjegan A, Aarons L. (2015) Combining the ‘bottom up’ and ‘top down’ approaches in pharmacokinetic modelling: fitting PBPK models to observed clinical data. Br J Clin Pharmacol 79:48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DC, Yin F, Kindt JT, Zhang H. (2012) Understanding pharmacokinetic food effects using molecular dynamics simulation coupled with physiologically based pharmacokinetic modeling. Biopharm Drug Dispos 33:510–521. [DOI] [PubMed] [Google Scholar]
- Varma MVS, Lai Y, Feng B, Litchfield J, Goosen TC, Bergman A. (2012) Physiologically based modeling of pravastatin transporter-mediated hepatobiliary disposition and drug-drug interactions. Pharm Res 29:2860–2873. [DOI] [PubMed] [Google Scholar]
- Varma MVS, Lin J, Bi Y, Kimoto E, Rodrigues D. (2015a). Quantitative rationalization of gemfibrozil drug interactions: consideration of transporters-enzyme interplay and the role of circulating metabolite gemfibrozil 1-O-β-glucuronide. Drug Metab Dispos 43:1108–1118. [DOI] [PubMed] [Google Scholar]
- Varma MVS, Lin J, Bi YA, Rotter CJ, Fahmi OA, Lam JL, El-Kattan AF, Goosen TC, Lai Y. (2013) Quantitative prediction of repaglinide-rifampicin complex drug interactions using dynamic and static mechanistic models: delineating differential CYP3A4 induction and OATP1B1 inhibition potential of rifampicin. Drug Metab Dispos 41:966–974. [DOI] [PubMed] [Google Scholar]
- Varma MV, Pang KS, Isoherranen N, Zhao P. (2015b) Dealing with the complex drug-drug interactions: towards mechanistic models. Biopharm Drug Dispos 36:71–92. [DOI] [PubMed] [Google Scholar]
- Varma MVS, Scialis RJ, Lin J, Bi YA, Rotter CJ, Goosen TC, Yang X. (2014) Mechanism-based pharmacokinetic modeling to evaluate transporter-enzyme interplay in drug interactions and pharmacogenetics of glyburide. AAPS J 16:736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira MLT, Zhao P, Berglund EG, Reynolds KS, Zhang L, Lesko LJ, Huang SM. (2012) Predicting drug interaction potential with a physiologically based pharmacokinetic model: a case study of telithromycin, a time-dependent CYP3A inhibitor. Clin Pharmacol Ther 91:700–708. [DOI] [PubMed] [Google Scholar]
- Vinks AA. (2013) The future of physiologically based pharmacokinetic modeling to predict drug exposure in pregnant women. CPT Pharmacometrics Syst Pharmacol 2:e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kleist M, Huisinga W. (2009) Pharmacokinetic-pharmacodynamic relationship of NRTIs and its connection to viral escape: an example based on zidovudine. Eur J Pharm Sci 36:532–543. [DOI] [PubMed] [Google Scholar]
- Wagner C, Jantratid E, Kesisoglou F, Vertzoni M, Reppas C, Dressman JB. (2012) Predicting the oral absorption of a poorly soluble, poorly permeable weak base using biorelevant dissolution and transfer model tests coupled with a physiologically based pharmacokinetic model. Eur J Pharm Biopharm 82:127–138. [DOI] [PubMed] [Google Scholar]
- Wang J, Xia S, Xue W, Wang D, Sai Y, Liu L, Liu X. (2013a) A semi-physiologically-based pharmacokinetic model characterizing mechanism-based auto-inhibition to predict stereoselective pharmacokinetics of verapamil and its metabolite norverapamil in human. Eur J Pharm Sci 50:290–302. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang Z, Zuo Z, Tomlinson B, Lee BTK, Bolger MB, Chow MSS. (2013b) Clinical pharmacokinetics of buffered propranolol sublingual tablet (Promptol™)-application of a new “physiologically based” model to assess absorption and disposition. AAPS J 15:787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH. (2010) Confidence assessment of the Simcyp time-based approach and a static mathematical model in predicting clinical drug-drug interactions for mechanism-based CYP3A inhibitors. Drug Metab Dispos 38:1094–1104. [DOI] [PubMed] [Google Scholar]
- Wang YH, Jones DR, Hall SD. (2004) Prediction of cytochrome P450 3A inhibition by verapamil enantiomers and their metabolites. Drug Metab Dispos 32:259–266. [DOI] [PubMed] [Google Scholar]
- Wu C, Sun L, Sun J, Yang Y, Ren C, Ai X, Lian H, He Z. (2013) Profiling biopharmaceutical deciding properties of absorption of lansoprazole enteric-coated tablets using gastrointestinal simulation technology. Int J Pharm 453:300–306. [DOI] [PubMed] [Google Scholar]
- Wu F, Gaohua L, Zhao P, Jamei M, Huang SM, Bashaw ED, Lee SC. (2014) Predicting nonlinear pharmacokinetics of omeprazole enantiomers and racemic drug using physiologically based pharmacokinetic modeling and simulation: application to predict drug/genetic interactions. Pharm Res 31:1919–1929. [DOI] [PubMed] [Google Scholar]
- Xia B, Heimbach T, Gollen R, Nanavati C, He H. (2013a) A simplified PBPK modeling approach for prediction of pharmacokinetics of four primarily renally excreted and CYP3A metabolized compounds during pregnancy. AAPS J 15:1012–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Heimbach T, Lin TH, Li S, Zhang H, Sheng J, He H. (2013b) Utility of physiologically based modeling and preclinical in vitro/in vivo data to mitigate positive food effect in a BCS class 2 compound. AAPS PharmSciTech 14:1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Chen Y, Pan Y, Skiles GL, Shou M. (2009) Prediction of human drug-drug interactions from time-dependent inactivation of CYP3A4 in primary hepatocytes using a population-based simulator. Drug Metab Dispos 37:2330–2339. [DOI] [PubMed] [Google Scholar]
- Yan GZ, Generaux CN, Yoon M, Goldsmith RB, Tidwell RR, Hall JE, Olson CA, Clewell HJ, Brouwer KLR, Paine MF. (2012) A semiphysiologically based pharmacokinetic modeling approach to predict the dose-exposure relationship of an antiparasitic prodrug/active metabolite pair. Drug Metab Dispos 40:6–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo KR, Kenny JR, Rostami-Hodjegan A. (2013) Application of in vitro-in vivo extrapolation (IVIVE) and physiologically based pharmacokinetic (PBPK) modelling to investigate the impact of the CYP2C8 polymorphism on rosiglitazone exposure. Eur J Clin Pharmacol 69:1311–1320. [DOI] [PubMed] [Google Scholar]
- Zane NR, Thakker DR. (2014) A physiologically based pharmacokinetic model for voriconazole disposition predicts intestinal first-pass metabolism in children. Clin Pharmacokinet 53:1171–1182. [DOI] [PubMed] [Google Scholar]
- Zhang H, Xia B, Sheng J, Heimbach T, Lin TH, He H, Wang Y, Novick S, Comfort A. (2014) Application of physiologically based absorption modeling to formulation development of a low solubility, low permeability weak base: mechanistic investigation of food effect. AAPS PharmSciTech 15:400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Quinney SK, Gorski JC, Jones DR, Hall SD. (2009) Semiphysiologically based pharmacokinetic models for the inhibition of midazolam clearance by diltiazem and its major metabolite. Drug Metab Dispos 37:1587–1597. [DOI] [PubMed] [Google Scholar]
- Zhao J, Cao Y, Jusko WJ. (2015) Across-species scaling of monoclonal antibody pharmacokinetics using a minimal PBPK model. Pharm Res [published ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Vieira MLT, Grillo JA, Song P, Wu TC, Zheng JH, Arya V, Berglund EG, Atkinson AJ, Jr, Sugiyama Y, et al. (2012a) Evaluation of exposure change of nonrenally eliminated drugs in patients with chronic kidney disease using physiologically based pharmacokinetic modeling and simulation. J Clin Pharmacol 52 (Suppl):91S–108S. [DOI] [PubMed] [Google Scholar]
- Zhao P, Ragueneau-Majlessi I, Zhang L, Strong JM, Reynolds KS, Levy RH, Thummel KE, Huang SM. (2009) Quantitative evaluation of pharmacokinetic inhibition of CYP3A substrates by ketoconazole: a simulation study. J Clin Pharmacol 49:351–359. [DOI] [PubMed] [Google Scholar]
- Zhao P, Rowland M, Huang SM. (2012b) Best practice in the use of physiologically based pharmacokinetic modeling and simulation to address clinical pharmacology regulatory questions. Clin Pharmacol Ther 92:17–20. [DOI] [PubMed] [Google Scholar]
- Zhao P, Zhang L, Grillo JA, Liu Q, Bullock JM, Moon YJ, Song P, Brar SS, Madabushi R, Wu TC, et al. (2011) Applications of physiologically based pharmacokinetic (PBPK) modeling and simulation during regulatory review. Clin Pharmacol Ther 89:259–267. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hu ZY. (2014) Physiologically based pharmacokinetic modelling and in vivo [I]/Ki accurately predict P-glycoprotein-mediated drug-drug interactions with dabigatran etexilate. Br J Pharmacol 171:1043–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Yang J, Zhang Y, Wang Y, Zhang J, Zhao Y, Dong W. (2015b) Prediction of pharmacokinetics and penetration of moxifloxacin in human with intra-abdominal infection based on extrapolated PBPK model. Korean J Physiol Pharmacol 19:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Zhang Y, Yang J, Wang Y, Zhang J, Zhao Y, Dong W. (2015a) Prediction of the pharmacokinetics and tissue distribution of levofloxacin in humans based on an extrapolated PBPK model. Eur J Drug Metab Pharmacokinet [published ahead of print]. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.